Abstract
Inhibitors of histone deacetylases (HDACs) have emerged as a new class of anticancer agents based on their actions in cancer cell growth and cell cycle arrest, terminal differentiation, and apoptosis. Previously, we rationally designed and developed a new class of hydroxamide- and mercaptoacetamide-bearing HDAC inhibitors. A subset of these inhibitors exhibited chemo-radiation sensitizing properties in various human cancer cells. Furthermore, some HDAC inhibitors protected normal cells from radiation-induced damage and extended the survival of mice following total body exposure to lethal dose radiation. Pathological analyses revealed that intestinal and bone marrow cellularities recovered significantly from radiation-induced damage by structural compartments restoration, suggesting the mechanism of action of these HDAC inhibitors. These findings support the hypothesis that epigenetic regulation may play a crucial role in the functional recovery of normal tissues from radiation injuries.
The epigenetic regulation governs gene expression, in part, through the balance of histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities. HATs acetylate lysine groups at the amino terminal tails of nuclear histones to neutralize positive charges on the histones, yielding a more open, transcriptionally active chromatin structure [1]. By contrast, HDAC family members deacetylate lysine residues on histones and induce transcription repression through chromatin condensation. Increased acetylation of histones and nonhistone proteins leads to changes in the chromatin architecture and accessibility for key cellular proteins to specific target sites [2, 3]. As such, a shift in the relative activities of these enzymes profoundly influences biological processes, including DNA repair, replication, cell cycle checkpoint activation, and cellular differentiation [4].
14.1 Histone Deacetylase Family Members
HDACs are initially identified as components of large multiprotein complexes that bind to promoters and repress transcription, and are found in the nuclear and cytoplasmic compartments [5, 6]. Eleven human HDACs have been identified and divided into four classes based on structure, sequence homology, and domain organization. Class I consists of HDACs 1, 2, 3, and 8, which are predominantly nuclear and play roles in cell proliferation [5]. Class II HDACs are further subdivided into IIa (HDACs 4, 5, 7, and 9) and IIb (HDAC6 and 10) [7]. These enzymes are characterized by a large NH2-terminal domain or a second catalytic site and their expression is more restricted, suggesting roles in cellular differentiation and development [8]. Class III enzymes include the SIRTs (sirtuins), and are NAD-dependent deacetylases [9], which are not inhibited by TSA or other hydroxamates. HDAC 11 is characterized as class IV, based on a phylogenetic analysis, and whose function is least known [10].
14.2 Structural Classifications of HDAC Inhibitors (HDACIs)
Inhibitors of HDACs have been discovered and developed as promising anticancer drugs [11]. On the basis of structural divergence, HDAC inhibitors are classified: (1) short-chain fatty acids (i.e., sodium phenyl butyrate, valproic acid, AN-9), (2) hydroxamic acids (i.e., suberoylanilide hydroxamic acid (SAHA; vorinostat), oxamflatin, trichostatin A (TSA), m-carboxycinnamic acid bis-hydroxamide (CBHA), LBH-589, LAQ-824, PCI-24781), (3) cyclic peptides (i.e., FR901228 (depsipeptide; romidepsin), apicidin, cyclic hydroxamic acid-containing peptides (CHAPS), trapoxin), (4) benzamides (i.e., MS-275, CI-994), (5) ketones (i.e., trifluoromethyl ketone), and (6) miscellaneous (i.e., MGCD-0103). A subset of HDACIs is reportedly as pan-HDAC inhibitors, such as TSA and SAHA, while MS-275 and depsipeptide confer isomer or/and class-specificities. Some of which are currently in clinical trials [12], and two drugs, vorinostat (Zolinza®) and romidepsin (Istodax®), have recently been approved by FDA for the treatment of cutaneous T-cell lymphoma (CTCL) in patients ([13], www.hhs.gov).
14.3 Development of Second-Generation HDACIs
Despite a generally acceptable toxicity profile for HDACIs, the lack of target specificity and the presence of the toxicophore hydroxamide group may still cause unpredictable toxicity problems. To improve physicochemical and pharmacological properties of first-generation HDACIs, we rationally designed and developed several new classes of HDACIs, including ligands bearing either a hydoxamate or mercaptoacetamide group [14, 15]. A subset of these candidates exhibited 50% HDAC inhibition activity within nanomolar ranges in vitro and radiosensitizing effects on various cancer cells [14, 15]. The candidate inhibitors potently repressed the growth of cancer cells: the GI50s for the hydroxamates ranged from 0.1 to >60 μM, while the mercaptoacetamides revealed weaker growth inhibitory activity against cancer cell lines. Table 14.1 presents the data demonstrating that the hydroxamates conferred potent cytotoxicity, while the mercaptoacetamides revealed less against all three cancer cell lines. However, normal human fibroblasts (NHP-5) and human primary skin fibroblasts (Hs-68) were found to be significantly resistant to these HDACIs, supporting the notion that candidate HDAC inhibitors preferentially suppress cancer cell proliferation.
Table 14.1.
Effects of hydroxamate (H6CAHA) and mercaptoacetamide (6MAQH) HDAC inhibitors on pan-HDACs and the growth of cancer and normal cells
| Cell proliferation (GI50 μM) |
||||||
|---|---|---|---|---|---|---|
| 50% pan-HDAC activity inhibition (nM) | Cancer cells |
Normal cells |
||||
| Compound | PC-3 | SQ-20B | MCF-7 | NHP-5 | Hs-68 | |
| H6CAHA | 176 | 1.77 | 2.88 | 3.98 | >300 | >500 |
| 6MAQH | 44 | 40 | 30 | 23 | >500 | >500 |
Note: Cell lines were obtained from the Tissue Culture Shared Resources of the Lombardi Comprehensive Cancer Center, Georgetown University Medical Center. PC-3 is a prostate cancer cell line; SQ-20B is a head and neck squamous carcinoma cell line; MCF-7 is a breast cancer cell line; NHP-5 and Hs-68 are nonmalignant human epithelial cell lines. Cell proliferation was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay after a 48-h incubation period, as previously described [16]. Growth inhibitory concentrations at 50% (GI50) were calculated graphically and the mean GI50 values were obtained. Results are the mean of at least three determinations. Statistical comparisons between treatment groups were made using GraphPad Prism 4.0 software
14.4 Cellular Target Specificity of HDACIs
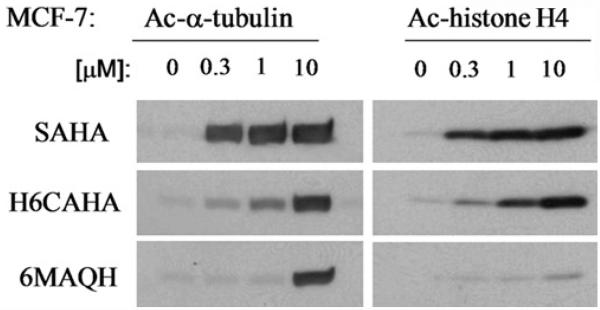
Given that 11 HDAC isomers are present in mammalian cells, whether isomer-specific HDACIs are better drugs has been an outstanding question and is actively in ongoing investigation. In general, the determination of acetylated histone and α-tubulin levels has been used as a measure of HDACI efficacy in vitro and in vivo. Pan-HDACIs, such as SAHA and TSA, appear to target both histones H3/H4 and α-tubulin evidenced by their enhanced acetylation levels, in a dose-dependent manner. In contrast, other HDACIs, such as trapoxin B, sodium butyrate, and FK-228, do not exhibit these characteristics, suggesting their selectivity at the cellular level [17]. Others have reported that depsipeptide exerts modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells [18]. Valproic acid, an anti-epileptic agent, has shown to be involved in the proteolysis of HDAC2 [19]. Hyperacetylations of α-tubulin and Hsp90 have also been tightly linked to class II HDAC6 inhibition. Our data, as shown in Fig. 14.1, demonstrate that mercaptoacetamide-based HDACI 6MAQH preferentially enhances acetylation of α-tubulin while hydroxamate HDACIs, SAHA and H6CAHA, enhance the acetylation levels of both histone H4 and α-tubulin in a dose-dependent manner, suggesting 6MAQH as a class II HDAC isomer-specific inhibitor. Therefore, understanding of the roles of various HDAC isoforms in cell processes and identifying HDACI-specific biomarkers in diseases may contribute to strategies for the identification of promising second-generation inhibitors that exhibit target specificity and that offer clinical relevance.
14.5 HDACs, DNA Damage Repair, and Radiation Sensitivity
Accumulated studies have shown that a particular subset of HDACIs induces radiosensitivity of various human cancer cells [14, 20–24]. Consistent with these findings, several recent reports have provided supporting evidence of specific HDAC isoforms’ roles in cell cycle regulation and intrinsic radiation sensitivity [25–29].
A significant line of evidence has demonstrated that a subset of HDAC family members plays a critical role in DNA damage response and repair [27, 30–32] and that post-translational modifications (e.g., acetylation) of histones or nonhistone proteins flanking DSBs affect accessibility of repair and signaling proteins to the damaged regions of DNA [33, 34]. Moreover, changes in the chromatin architecture by HDACI have shown to enhance the activation of nonhis-tone proteins, including p53, pRB, and transcriptional factors that are involved in cell cycle regulation and affect cell proliferation and apoptosis, processes that are frequently aberrant in cancers [11, 35]. With their intrinsic anticancer properties, a subset of HDAC inhibitors has exhibited radiation sensitizing properties in certain cancer cell lines and in preclinical models [20–24, 36, 37]. However, underlying mechanisms are not fully understood.
Previously, we have shown that an HDAC inhibitor, trichostatin A (TSA), increased the amount of radiation-induced DNA damage and slowed the repair kinetics [31]. Gene expression profiling also revealed that a majority of genes that control cell cycle, DNA replication, and damage repair processes were down-regulated following TSA exposure. HDACI treatment modulated the radiation-induced DNA damage repair process, in part, by suppressing BRCA1 gene expression [31]. Further studies have demonstrated prolonged appearance of γH2AX and Rad51 foci and suppression of DNA damage repair genes (ATM, Brca1, and Brca2). Functions of damage sensing and repair proteins, including ATM, MRE11, γ-H2AX, and 53BP1, are also affected by changes in chromatin structure conformation [38–40].
A sensor of DNA damage, ATM has been shown to interact with HDAC1, to play a role with HDAC4 in the DNA damage response pathway, and to be associated with ionizing radiation-induced changes in chromatin structure [25, 28]. Inactivation of HDAC3 affects S phase progression and enhances DNA damage [26]. HDAC4 co-localizes with 53BP1 (a protein which participates in the phosphorylation of p53 and Chk2 and in the maintenance of S and G2 cell cycle checkpoints) in DNA damage-induced nuclear foci [25]. Finally, other studies have suggested that butyrates, TSA, and inhibitory HDAC RNAs are capable of sensitizing cancer cells to ionizing radiation [27, 30–32, 41].
14.6 HDACI and Radioprotection In Vitro
One of the hallmarks of HDACI action is to preferentially sensitize actively proliferating cancer cells [42–45]. Normal cells are considerably more resistant to HDAC inhibitors than are tumor cells [45]. To determine underlying mechanisms of biological discrepancy between cancer and normal cells, we examined the synergistic effect of HDACI on DNA damage response by scoring phosphorylated H2AX (γH2AX) foci, one of the first events occurring at DNA double-strand breaks (DSB). The data demonstrated that the foci formation in cancer cells (PC3, LNCaP, and DU145) was markedly increased within 0.5 h and gradually decreased by 3 h after irradiation (5 Gy) (Fig. 14.2). HDAC inhibitor treatment alone resulted in the accumulation of γH2AX foci formation. However, after pretreatment with H6CAHA for 16 h followed by irradiation (5 Gy), γH2AX foci/cell growth increased significantly and sustained these levels up to 6 h and persisted for at least 24 h. This trend was not observed in normal cells (data not shown). These results indicate that disruption of the radiation-induced DSB damage repair mechanism may account for H6CAHA-induced radiosensitization of cancer cells and also indicate that H6CAHA may have the potential to facilitate DNA damage repair in normal cells.
Fig. 14.2.
Effect of H6CAHA on radiation-induced γH2AX foci; PC3, LNCaP, and DU145 cells were incubated with vehicle or 1-μM H6CAHA for 16 h and then exposed to 5 Gy of γ-radiation. The 0-h samples were not exposed to radiation. γH2AX foci were evaluated in 75 nuclei per treatment per experiment. Columns represent mean values from three independent experiments; bars represent SD. After staining, foci were visualized by Olympus FV300 equipped with FluoView software. Statistical comparisons between treatment groups were made using GraphPad Prism 4.0 software by Student's t-test. A probability value of p < 0.05 was considered to be significant
14.7 HDACI and Radioprotection In Vivo
Given that the ability to selectively kill cancer cells while limiting damage of normal cells is the ultimate therapeutic goal and that HDACIs (phenylbutyrate, trichostatin A, and valproic acid) could suppress cutaneous radiation syndrome [42, 43], understanding the mechanisms of cancer cell radiation sensitization and normal cell protection by HDACIs is important.
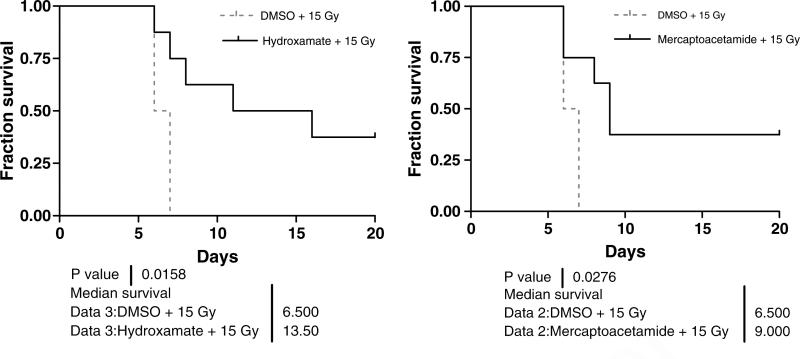
To address how the candidate HDACIs affect normal cells in response to ionizing radiation in vivo, C57BL/6 mice were treated with an i.p. subtoxic dose (4 mg/kg) of either hydoxamate H6CAHA or mercaptoacetamide 6MAQH, 4 h prior to total-body irradiated (TBI) with 15 Gy of gamma radiation. The survival rate and body weight were monitored daily. The data demonstrated that 100% of animals undergoing TBI died within 6–7 days. The loss of body weight was evident, possibly indicating intestinal damage. However, HDACI treatment extended the survival of animals; 40% of animals treated with hydroxamide H6CAHA or mercaptoacetamide 6MAQH survived until the end of the study at 25 days. The data also showed that the duration of survival of animals pretreated with hydoxamate H6CAHA (median survival: 14 days) extended further than those treated with mercaptoacetamide 6MAQH (median survival: 9 days) (Fig. 14.3). These animals eventually gained weight and continued to survive until termination of the experiments.
Fig. 14.3.
The effects of each potential HDAC inhibitor, hydoxamate (H6CAHA) and mercaptoacetamide (6MAQH), on survival of mice undergoing total body irradiation of 15 Gy gamma. Each cohort (10 mice per group) received i.p. administered HDACI (4 mg/kg body weight) 4 h prior to irradiation. Survivals of animals were monitored and the body weights were measured daily for 25 days until the end of experiments. C57BL/6 mice (18–22 g) were treated with an intraperitoneal dose (4 mg/kg) of hydoxamate or mercaptoacetamide, 4 h prior to total-body irradiation (TBI). The animal protocol was reviewed and approved by the Georgetown University Animal Welfare Committee. The body weights of control and treated mice were measured at weekly intervals. Total body irradiation (TBI) was performed using a Mark-30 irradiator with a 137Cs source at a fixed dose rate of 2.71 Gy/min. The Kaplan-Meier method was used to analyze survival data. All statistical analyses were performed on relative survival data (percentage change from initial survival). Student's t-tests were performed to determine if the differences between the treatments were significant. p < 0.05 was considered statistically significant
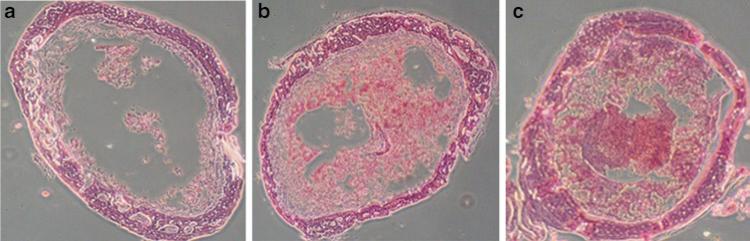
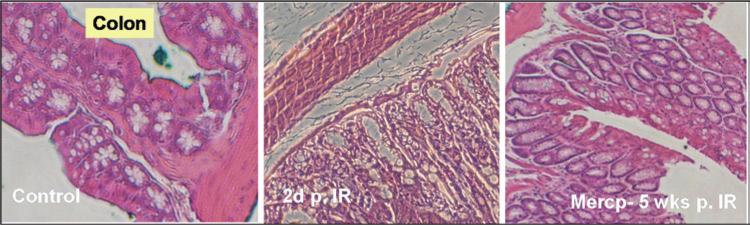
To understand underlying mechanisms, histological analyses were performed using various tissues, including the pelvic bone marrow (BM) and the intestine. The data demonstrated that HDACI pretreatment resulted in less BM cell loss within 2 days compared with treatment with IR alone (Fig. 14.4). Structural recovery accompanied by an increase in small intestine crypts with villus height was also interpreted as an indicator of radioprotection (Fig. 14.5).
Fig. 14.4.
Mouse bone marrow (BM) 2 days after exposure to 15 Gy of Cs-137 total body irradiation (IR). Femur cross sections: (a) DMSO (control); (b) 4 mg/kg of mercaptoacetamide 6MAQH; (c) 4 mg/kg of hydroxamate H6CAHA
Fig. 14.5.
Mouse intestine (colon) after exposure to 15 Gy of Cs-137 total body irradiation (IR). Left: DMSO control; Middle: 2 days post-IR; Right: merceptoacetamide 6MAQH, 5 weeks post-IR
Although the underlying mechanisms responsible for survival remains unclear, these results suggest that the candidate HDACIs protect mice from BM and intestinal injuries after total-body irradiation (TBI), supporting the clinical potential of HDACI in protecting the damage of normal cells/tissues during radiation treatment. It is attempted to speculate that candidate HDAC inhibitors may trigger the activation of BM progenitors and promote their infiltration into the damaged sites and that suppression of cytokines by candidate compounds may contribute to reduced inflammation and/or cell death rates, thereby facilitating recovery from radiation-induced injuries.
14.8 Conclusion
Radiation-induced acute and late injuries often cause dysfunction of organs and generate secondary effects on functional impairment [46, 47]. Limitations and efficacy of radiation therapy in cancer treatment are contingent upon tolerances of normal tissues encompassed in the irradiated fields. Our novel and structurally distinct HDACIs, hydroxamide H6CAHA and merceptoacetamide 6MAQH, exhibited modulating effects on radiation responses by which preferentially killing cancer cells in vitro and in vivo, and enhancing the survival rate of mice following lethal doses of radiation. The data suggest that significant recovery of intestinal and BM cellularities and restoring structural compartments from radiation-induced damage is a mechanism of action of these HDAC inhibitors. Therefore, the use of HDAC inhibitors for radiosensitizing cancer cells and radioprotection of normal tissues identifies an entirely new category of drugs with clinical translation potential. Further studies are warranted for its precise evaluation in which these candidate compounds can be advanced as a treatment agent for normal cell protection from radiation-induced injury.
Fig. 14.1.
Dose-dependent and differential effects of hydroxamate H6CAHA and mercaptoacetamide 6MAQH on histone H4 and α-tubulin acetylation levels in MCF7 cells were analyzed after 4-h treatment with indicated concentrations of HDAC inhibitors. Cells lysates were prepared in a lysis buffer containing 20-mM Tris–HCl (pH 7.4), 2-mM EDTA (pH 7.4), 2-mM EGTA (pH 7.4), 6-mM mercaptoethanol, 10 μg/ml leupeptin, 2 μg/ml aprotinin, and 1% NP40. The samples were subjected to SDS-polyacrylamide gel electrophoresis. The indicated proteins were probed with a polyclonal rabbit antibody against acetylated histone H4 or α-tubulin (Upstate, Inc). The immune-reactive bands were detected by enhanced chemiluminescence (Amersham Biosciences)
Acknowledgments
We thank S. Lee and J. Tuturea for technical support. This work was supported in part by USMRC grants PC030471 (M. Jung) as well as the Lombardi Comprehensive Cancer Center Microscopy and Imaging Shared Resource, US Public Health Service Grant 2P30-CA-51008 and 1S10 RR15768-01.
References
- 1.Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 2.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 3.Katan-Khaykovich Y, Struhl K. Dynamics of global histone acetylation and deacetylation in vivo: rapid restoration of normal histone acetylation status upon removal of activators and repressors. Genes Dev. 2002;16:743–752. doi: 10.1101/gad.967302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opinion Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 5.De Ruijter AJ, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 7.Fischle W, Kiermer V, Dequiedt F, Verdin E. The emerging role of class II histone deacetylases. Biochem Cell Biol. 2001;79:337–348. [PubMed] [Google Scholar]
- 8.Verdin E, Dequiedt F, Kasler H. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;5:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 9.Michan S, Sinclair D. Sirtulins in mammals: insight into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. JMC. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 12.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 13.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 14.Jung M, Velena A, Chen B, Petukhov PA, Kozikowski AP, Dritschilo A. Novel HDAC inhibitors with radiosensitizing properties. Radiat Res. 2005;163:488–493. doi: 10.1667/rr3345. [DOI] [PubMed] [Google Scholar]
- 15.Chen B, Petukhov PA, Jung M, Velena A, Eliseeva E, Dritschilo A, Kozikowski AP. Chemistry and biology of mercaptoacetamides as novel histone deacetylase inhibitors. Bioorg Med Chem Let. 2005;15:1389–1392. doi: 10.1016/j.bmcl.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Konsoula Z, Cao H, Velena A, Jung M. Pharmacokinetics-pharmacodynamics and antitumor activity of mercaptoacetamide-based histone deacetylase inhibitors. Mol Cancer Ther. 2009;8:2844–2851. doi: 10.1158/1535-7163.MCT-09-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung M, Yong KJ, Velena A, Lee SA. Epigenetic Targets in drug discovery: cell-based assays for HDAC inhibitor hit validation. The Wiley-VCH series “Methods and Principles in Medicinal Chemistry”. 2009;42:119–137. [Google Scholar]
- 18.Yu X, Guo ZS, Marcu MG, et al. Modulation of p53, ErbB1, ErbB2, and Raf-1 expression in lung cancer cells by depsipeptide FR901228. J Natl Cancer Inst. 2002;94:504–513. doi: 10.1093/jnci/94.7.504. [DOI] [PubMed] [Google Scholar]
- 19.Kramer OH, Zhu P, Ostendorff HP, et al. the histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDA2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biade S, Stobbe CC, Boyd JT, Chapman JD. Chemical agents that promote chromatin compaction radiosensitize tumor cells. Int J Radiat Biol. 2001;77:1033–1042. doi: 10.1080/09553000110066068. [DOI] [PubMed] [Google Scholar]
- 21.Karagiannis TC, El-Osta A. Modulation of cellular radiation responses by histone deacetylase inhibitors. Oncogene. 2006;25:3885–3893. doi: 10.1038/sj.onc.1209417. [DOI] [PubMed] [Google Scholar]
- 22.Arundel CM, Glicksman AS, Leith JT. Enhancement of radiation injury in human colon tumor cells by the maturational agent Sodium Butyrate (NaB). Radiation Res. 1985;104:443–448. [PubMed] [Google Scholar]
- 23.Kim JH, Shin JH, Kim IH. Susceptibility and radiosensitization of human glioblastoma cells to trichostatin A, a histone deacetylase inhibitor. Int J Radiat Oncol Biol Phys. 2004;59:1174–1180. doi: 10.1016/j.ijrobp.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Jung M, Dritschilo A, Jung M. Enhancement of radiation sensitivity of human squamous carcinoma cells by histone deacetylase inhibitors. Radiat Res. 2004;161:667–674. doi: 10.1667/rr3192. [DOI] [PubMed] [Google Scholar]
- 25.Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. JCB. 2003;160:1017–1027. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhaskara S, Chyla BJ, Amann JM, Knutson SK, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30:61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller KM, Tjeertes JV, Coates J, Legube G, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1152. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim GD, Choi YH, Dimtchev A, Jeong SJ, Dritschilo A, Jung M. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J Biol Chem. 1999;274:31127–31130. doi: 10.1074/jbc.274.44.31127. [DOI] [PubMed] [Google Scholar]
- 29.Ju R, Muller MT. Histone deacetylase inhibitors activate p21 (WAF1) expression via ATM. Cancer Res. 2003;63:2891–2897. [PubMed] [Google Scholar]
- 30.Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang Y, Carr T, Dimtchev A, Zaer N, Dritschilo A, Jung M. Attenuated DNA damage repair by trichostatin A through BRCA1 suppression. Radiat Res. 2007;168:115–124. doi: 10.1667/RR0811.1. [DOI] [PubMed] [Google Scholar]
- 32.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lo brich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 34.Kouzarides T. Chromatin modifcations and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Johnstone RW. Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov. 2002;1:287–299. doi: 10.1038/nrd772. [DOI] [PubMed] [Google Scholar]
- 36.Camphausen K, Scott T, Sproull M, Tofilon PJ. Enhancement of xenograft tumor radiosensitivity by the histone deacetylase inhibitor MS-275 and correlation with histone hyperacetylation. Clin Cancer Res. 2004;10:6066–6071. doi: 10.1158/1078-0432.CCR-04-0537. [DOI] [PubMed] [Google Scholar]
- 37.Folkvord S, Ree AH, Furre T, et al. Radiosensitization by SAHA in experimental colorectal carcinoma models-in vivo effects and relevance of histone acetylation status. Int J Radiat Oncol Biol Phys. 2009;74:546–552. doi: 10.1016/j.ijrobp.2009.01.068. [DOI] [PubMed] [Google Scholar]
- 38.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 39.Keogh MC, Kim JA, Downey M, Fillingham J, et al. A phosphase complex that dephosphorylates gH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 40.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nusssenzwig A. Histone H2Ax phoshporylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 41.Glaser KB, Li J, Staver MJ, Wei RQ, Albert DH, Davidsen SK. Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochem Biophys Res Commun. 2003;310:529–536. doi: 10.1016/j.bbrc.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 42.Chung YL, Wang AJ, Yao LF. Antitumor histone deacetylase inhibitos suppress cutaneous radiation syndrome: implications for increasing therapeutic gain in cancer radiotherapy. Mol Cancer Ther. 2004;3:317–325. [PubMed] [Google Scholar]
- 43.Paoluzzi L, Figg WD. Histone deacetylase inhibitors are potent radiation protectants. Cancer Biol Ther. 2004;3:612–613. doi: 10.4161/cbt.3.7.931. [DOI] [PubMed] [Google Scholar]
- 44.Papeleu P, Vanhaecke T, Elaut G, et al. Differential effects of histone deacetylase inhibitors in tumor and normal cells-what is the toxicological relevance? Crit Rev Toxicol. 2005;35:363–378. doi: 10.1080/10408440590935639. [DOI] [PubMed] [Google Scholar]
- 45.Ungerstedt JS, Sowa Y, Xu WS, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang BK, Timmerman RB. Stereotactic body radiation therapy: a comprehensive review. Am J Clin Onc. 2007;30:637–644. doi: 10.1097/COC.0b013e3180ca7cb1. [DOI] [PubMed] [Google Scholar]
- 47.Timmerman R, McGarry R, Yiannoutsos C, Papiez L, et al. Excessive toxicity when treating central tumors in a Phase II study of stereotactic body radiation therapy for medically inoperable early-stage lung cancer. J Clin Onc. 2006;24:4833–4839. doi: 10.1200/JCO.2006.07.5937. [DOI] [PubMed] [Google Scholar]