Abstract
The pac gene, encoding the penicillin G acylase from Escherichia coli W, is regulated by the PaaX repressor of the phenylacetate catabolic pathway. pac expression depends on the synthesis of phenylacetyl-coenzyme A. PaaX and the cyclic AMP receptor protein (CRP) bind in vitro to the Ppac promoter region. A palindromic sequence proposed as the PaaX operator is located upstream of the −35 box overlapping a CRP binding site, an unusual position that suggests a novel regulatory mechanism.
Penicillin G acylase (PAC) (penicillin G aminohydrolase, EC 3.5.1.11) is a member of a large enzyme family conventionally known as β-lactam acylases because they are used for the semisynthesis of β-lactam antibiotics (30). PAC has a broad substrate range and is able to hydrolyze different esters and amides of phenylacetic acid (PA) and other aromatic and aliphatic acids, which has made it one of the most important enzymes used at the industrial scale worldwide (1, 5, 6, 11, 33). Although many PAC enzymes have been identified and characterized in different microorganisms, the PAC from Escherichia coli W is by far the best-studied enzyme of this family (30, 33).
Despite the ability of PAC to hydrolyze penicillin G, it does not have a function in bacterial antibiotic resistance, and its physiological role still remains unclear. Nevertheless, its broad substrate range together with the fact that its synthesis is activated by PA has favored the proposal of PAC as a scavenger enzyme for natural compounds containing a phenylacetate or hydroxyphenylacetate residue in ester or amide linkage (10, 21). Supporting this hypothesis, we have demonstrated that the pac gene encoding the PAC is located in the vicinity of the hpa cluster responsible for the degradation of 3- and 4-hydroxiphenylacetate in the chromosome of E. coli W (Fig. 1) (10, 23, 24). This observation suggested the implication of evolutionary selective forces which favored the clustering of physiologically interdependent genes, like pac and hpa. However, the recently identified paa cluster responsible for the catabolism of PA was located very far from the pac gene (Fig. 1) (13). Therefore, new data are needed to solve this apparent paradox.
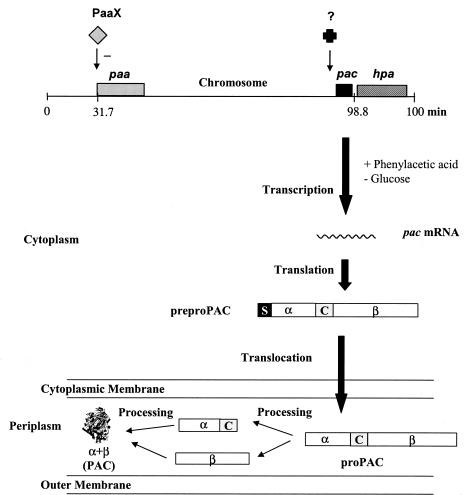
FIG. 1.
General scheme of the PAC regulatory system. The top of the figure shows the relative localizations of the pac gene (black block), the paa cluster (gray block) for the degradation of PA, and the hpa cluster (cross-hatched block) for the degradation of 4-hydroxyphenylacetate in the E. coli chromosome, referred to as the E. coli K-12 linkage map (10). The regulatory protein PaaX is represented by a gray diamond. The − indicates transcriptional repression. The black cross represents an undetermined factor that controls the dependent PA expression of the pac gene. The synthesis and maturation processes of the PAC protein are shown. The pac gene encodes a polypeptide precursor (preproPAC) which has a molecular mass of 92 kDa and is composed of, in the direction of N to C, a signal peptide (S), an α subunit (α), a connecting peptide (C), and a β subunit (β). The signal peptide directs the export of a polypeptide precursor into the periplasm and is removed to form another precursor (proPAC) after the translocation. Periplasmic processing steps consist of autoproteolysis (α+β) of the precursor and folding of the mature PAC.
It is well known that the synthesis of PAC is probably one of the most complex processes for bacterial proteins described so far since it is subjected to sophisticated regulatory controls at both the transcriptional and translational levels (Fig. 1) (16, 18, 21, 22, 27, 29, 31). Despite the fact that this system was deeply studied, the regulatory proteins involved in PA induction still remained unclear. Although a pacR regulatory gene located inside the pac gene has recently been identified (7), several data obtained by using Ppac::lacZ fusions suggested that other factors might be involved in the induction process (21).
Using different genetic and biochemical approaches, both in vivo and in vitro, we have demonstrated that the PaaX regulator that controls the expression of the PA-coenzyme A (CoA) catabolon is also involved in the transcriptional regulation of the pac gene. This work not only settles the basis for clarifying the puzzling data about this complex regulatory system obtained so far but definitively supports the implication of PAC in the PA-CoA catabolon and provides interesting evolutionary evidence that helps to explain how the cell is able to integrate and tune the regulation of genes that are involved in the same catabolic processes.
Localization of a PaaX binding site in the Ppac promoter region.
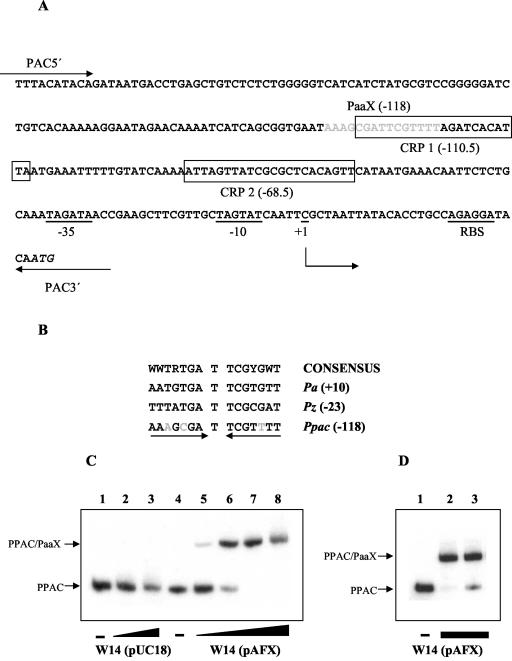
It has been reported that PaaX negatively controls the expression of the paa catabolic cluster and that the PA-CoA generated by the PA-CoA ligase (PaaK) acts as the true inducer of the system (12, 13). Since PAC synthesis is induced by PA, we analyzed the possibility that PaaX might directly or indirectly control the transcription of the pac gene. A detailed analysis of the Ppac promoter region showed a potential PaaX binding site located between positions −125 and −111 with respect to the transcriptional start site described for the pac gene (27) (Fig. 2). This putative operator has a palindromic sequence with a pseudodyad axis through a central T base, in perfect agreement with the consensus sequence for the PaaX operators described so far (12) (Fig. 2). The inverted sequences of the consensus PaaX binding region are well conserved in the Ppac promoter since only one and two nucleotide changes are detected on the right and left half of the binding site, respectively (Fig. 2).
FIG. 2.
In vitro demonstration of the PaaX function on the Ppac promoter. (A) Nucleotide sequence of the Ppac promoter of E. coli W. The putative −35 and −10 boxes, the ribosome binding site (RBS), and the transcription start site (+1) are underlined. The ATG start codon of pac is shown in italics. The PaaX putative operator located at −118 relative to the transcription start site is indicated by gray letters. Putative CRP binding motifs (CRP 1 [−110.5] and CRP 2 [−68.5]) are boxed. The locations of the primers PAC5′ and PAC3′ used to amplify the PPAC DNA fragment are shown. (B) Alignment of the nucleotide sequences of the described operators for PaaX. The Pz promoter drives the expression of the paaZ gene, while Pa controls the expression of the operon paaABCDEFGHIJK; both promoters control expression from the paa cluster of the PA degradation pathway in E. coli. The promoters are numbered with respect to the transcription start site (+1) of each respective promoter. Nucleotides that are not conserved relative to the consensus site (12) are indicated by gray letters. (C) In vitro binding of PaaX to the Ppac promoter by gel retardation assay. Lanes 2 and 3 contain 31 and 630 ng, respectively, of total protein of PaaX-free extracts from strain W14(pUC18). Lanes 5 to 8 contain 6.3, 12.6, 31, and 630 ng, respectively, of total protein of extracts obtained from E. coli W14(pAFX). A − indicates that no extract was added to the reaction mixture. (D) PaaX released by PA-CoA. Lanes 2 and 3 contain 31 ng of total protein extracts from the strain E. coli W14(pAFX). PA-CoA (125 μM) was added to the reaction mixture in lane 3. The − indicates that no extract was added to the reaction mixture. The DNA probe used for the experiments whose results are shown in panels C and D was PPAC at 1 nM.
To test the ability of PaaX to bind a Ppac promoter, we performed gel retardation assays (15) using a DNA fragment of 269 bp (PPAC) that covers the entire Ppac promoter as a probe (Fig. 2). The PPAC fragment was generated by PCR with 10 ng of the plasmid pPGA1 (17) and the primers PAC5′ (5′-CGGAATTCTTTACATACAGATAATGACCTGAGC-3′) and PAC3′ (5′-CGGGATCCTCTATTTTTCATTGTATCCTCTGGC-3′). Cell extracts from E. coli W14(pAFX), which expresses the paaX gene, and from E. coli W14(pUC18), which does not produce PaaX (12), were used for the assays. The strain E. coli W14 (14) is a derivative of E. coli W (9), which lacks the whole paa cluster, including the paaX repressor and the paaK ligase coding genes. As expected, whereas the cell extract containing PaaX was able to retard the migration of the PPAC probe in a protein concentration-dependent manner, no protein-DNA complexes were detected when the control extract was tested (Fig. 2). Furthermore, we proved that the interaction of PaaX with Ppac was specific, since a 100-fold excess of unlabeled PPAC fragment prevented the formation of the PaaX-DNA complex, whereas an excess of salmon DNA did not (data not shown). More important, PA-CoA inhibited the binding of PaaX to the Ppac promoter, suggesting that it should be the true inducer of the system (Fig. 2).
Demonstration that PaaX regulates in vivo the expression of the pac gene.
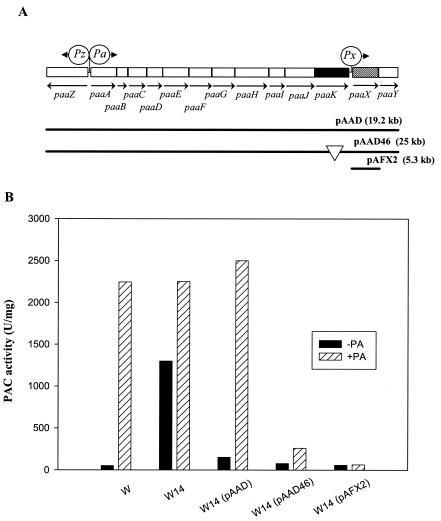
To establish in vivo the role of PaaX in pac expression, we first compared the PAC activity (2) produced by the mutant E. coli W14 (Δpaa) (14) with that of the wild-type E. coli W (Fig. 3). Remarkably, the paa mutant W14 produced a large amount of PAC in the absence of PA, strongly suggesting that the paa cluster is involved in the regulation of the Ppac promoter and that, most likely, PaaX behaves as a repressor. More interesting, when E. coli W14 was transformed with the low-copy-number plasmid pAAD, which harbors the paa cluster (13), the expression of the Ppac promoter recovered its normal PA dependence (Fig. 3), confirming that the Ppac promoter was regulated by a gene of the paa cluster. To unequivocally ascribe a function to PaaX in Ppac regulation, we compared the PAC activities of three strains: E. coli W14(pAAD46), carrying a pAAD derivative plasmid lacking the PaaK ligase-coding gene (13); E. coli W14(pAFX2), harboring a plasmid that expresses in trans the paaX gene under the control of the Plac promoter (13); and the wild type, E. coli W (Fig. 3). The PAC activity was strongly reduced in the absence of PaaK ligase, suggesting that PA-CoA is the true inducer of PAC synthesis. In addition, the overexpression of paaX produced a hyperrepression of the Ppac promoter.
FIG. 3.
In vivo demonstration of the role of PaaX in the expression driven by the Ppac promoter. (A) Genetic organization of the paa pathway for the catabolism of PA in E. coli. Relevant genes are indicated by blocks. The regulatory gene paaX and the PA-CoA ligase-coding gene paaK are represented by hatched and black blocks, respectively. The arrows show the direction of gene transcription. Circles indicate Pa, Pz, and Px promoters. Plasmids containing different subcloned DNA fragments are indicated with continuous lines. The location of the insertion of the Tn1000 transposon in plasmid pAAD46 is indicated by a white triangle. (B) Diagram showing the PAC activity produced in E. coli W and W14 cells containing different plasmids. Cells were grown overnight in Luria broth at 28°C in the absence (black columns) or in the presence (hatched columns) of 1 mM PA. The crude protein extracts were obtained as described previously (12). PAC activity was measured as described previously (2), with 1 U of PAC activity being defined as the amount of enzyme required to form 1 nmol of 6-aminopenicillanic acid per min at 37°C. Protein concentrations were determined by the method of Bradford (4).
In vitro analysis of CRP and IHF binding to the Ppac promoter.
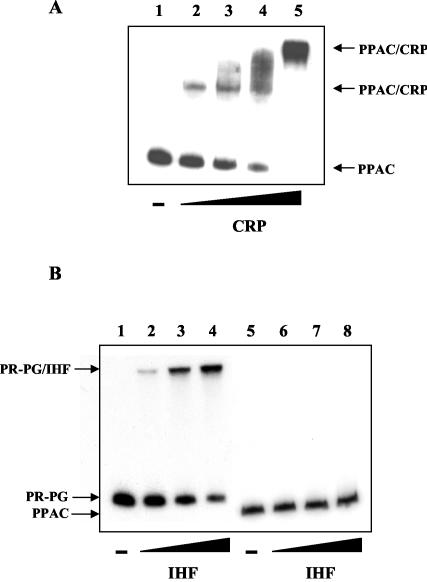
The implication of the presence of the global regulators cyclic AMP receptor protein (CRP) and integration host factor (IHF) in the pac regulatory apparatus has been demonstrated by in vivo experiments (27, 29, 32), but direct evidence of the binding of these proteins to the Ppac promoter has not been obtained. Using the PPAC fragment as a probe, we tested the binding of purified CRP (kindly provided by A. Kolb) and IHF (kindly provided by F. Boccard) to the Ppac region by use of a gel retardation assay (Fig. 4). These analyses have demonstrated that CRP interacts directly with the Ppac promoter. Remarkably, inasmuch as the amount of the CRP increased, two distinct CRP-PPAC complexes were detected, a result that is in perfect agreement with those for the two putative CRP binding sites predicted in the Ppac region (Fig. 2). These results also suggest that CRP binds with different affinities to both sites.
FIG. 4.
In vitro binding of purified global regulators CRP and IHF to the Ppac promoter. (A) CRP binding to the Ppac promoter. A gel retardation assay was carried out in the presence of 200 μM cyclic AMP in an electrophoresis buffer. The DNA probe used was PPAC at 1 nM. The − indicates the absence of purified CRP. Lanes 2 to 5 contain 50, 100, 150, and 300 nM of purified CRP, respectively. (B) IHF binding to the Ppac promoter. The DNA probes used were PR-PG at 1 nM and PPAC at 1 nM. Lanes 2 and 6 contain 100 nM protein, lanes 3 and 7 contain 200 nM protein, and lanes 4 and 8 contain 300 nM protein.
The role of IHF as a positive transcriptional regulator of the pac gene has been demonstrated by using an IHF− strain (29). Furthermore, two putative sites for IHF binding in the Ppac promoter between the positions −76 and −88 and positions −137 and −149 have been proposed (29). This finding appears to be in agreement with that of the paa regulatory system, where CRP, IHF, and PaaX coregulate the catabolic operons (12). However, our assay showed that IHF was not able to retard the migration of the PPAC probe, whereas it was able to retard our control IHF probe, PR-PG (15). This result suggests that IHF does not bind to the Ppac promoter region, and therefore, the response observed in vivo is most likely due to an indirect effect on other global regulatory systems.
Occurrence of paaX-homologous genes in several strains used for the industrial production of PAC.
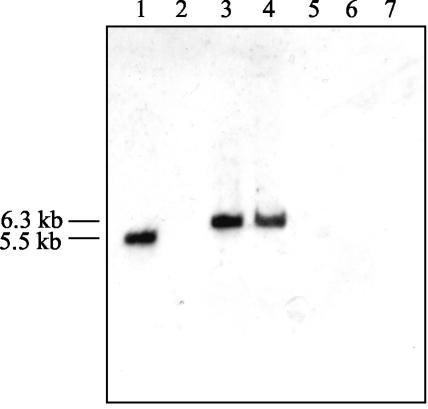
The PA induction of PAC synthesis is not restricted to E. coli W, since other PAC producer bacteria, like Bacillus megaterium, have shown similar responses to PA (34, 35). Taking into account that many other bacteria mineralize PA by homologous paa clusters (10), we have investigated by Southern blot analysis the presence of paaX-homologous genes in several PAC producer microorganisms (19). The paaX DNA fragment (950 bp) used as a probe containing the complete paaX gene was generated by PCR with primers X5-Bam (5′-TCGGATCCGTAAACTTGTTACTTTTATCC-3′) and X3-Sac (5′-CCGGAGCTCGACCATCTATCTG-3′) with plasmid pAAD as a template. The PAAX probe was labeled by the random primer method using the digoxigenin system (Boehringer Mannheim). The results shown in Fig. 5 suggest that the three PAC producer strains tested, Kluyvera citrophila ATCC 21285, a close relative of E. coli (3); B. megaterium ATCC 14945 (20); and Providencia rettgeri ATCC 31052 (8), did not contain DNA sequences homologous to paaX.
FIG. 5.
Presence of paaX-homologous genes in heterologous microorganisms. Results of Southern blot analysis of chromosomal DNAs from E. coli W, E. coli W14, E. coli HB101 (28), E. coli MC4100 (25), K. citrophila, B. megaterium, and P. rettgeri are shown. Total DNAs were digested with EcoRI and were probed with fragment PAAX. The sizes of the bands corresponding to the EcoRI fragments that contain sequences homologous to the paaX gene are indicated.
The finding of the lack of a paaX-homologous gene in K. citrophila is in agreement with the fact that it is not able to mineralize PA (unpublished data) and that PAC production is constitutive in this strain (17). Nevertheless, a detailed analysis of the Ppac promoter from K. citrophila revealed a nucleotide sequence (5′-GAAATGATTCGCTTT-3′) located between positions −64 and −78 of its corresponding transcriptional start site (26), which shows only three nucleotide mismatches compared with the consensus PaaX operator binding site (Fig. 2). This finding might explain the unexpected PA-dependent expression of the pac gene from K. citrophila when it was cloned and expressed in E. coli K-12 strains (26). Moreover, as shown in Fig. 5, some E. coli K-12 strains harbor the paaX gene, and therefore, we assumed that we could ascribe to this gene a role in the expression of the cloned heterologous pac gene. To determine if PaaX might also control the expression of the pac gene from K. citrophila, we transformed this strain with the plasmid pAFX2 expressing in trans the paaX gene from E. coli W. Remarkably, the PAC activity of K. citrophila (pAFX2) was 4.5-fold lower than that of the wild-type strain (data not shown), strongly suggesting that PaaX is also able to recognize the homologous operator sequence found in K. citrophila and repress pac expression. This result poses the intriguing question of why a pac gene that can be regulated by PaaX still survives in a strain which does not have PaaX and which does not use PA as a carbon source. Perhaps this is because PAC may not be exclusively dedicated to the generation of PA, and it may fulfill other missions not yet defined. In summary, the results presented above allow us to unequivocally establish that the pac gene of E. coli W is regulated by the PaaX repressor of the paa catabolic cluster. The induction by PA of PAC activity is mediated by the synthesis of the PA-CoA derivative, the first intermediate of the PA catabolic pathway. Moreover, it is worth mentioning the finding that the palindromic sequence proposed as the PaaX operator for the Ppac promoter is located upstream of the −35 box, overlapping one of the CRP binding sites. This location is certainly very unusual and suggests a novel mechanism of repression that will require more-sophisticated analyses. Finally, our results reveal a quite complex evolutionary scheme for the PA-CoA catabolon. Taking into account both the role of PAC as a PA scavenger enzyme and the fact that both systems, the pac and paa genes, are controlled by the same regulator (PaaX), one might expect that these genes evolved in a coordinated way. However, other data suggest that these genes have followed independent evolutionary tracks, since pac is located far from the paa cluster in E. coli W, it is absent in E. coli K-12 but contains the paa cluster, and it is alone in K. citrophila. These observations suggest that the pac gene has been acquired by E. coli W as a peripheral pathway to funnel the PA esters and amides to a resident PA central pathway. Thereafter, it was further subjugated to the discipline of PaaX repression by introducing the PaaX palindromic sequences within the Ppac promoter.
Acknowledgments
We thank E. Díaz for helpful discussions. We are indebted to F. Boccard and A. Kolb for the kind gift of purified IHF and CRP proteins, respectively. We gratefully acknowledge the technical assistance of E. Cano and I. Alonso.
This work was supported by the Comisión Interministerial de Ciencia y Tecnología (grants ABM97-603-C02-02, BIO2003-05309-C04-02, and CICYT-P4 [Optimización de la producción de cefalosporina]).
REFERENCES
- 1.Arroyo, M., I. de la Mata, C. Acebal, and P. Castillón. 2003. Biotechnological applications of penicillin acylases: state-of-the-art. Appl. Microbiol. Biotechnol. 60:507-514. [DOI] [PubMed] [Google Scholar]
- 2.Balasingham, K., D. Warburton, P. Dunnill, and M. D. Lilly. 1972. The isolation and kinetics of immobilised penicillin amidases from Escherichia coli. Biochim. Biophys. Acta 276:250-256. [DOI] [PubMed] [Google Scholar]
- 3.Barbero, J. L., J. M. Buesa, G. González de Buitrago, E. Menéndez, A. Pérez-Aranda, and J. L. García. 1986. Complete nucleotide sequence of the penicillin acylase gene from Kluyvera citrophila. Gene 49:69-80. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cole, B. M. 1969. Hydrolysis of penicillins and related compounds by the cell-bound penicillin acylase of Escherichia coli. Biochem. J. 115:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, B. M. 1969. Penicillins and other acylamino compounds synthesized by the cell-bound penicillin acylase of Escherichia coli. Biochem. J. 115:747-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, M., Y. Zhu, Y. Yang, E. Wang, Y. Xie, G. Zhao, and W. Jiang. 2001. Expression of penicillin G acylase from the cloned pac gene of Escherichia coli ATCC 11105. Effects of PacR and temperature. Eur. J. Biochem. 268:1298-1303. [DOI] [PubMed] [Google Scholar]
- 8.Daumy, G. O., J. A. Williams, A. S. McColl, T. J. Zuzel, and D. Danley. 1986. Expression and regulation of the penicillin G acylase gene from Proteus rettgeri cloned in Escherichia coli. J. Bacteriol. 168:431-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, B. D., and E. S. Mingioli. 1950. Mutants of Escherichia coli requiring methionine or vitamin B12. J. Bacteriol. 60:17-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Díaz, E., A. Ferrández, M. A. Prieto, and J. L. García. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol. Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elander, R. P. 2003. Industrial production of β-lactam antibiotics. Appl. Microbiol. Biotechnol. 61:385-392. [DOI] [PubMed] [Google Scholar]
- 12.Ferrández, A., J. L. García, and E. Díaz. 2000. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. J. Biol. Chem. 275:12214-12222. [DOI] [PubMed] [Google Scholar]
- 13.Ferrández, A., B. Miñambres, B. García, E. R. Olivera, J. M. Luengo, J. L. García, and E. Díaz. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 14.Ferrández, A., M. A. Prieto, J. L. García, and E. Díaz. 1997. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 406:23-27. [DOI] [PubMed] [Google Scholar]
- 15.Galán, B., A. Kolb, J. L. García, and M. A. Prieto. 2001. Superimposed levels of regulation of the 4-hydroxyphenylacetate catabolic pathway in Escherichia coli. J. Biol. Chem. 276:37060-37068. [DOI] [PubMed] [Google Scholar]
- 16.Gang, D. M., and K. Shaikh. 1976. Regulation of penicillin acylase in Escherichia coli by cyclic AMP. Biochim. Biophys. Acta 425:110-114. [DOI] [PubMed] [Google Scholar]
- 17.García, J. L., and J. M. Buesa. 1986. An improved method to clone penicillin acylase genes: cloning and expression in Escherichia coli of penicillin G acylase from Kluyvera citrophila. J. Biotechnol. 3:187-195. [Google Scholar]
- 18.Hewitt, L., V. Kasche, K. Lummer, R. J. Lewis, G. N. Murshudov, C. S. Verma, G. G. Dodson, and K. S. Wilson. 2000. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J. Mol. Biol. 302:887-898. [DOI] [PubMed] [Google Scholar]
- 19.Mahajan, P. B. 1984. Penicillin acylases. An update. Appl. Biochem. Biotechnol. 9:537-554. [DOI] [PubMed] [Google Scholar]
- 20.Martín, L., M. A. Prieto, E. Cortés, and J. L. García. 1995. Cloning and sequencing of the pac gene encoding the penicillin G acylase of Bacillus megaterium ATCC 14945. FEMS Microbiol. Lett. 125:287-292. [DOI] [PubMed] [Google Scholar]
- 21.Merino, E., P. Balbas, F. Recillas, B. Becerril, F. Valle, and F. Bolivar. 1992. Carbon regulation and the role in nature of the Escherichia coli penicillin acylase (pac) gene. Mol. Microbiol. 6:2175-2182. [DOI] [PubMed] [Google Scholar]
- 22.Oh, S. J., Y. C. Kim, Y. W. Park, S. Y. Min, I. S. Kim, and H. S. Kang. 1987. Complete nucleotide sequence of the penicillin G acylase gene and the flanking regions, and its expression in Escherichia coli. Gene 56:87-97. [DOI] [PubMed] [Google Scholar]
- 23.Prieto, M. A., A. Perez-Aranda, and J. L. Garcia. 1993. Characterization of an Escherichia coli aromatic hydroxylase with a broad substrate range. J. Bacteriol. 175:2162-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prieto, M. A., E. Díaz, and J. L. García. 1996. Molecular characterization of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli W: engineering a mobile aromatic degradative cluster. J. Bacteriol. 178:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prieto, M. A., and J. L. García. 1997. Identification of a novel positive regulator of the 4-hydroxyphenylacetate catabolic pathway of Escherichia coli. Biochem. Biophys. Res. Commun. 232:759-765. [DOI] [PubMed] [Google Scholar]
- 26.Roa, A., and J. L. García. 1997. Identification of the pac promoter from Kluyvera citrophila. FEMS Microbiol. Lett. 151:9-16. [DOI] [PubMed] [Google Scholar]
- 27.Roa, A., and J. L. García. 1999. New insights into the regulation of pac gene from Escherichia coli W ATCC 11105. FEMS Microbiol. Lett. 177:7-14. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Rusell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Stojcevic, N., I. Moric, J. Begovic, S. Radoja, and M. Konstantinovic. 2001. DNA architecture and transcriptional regulation of the Escherichia coli penicillin amidase (pac) gene. Biomol. Eng. 17:113-117. [DOI] [PubMed] [Google Scholar]
- 30.Sudhakaran, V. K., B. S. Deshpande, S. S. Ambedkar, and J. G. Shewale. 1992. Molecular aspects of penicillin and cephalosporin acylases. Process Biochem. 27:131-143. [Google Scholar]
- 31.Szentimai, A. 1964. Production of penicillin acylase. Appl. Microbiol. 12:185-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valle, F., G. Gosset, B. Tenorio, G. Oliver, and F. Bolivar. 1986. Characterization of the regulatory region of the Escherichia coli penicillin acylase structural gene. Gene 50:119-122. [DOI] [PubMed] [Google Scholar]
- 33.Valle, F., P. Balbas, E. Merino, and F. Bolivar. 1991. The role of penicillin amidases in nature and in industry. Trends Biochem. Sci. 16:36-40. [DOI] [PubMed] [Google Scholar]
- 34.Vandamme, E. J., and J. P. Voets. 1974. Microbial penicillin acylases. Adv. Appl. Microbiol. 17:311-369. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, L. F., Z. W. Li, and Q. J. Zhang. 1991. Cloning and expression of penicillin G acylase gene in Bacillus megaterium. Chin. J. Biotechnol. 7:63-72. [PubMed] [Google Scholar]