Abstract
Replication of rolling-circle replicating (RCR) plasmids in gram-positive bacteria requires the unwinding of initiator protein-nicked plasmid DNA by the PcrA helicase. In this report, we demonstrate that heterologous PcrA helicases from Bacillus anthracis and Bacillus cereus are capable of unwinding Staphylococcus aureus plasmid pT181 from the initiator-generated nick and promoting in vitro replication of the plasmid. These helicases also physically interact with the RepC initiator protein of pT181. The ability of PcrA helicases to unwind noncognate RCR plasmids may contribute to the broad-host-range replication and dissemination of RCR plasmids in gram-positive bacteria.
PcrA is an essential helicase in gram-positive bacteria that is also required for the rolling-circle (RC) replication of small, multicopy plasmids (5, 6, 12, 13, 21, 29, 30, 33). PcrA from Staphylococcus aureus and Bacillus anthracis have both 5′→3′ and 3′→5′ helicase activities (5, 27). PcrA from Bacillus stearothermophilus appears to be predominantly a 3′→5′ helicase (2, 33-35). Although most rolling-circle replicating (RCR) plasmids have been identified from gram-positive bacteria, they are also found in gram-negative bacteria and in archaea (for reviews, see references 6, 10, 16, 20, 21, and 28). Many RCR plasmids have a broad host range and can be established in diverse bacteria (6, 10, 20, 22, 28). The pcrA gene, which was originally identified as being required for plasmid pT181 replication, has been identified in all the gram-positive bacteria whose genomes have been sequenced so far (12-14). Some RCR plasmids, such as pC194, have also been shown to replicate, although at a lower copy number, in Escherichia coli that lacks the PcrA helicase (3, 29). In this host, pC194 replication is supported by DNA helicase II (UvrD) which has 40% identity to PcrA (3, 11, 26, 29). Recent in vitro studies have shown a direct requirement for the PcrA helicase in the replication of the pT181 plasmid of S. aureus (5). Purified PcrA was found to stimulate plasmid pT181 replication in vitro when added to cell extracts made from the pcrA3 mutant of S. aureus that is defective in pT181 replication (5). These and other studies also showed that PcrA promotes unwinding of pT181 and the related pC221 DNA nicked at the origin by the plasmid initiator protein (5, 33). Also, PcrA-driven DNA unwinding of pT181 required the presence of RepC protein covalently attached to the 5′ phosphate end at the nick site, since nicked open circular (OC) DNA from which RepC was removed was not unwound by RepC (5). Finally, pull-down studies showed that RepC and PcrA directly interact (5). Taken together, the results of the pull-down studies and in vivo studies (13) suggest that an interaction between the plasmid initiator proteins and heterologous PcrA helicases may be critical in establishing RCR plasmids in different hosts. In this study, we demonstrate that heterologous PcrA helicases from two gram-positive bacteria B. anthracis and Bacillus cereus can functionally interact with the initiator protein of plasmid pT181 and support pT181 replication in vitro. We also demonstrate that the pT181 replicon can support plasmid replication in vivo in B. anthracis and B. cereus.
Purification of the PcrA helicases of S. aureus, B. anthracis, and B. cereus.
The purification of the PcrA helicases from S. aureus and B. anthracis fused at their amino-terminal end to six histidine residues (His6) using nickel affinity chromatography has been described previously (5, 27). To purify the PcrA helicase from B. cereus, chromosomal DNA was prepared from strain ATCC 10987 by incubating the cells in a cetyltrimethylammonium bromide (CTAB) solution at 65°C, followed by chloroform extraction and isopropanol precipitation as described previously (27). Preliminary sequence data of the B. cereus genome were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org). The pcrA gene of B. cereus encodes 747 amino acids and contains the ATG sequence at the first and third codons. The pcrA open reading frame was amplified by PCR such that it included amino acids 4 to 745 and was fused in frame (using vector pQE30) to His6 residues at its amino-terminal end. The first three amino acids were not included to rule out the possibility of initiation from the ATG codon of pcrA instead of the ATG codon of the His6 epitope. The genomic DNA of B. cereus isolated by the procedure described above was used as a template for the amplification of the pcrA gene (2.2 kb). The sequences of the primers used follow: 5′-CCGGATCCACAGATAGATTATATTAAATGGTTTAAATCCGCAGCAAC-3′ for the forward primer and 5′-CCGGATCCCGTTTTTTTGCTATCTCTTTTGACATATCCTCATTCC-3′ for the reverse primer. The PCR primers contained BamHI linkers at their ends. The reaction mixtures contained a 200 μM concentration of each deoxynucleoside triphosphate, 200 ng of B. cereus genomic DNA, a 1 μM each concentration of primer, and 5 U of the Pfu polymerase (Stratagene, La Jolla, Calif.). The conditions of amplification were as follows: (i) 94°C for 3 min; (ii) 35 cycles, with 1 cycle consisting of 1 min at 94°C, 1 min at 55°C, and 6 min at 72°C; and (iii) 72°C for 10 min. The amplified product was gel purified and digested with BamHI. The pcrA gene was then fused in frame to the His6 epitope at the BamHI site of the pQE30 vector from Qiagen. The ligation mixture was then introduced into E. coli M15 by electroporation, and the appropriate clones were isolated for protein overexpression. The sequence of the cloned B. cereus pcrA gene was confirmed by automated DNA sequencing.
A single colony of the E. coli strain expressing B. cereus PcrA was used for the overexpression of His-PcrA by induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C for 2 h. The His-PcrA fusion protein was purified by nickel affinity chromatography as described (5) using protease inhibitors throughout the purification procedures. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Fig. 1) showed that the full-length protein was approximately 90% pure. To confirm that the purified B. cereus PcrA was enzymatically active, we measured its ATPase activity by hydrolysis of [α-32P]ATP as described earlier (5). These studies showed that, as observed with the PcrA helicases of S. aureus and B. anthracis (5, 27), the B. cereus PcrA had an ATPase activity that was stimulated by single-stranded DNA (ssDNA) (data not shown).
FIG. 1.
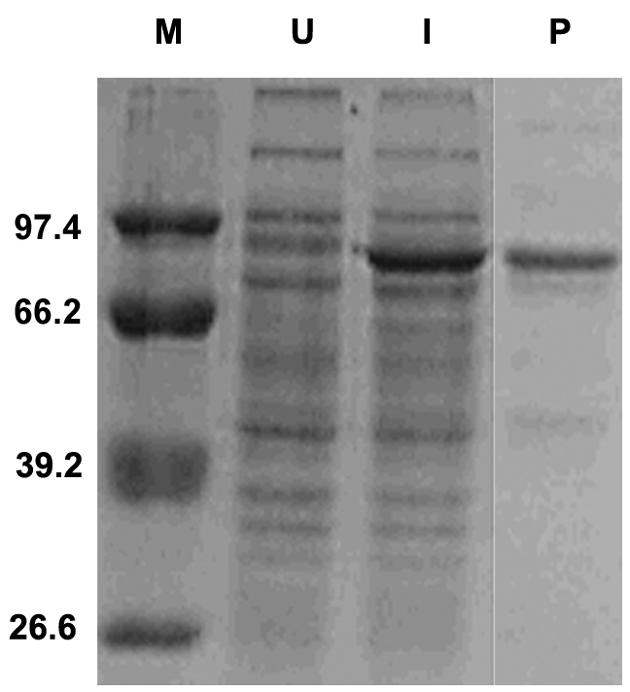
SDS-PAGE analysis of the purified B. cereus PcrA protein. Lanes: U, lysates from uninduced cells; I, lysates from IPTG-induced cells overexpressing the His-PcrA protein; P, B. cereus PcrA protein purified by nickel affinity column chromatography; M, protein molecular mass standards (in kilodaltons).
Heterologous PcrA helicases promote unwinding of RepC-nicked pT181 DNA.
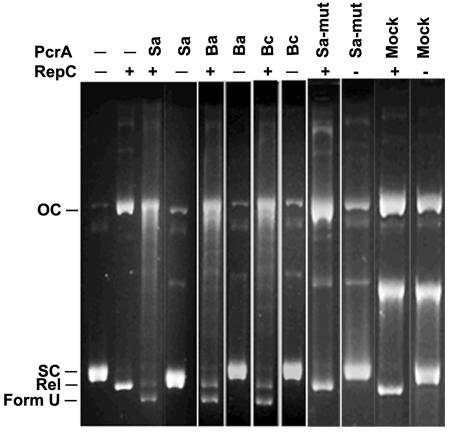
Experiments were performed to determine whether the PcrA helicases of B. anthracis and B. cereus can unwind supercoiled (SC) pT181cop608 DNA nicked by the initiator RepC protein. pT181cop608 DNA (0.5 μg) was incubated in TEKEM buffer containing 5 mM ATP in the presence or absence of RepC (200 ng) and/or the PcrA helicases (10 ng) at 32°C for 30 min (25). The reaction products were subjected to electrophoresis on 1% agarose gels with Tris-borate-EDTA buffer containing 0.5 μg of ethidium bromide per ml (5). Incubation of pT181 DNA with RepC results in the formation of covalently closed, relaxed pT181 DNA that migrates faster than SC DNA in the presence of ethidium bromide (Fig. 2) (7, 23, 25).
FIG. 2.
B. anthracis and B. cereus PcrA helicases can unwind plasmid pT181 DNA from a RepC-generated nick. Plasmid pT181cop608 DNA was incubated with 10 ng of different PcrA helicases from different species in the absence (−) or presence (+) of RepC, and the reaction products were analyzed by agarose gel electrophoresis in the presence of ethidium bromide. The PcrA helicases were from S. aureus (Sa), B. anthracis (Ba), B. cereus (Bc), and the K33AQ250R mutant of S. aureus PcrA (Sa-mut). The nickel affinity column fraction obtained by mock purifying proteins from the wild-type E. coli M15 cells (Mock) was tested. The positions of nicked open-circular DNA (OC), supercoiled plasmid DNA (SC), covalently closed relaxed DNA (Rel), and unwound plasmid DNA (U) are indicated to the left of the gel.
As observed previously (5), the S. aureus PcrA helicase unwound RepC-nicked DNA, which resulted in the formation of the faster-migrating unwound form of the DNA (Fig. 2). We have previously shown that the highly unwound form of the DNA contains extensive ssDNA regions that are digested by the S1 nuclease (5). This form of DNA is expected to contain RepC covalently attached to the 5′ phosphate end and a free 3′ OH end that serves as a primer for the initiation of plasmid RC replication (5). This form of DNA differs from the U form of DNA generated during theta-type replication which contains unwound but covalently closed DNA (17). As expected, treatment of SC pT181 DNA with different PcrA helicases alone in the absence of RepC did not affect the migration pattern of the DNA (Fig. 2). However, when RepC was added along with 10 ng (each) of the B. anthracis and B. cereus helicases to SC pT181 DNA, the faster-migrating unwound form of the DNA was observed (Fig. 2). Several faint bands migrating between the SC and OC forms of DNA were also visible in these samples. These bands presumably represent DNA that has been unwound to different degrees by PcrA in the presence of RepC, since we have previously shown that these bands are sensitive to degradation by the S1 nuclease (5). Incubation of PcrA helicases with nicked pT181 DNA from which RepC had been removed by proteinase K treatment and phenol extraction did not generate the unwound DNA (data not shown).
To rule out the possibility that a contaminating E. coli protein may be responsible for unwinding DNA, we purified a His6 fusion of the K33AQ250R mutant of the S. aureus PcrA helicase which is defective in its ATPase and helicase activities (data not shown) and used it in this assay. As shown in Fig. 2, this mutant was unable to unwind RepC-nicked pT181 DNA. We also mock purified proteins from the host E. coli M15 cells lacking PcrA using a nickel affinity resin under conditions identical to those used for purifying PcrA. Addition of 10 ng of mock-purified protein from the fraction corresponding to where PcrA elutes did not generate the unwound form of pT181 DNA in the presence of RepC (Fig. 2). Instead, a new band presumably corresponding to linear DNA was observed in these lanes. This band is likely to be generated due to the presence of nonspecific endonucleases in the mock-purified fraction, since a very large volume of this fraction was added to the reaction mixtures (since extremely low levels of nonspecific proteins bind to the nickel affinity resin). The above results demonstrate that heterologous PcrA helicases can unwind the noncognate pT181 DNA that had been nicked at the origin by RepC. These results further suggest a functional interaction between the heterologous PcrA helicases and RepC, since the presence of the initiator protein is required for PcrA-catalyzed SC pT181 DNA unwinding (5).
B. anthracis and B. cereus PcrA helicases support plasmid pT181 replication in vitro.
While DNA unwinding by PcrA is essential during the initiation of plasmid RC replication, the ability of this helicase to interact with other components of the replisome may also be critical during the elongation and termination of replication. Therefore, we tested whether the heterologous helicases can promote replication of plasmid pT181 DNA in vitro. For these studies, cell extracts were prepared from the pcrA3 mutant of S. aureus as described earlier (4, 5). The mutant PcrA3 helicase is inactive in plasmid pT181 replication but retains functions required for cell growth and viability (5, 12, 29). Cell-free replication extracts were prepared from the S. aureus strain RN4220 and the pcrA3 mutant as described previously (5, 24). Replication reaction mixtures (30 μl) contained 600 μg of protein extracts, 500 ng of pT181cop608 DNA, 200 ng of RepC protein, and 100 ng of the different PcrA helicases. Replication products were labeled with [α-32P]dATP. Reaction mixtures were incubated at 32°C for 1 h and treated with proteinase K, and DNA was isolated by phenol-chloroform extraction and alcohol precipitation (4, 5). The reaction products were subjected to electrophoresis on 1% agarose gels using Tris-borate-EDTA buffer containing 1 μg of ethidium bromide per ml (32). Gels were dried and subjected to autoradiography.
Incubation of plasmid pT181 DNA with wild-type cell extract along with RepC generated labeled DNA corresponding to the SC and OC forms of the DNA, as well as replication intermediates and low levels of ssDNA (Fig. 3). In the presence of the pcrA3 cell extract, a very faint band corresponding to the nicked OC form of pT181 DNA was observed (Fig. 3). This band reflects the incorporation of a few nucleotides immediately downstream of the RepC nick site in the absence of any PcrA activity (14, 18, 19). As observed earlier (5), addition of the cognate S. aureus PcrA to the mutant pcrA3 extract restored replication of the pT181 DNA (Fig. 3). Addition of purified His-PcrA from B. anthracis and B. cereus to the mutant extract also resulted in the restoration of plasmid pT181 replication (Fig. 3). These results showed that the heterologous PcrA helicases can substitute for the S. aureus PcrA helicase, presumably playing a role during both the initiation and elongation phases of pT181 replication. Furthermore, the B. anthracis and B. cereus PcrA helicases must be able to interact with S. aureus replication proteins that are involved in pT181 RC replication.
FIG. 3.
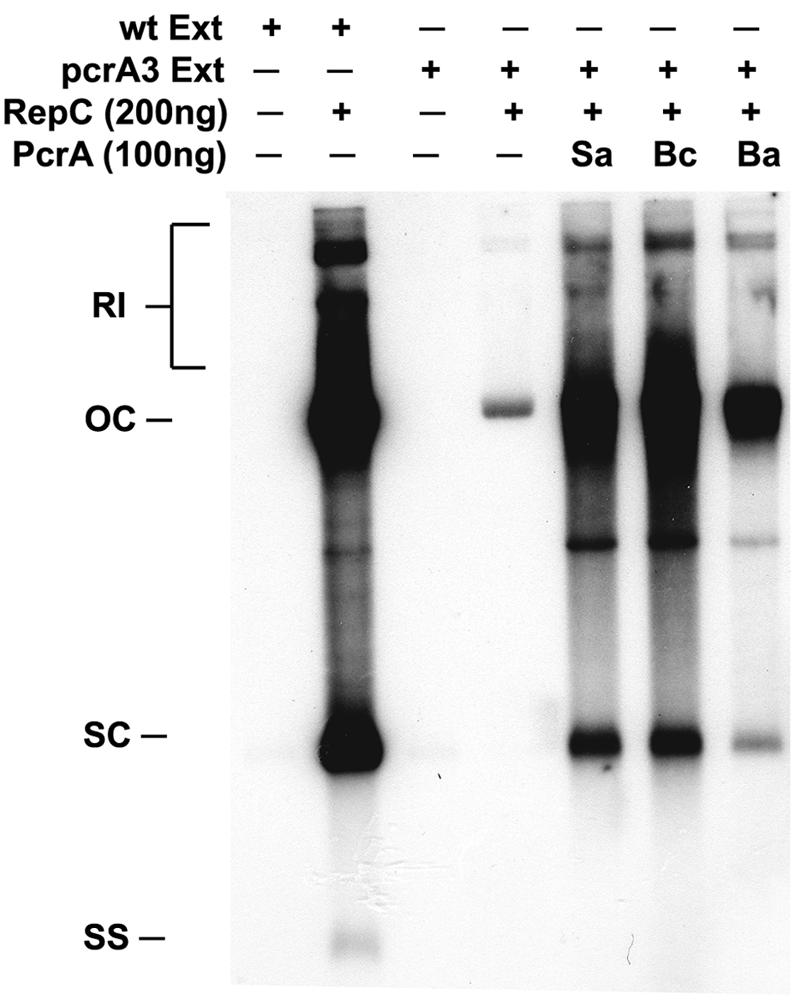
B. anthracis and B. cereus PcrA helicases support in vitro replication of the heterologous pT181 plasmid. In vitro replication was performed using the RepC protein (+), and cell extracts (Ext) were made from either wild-type (Wt) or pcrA3 mutant S. aureus cells. PcrA helicases from S. aureus (Sa), B. cereus (Bc), and B. anthracis (Ba) were tested. The positions of replication intermediates (RI), open circular DNA (OC), supercoiled pT181cop608 DNA (SC), and single-stranded DNA (SS) are shown to the left of the gel.
Direct physical interaction between B. anthracis and B. cereus PcrA helicases and RepC.
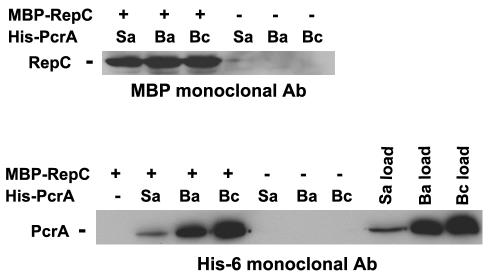
Previous studies have shown that the S. aureus PcrA physically interacts with the RepC initiator protein of pT181 and that this interaction is important for the recruitment of this helicase to the plasmid origin of replication (5). Since B. anthracis and B. cereus PcrA helicases supported RepC-dependent pT181 DNA unwinding and replication, we tested whether these helicases also interact directly with the RepC protein. To test this, we made use of the different epitope tags present on the PcrA and RepC proteins. Cell lysates from E. coli expressing the maltose binding protein (MBP)-RepC fusion protein were absorbed to 50 μl of amylose resin as described previously (4) and washed with 1× TEKEM buffer containing 1% bovine serum albumin (BSA). Subsequently, equal amounts of cell lysates containing different His-PcrA helicases were mixed with MBP-RepC bound to the resin and incubated at 4°C for 1 h. The suspension was then washed three times with 1× TEKEM buffer, and the proteins eluted directly in SDS-PAGE sample buffer (32). In control experiments, the His-PcrA proteins were mixed with amylose resin treated with 1% BSA but not containing any bound MBP-RepC. The eluted proteins were blotted onto membranes (32) and hybridized with either an anti-MBP monoclonal antibody (Santa Cruz Biotechnology) or anti-His6 monoclonal antibody (Sigma) and visualized by using an enhanced chemiluminescence (ECL) kit from Amersham according to the manufacturer's instructions. Western blot analysis using the anti-MBP monoclonal antibody showed that MBP-RepC was bound to the amylose resin as expected (Fig. 4). Furthermore, while His-PcrA proteins did not bind to the amylose resin incubated with BSA, they were retained on the resin to which MBP-RepC was bound (Fig. 4). These results suggest that heterologous PcrA proteins of B. anthracis and B. cereus can interact with RepC.
FIG. 4.
Interaction between B. anthracis and B. cereus PcrA helicases and RepC. The physical interaction between PcrA and RepC proteins was analyzed by a pull-down assay as described in the text. The eluted fractions from amylose resin columns (either containing [+] or lacking [−] bound MBP-RepC) were probed with either anti-MBP or anti-His6 monoclonal antibodies (Ab). His-PcrA helicases from S. aureus (Sa), B. anthracis (Ba), and B. cereus (Bc) were used. Equal amounts of protein extracts containing His-PcrA were deposited in the load control lanes (Sa, Ba, and Bc load).The protein extracts were bound to a Ni-nitrilotriacetic acid column, eluted, and then analyzed by SDS-PAGE and immunoblotting. The same amounts of protein extracts were used in the pull-down assays. In lanes lacking MBP-RepC, 1% BSA was used instead.
Replication of a pT181 derivative plasmid in B. anthracis and B. cereus.
Since heterologous PcrA helicases supported RepC-dependent unwinding and in vitro replication of the pT181 plasmid, we tested whether this plasmid can be established in B. anthracis and B. cereus. Plasmid pT181 encodes resistance to tetracycline. Since current regulations prohibit the introduction of tetracycline resistance genes into B. anthracis, we utilized plasmid pSA5000 in these studies. pSA5000 is a derivative of pT181 in which the tetracycline resistance gene has been replaced by the chloramphenicol resistance gene of plasmid pC221 (15). Plasmid pSA5000 was introduced into the plasmid-less B. anthracis strain UM23C1-1 (9) and B. cereus strain ATCC 10987 by electroporation (8) with selection for the Cmr marker. Chloramphenicol-resistant colonies were isolated, and agarose gel analysis of sheared whole-cell lysates (31) showed the presence of plasmid DNA of the appropriate size in B. anthracis (Fig. 5A). Since the plasmid band in B. cereus transformants was very faint, Southern blot analysis was performed using 32P-labeled pSA5000 DNA as the probe (32). The B. cereus transformants contained pSA5000 DNA, which was present mostly in the OC form (Fig. 5B). These results showed that the plasmid pT181 replicon can be established in B. anthracis and B. cereus. Furthermore, pT181 appeared to be maintained at a much lower copy number in B. cereus than in B. anthracis.
FIG. 5.
Agarose gel analysis of lysed B. anthracis and B. cereus cells containing the pSA5000 plasmid. (A) Ethidium bromide staining of B. anthracis transformed with pSA5000 DNA. (B) Southern blot analysis of a B. cereus transformant containing plasmid pSA5000. The positions of supercoiled plasmid (SC) and open-circular plasmid (OC) are indicated to the left of the gel.
RCR plasmids can be broadly classified into five major families in which individual members show conservation in their initiator proteins and leading strand origins of replication (6, 20). Several studies have suggested that the ss origins of RCR plasmids play a role in determining the host ranges of these plasmids (6, 10, 21). Since an interaction between PcrA and RCR plasmid initiator proteins is critical for replication, this event may also, at least in part, be responsible for narrow- versus broad-host-range replication of RCR plasmids. The B. anthracis and B. cereus helicases share approximately 58% identity and 72% similarity with the PcrA of S. aureus based on BLAST analysis (1). Several studies suggest that PcrA has multiple functions in cellular DNA metabolism, such as DNA repair and possibly the resolution of stalled replication forks and blocked recombination structures generated by the RecFOR pathway (29, 30). The pcrA3 mutation results in a Thr-to-Ile change at position 61 of S. aureus PcrA (12). Interestingly, the pcrA3 mutant is inactive in pT181 replication but supports replication of other RCR plasmids (12, 29). The above series of studies suggest that PcrA contains multiple domains that are likely involved in its various functions. A specific region(s) of PcrA may be involved in their functional interactions with the replication initiator proteins of RCR plasmids.
Our results have shown that the PcrA helicases of B. anthracis and B. cereus can interact with RepC and support RepC-dependent unwinding and in vitro replication of pT181 DNA (Fig. 2 to 4). These data are consistent with our observations that plasmid pT181 can replicate in vivo in B. anthracis and B. cereus (Fig. 5), as well as in many other gram-positive bacteria (10, 16, 20, 29). Our results have shown that the B. anthracis and B. cereus PcrA proteins interact equally well with RepC (Fig. 4). However, pT181 replication levels observed in vitro with the B. cereus PcrA helicase were much higher than those with the B. anthracis PcrA helicase (Fig. 3). It is possible that the B. cereus PcrA interacts more efficiently with the heterologous S. aureus replication proteins than the B. anthracis PcrA does. Interestingly, the copy number of the pT181 derivative plasmid pSA5000 was much lower in B. cereus than in B. anthracis (Fig. 5). Thus, multiple factors are likely to contribute to the broad-host-range replication of RCR plasmids. These factors include Rep-PcrA interactions, the ability of the Rep-PcrA complex to recruit host replication proteins, functionality of the lagging strand origins, and differences in the expression levels of plasmid replication genes in different organisms.
Acknowledgments
We thank members of our laboratory for helpful discussions.
This work was supported in part by grants GM31685 and AI55929 from the National Institutes of Health (to S.A.K.). Sequencing of the B. cereus genome at TIGR was accomplished with financial support from NIH, ONR, DOE, and DERA.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bird, L. E., S. Subramanya, and D. B. Wigley. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14-18. [DOI] [PubMed] [Google Scholar]
- 3.Bruand, C., and S. D. Ehrlich. 2000. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol. Microbiol. 35:204-210. [DOI] [PubMed] [Google Scholar]
- 4.Chang, T.-L., M. G. Kramer, R. A. Ansari, and S. A. Khan. 2000. Role of individual monomers of a dimeric initiator protein in the initiation and termination of plasmid rolling circle replication. J. Biol. Chem. 275:13529-13534. [DOI] [PubMed] [Google Scholar]
- 5.Chang, T.-L., A. Naqvi, S. P. Anand, M. G. Kramer, R. Munshi, and S. A. Khan. 2002. Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling-circle replication. J. Biol. Chem. 277:45880-45886. [DOI] [PubMed] [Google Scholar]
- 6.del Solar, G., R. Giraldo, M. J. Ruiz-Echevarria, M. Espinosa, and R. Diaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dempsey, L. A., P. Birch, and S. A. Khan. 1992. Uncoupling of the DNA topoisomerase and replication activities of an initiator protein. Proc. Natl. Acad. Sci. USA 89:3083-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, B. D., L. Battisti, T. M. Koehler, and C. B. Thorne. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruss, A., and S. D. Ehrlich. 1989. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol. Rev. 53:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 12.Iordanescu, S. 1993. Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol. Gen. Genet. 241:185-192. [DOI] [PubMed] [Google Scholar]
- 13.Iordanescu, S. 1993. Plasmid pT181-linked suppressors of the Staphylococcus aureus pcrA3 chromosomal mutation. J. Bacteriol. 175:3916-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iordanescu, S., and R. Basheer. 1991. The Staphylococcus aureus mutation pcrA3 leads to the accumulation of pT181 replication initiation complexes. J. Mol. Biol. 221:1183-1189. [DOI] [PubMed] [Google Scholar]
- 15.Iordanescu, S., and M. Surdeanu. 1980. Complementation of a replication defect by autonomous incompatible plasmids in Staphylococcus aureus. Plasmid 4:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Janniere, L., A. Gruss, and S. D. Ehrlich. 1993. Plasmids, p. 625-644. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology and molecular genetics. American Society for Microbiology, Washington, D.C.
- 17.Jiang, Y., J. Pogliano, D. R. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44:971-979. [DOI] [PubMed] [Google Scholar]
- 18.Jin, R., M.-E. Fernandez-Beros, and R. P. Novick. 1997. Why is the initiation site of an AT-rich rolling circle plasmid at the tip of a GC-rich cruciform? EMBO J. 16:4456-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, R., and R. P. Novick. 2001. Role of the double-strand origin cruciform in pT181 replication. Plasmid 46:95-105. [DOI] [PubMed] [Google Scholar]
- 20.Khan, S. A. 1997. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 61:442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan, S. A. 2000. Plasmid rolling-circle replication: recent developments. Mol. Microbiol. 37:477-484. [DOI] [PubMed] [Google Scholar]
- 22.Koehler, T. M. 2002. Bacillus anthracis genetics and virulence gene regulation. Curr. Top. Microbiol. Immunol. 271:143-164. [DOI] [PubMed] [Google Scholar]
- 23.Koepsel, R. R., R. W. Murray, and S. A. Khan. 1986. Sequence-specific interaction between the replication initiator protein of plasmid pT181 and its origin of replication. Proc. Natl. Acad. Sci. USA 83:5484-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koepsel, R. R., R. W. Murray, W. D. Rosenblum, and S. A. Khan. 1985. Purification of pT181-encoded RepC protein required for the initiation of plasmid replication. J. Biol. Chem. 260:8571-8577. [PubMed] [Google Scholar]
- 25.Koepsel, R. R., R. W. Murray, W. D. Rosenblum, and S. A. Khan. 1985. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc. Natl. Acad. Sci. USA 82:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 27.Naqvi, A., E. Tinsley, and S. A. Khan. 2003. Purification and characterization of the PcrA helicase of Bacillus anthracis. J. Bacteriol. 185:6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novick, R. P. 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43:537-565. [DOI] [PubMed] [Google Scholar]
- 29.Petit, M. A., E. Dervyn, M. Rose, K. D. Entian, S. McGovern, S. D. Ehrlich, and C. Bruand. 1998. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 29:261-273. [DOI] [PubMed] [Google Scholar]
- 30.Petit, M. A., and S. D. Ehrlich. 2002. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 21:3137-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Projan, S. J., S. Carleton, and R. P. Novick. 1983. Determination of plasmid copy number by fluorescence densitometry. Plasmid 9:182-190. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Soultanas, P., M. S. Dillingham, F. Papadapoulos, S. E. Philips, C. D. Thomas, and D. B. Wigley. 1999. Plasmid replication initiator protein RepD increases the processivity of PcrA DNA helicase. Nucleic Acids Res. 27:1421-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soultanas, P., and D. B. Wigley. 2001. Unwinding the “Gordian knot” of helicase action. Trends Biochem. Sci. 26:47-54. [DOI] [PubMed] [Google Scholar]
- 35.Velankar, S. S., P. Soultanas, M. S. Dillingham, H. S. Subramanya, and D. B. Wigley. 1999. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97:75-84. [DOI] [PubMed] [Google Scholar]