SUMMARY
Lung squamous cell carcinoma (SqCC) is the second most common subtype of non-small-cell lung cancer and leads to 40,000–50,000 deaths per year in the USA. Management of non-small-cell lung cancer has dramatically changed over the past decade with the introduction of targeted therapeutic agents for genotypically selected individuals with lung adenocarcinoma. These agents lead to improved outcomes, and it has now become the standard of care to perform routine molecular genotyping of lung adenocarcinomas. By contrast, progress in lung SqCC has been modest, and there has yet to be a successful demonstration of targeted therapy in this disease. Here, we review exciting work from ongoing genomic characterization and biomarker validation efforts that have nominated several likely therapeutic targets in lung SqCCs. These studies suggest that targeted therapies are likely to be successful in the treatment of lung SqCCs and should be further explored in both preclinical models and in clinical trials.
Despite the recent development of several new and effective therapeutic agents, lung cancer remains the leading cause of cancer-related mortality in the USA [101]. Non-small-cell lung cancer (NSCLC) is far more prevalent than small-cell lung cancer and is comprised of several histologic subtypes, of which adenocarcinoma and squamous cell carcinoma (SqCC) are the most common. The treatment of lung adenocarcinoma, which now routinely includes the use of targeted therapeutic agents, has been extensively reviewed elsewhere [1–3]. The focus of this review will be to highlight recent advances in the discovery of potential therapeutic targets in lung SqCC.
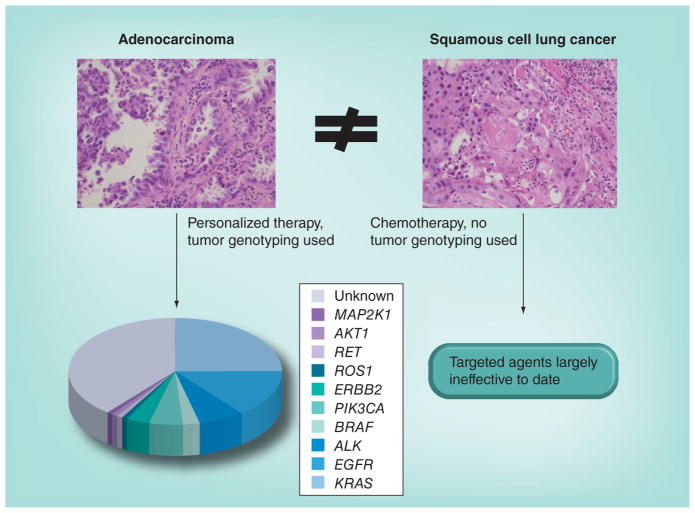
Lung SqCC comprises 20–30% of newly diagnosed NSCLC in the USA and a greater proportion of new cases in Europe and Asia. In contrast to lung adenocarcinoma, which has been the focus of several studies aimed at identifying recurrent genomic alterations that are likely therapeutic targets, laboratory characterization of lung SqCC has only recently begun in earnest. The majority of targetable alterations described in lung adenocarcinomas, including mutations in EGFR [4–6] and EML4–ALK fusions [7], have not been identified as recurrent events in lung SqCC, and clinical trials with pharmacologic agents targeting these kinases have been disappointing in patients with lung SqCC compared with adenocarcinoma. This has led to a tremendous disparity in the way in which patients with advanced NSCLC are currently treated. Patients with lung adenocarcinoma undergo routine genotyping to identify genetic alterations in their tumors that predict response to targeted therapies, given that targeted agents have demonstrated superior efficacy compared with chemotherapy in selected populations [8–11]. However, these personalized treatments are not typically effective for individuals with lung SqCC given the lack of overlapping genomic alterations with lung adenocarcinoma. Patients with lung SqCCs ostensibly receive the same conventional chemotherapy today as they would have received a decade ago (Figure 1).
Figure 1. Current diagnostic and treatment paradigms for lung adenocarcinoma and squamous cell lung cancer.
Tumor genotyping and the use of targeted agents are now routine in lung adenocarcinoma, but not for squamous cell lung cancer.
Because of this disparity, and based on the successes in lung adenocarcinoma, there has been growing interest in understanding the alterations that drive lung SqCC and identifying novel therapeutic targets in this disease. Initial studies, performed using single-nucleotide polymorphism (SNP) arrays, identified several genes, including FGFR1, which are frequently amplified in lung SqCC and represent potential therapeutic targets [12,13]. Sanger sequencing studies aimed at specifically characterizing mutated kinase genes have demonstrated mutations in DDR2 [14]. Tremendous advances in technology have recently enabled more comprehensive analyses of lung SqCC, notably the work performed by The Cancer Genome Atlas (TCGA) Network, which reported SNP array profiling, whole-exome sequencing, RNA and miRNA sequencing and methylation profiling for 178 lung SqCCs, and whole-genome sequencing of 19 tumor/normal pairs [15]. This comprehensive effort has outlined the landscape of genomic alterations in lung SqCC and suggested a number of additional targets for therapy (Figure 2). Here, we review these findings and understanding of SqCCs and therapeutic target development.
Figure 2. Targetable alterations in lung squamous cell carcinomas.
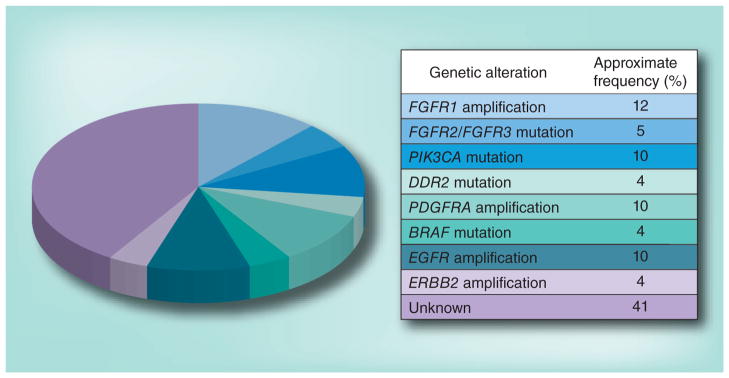
The pie chart indicates potential targetable genomic alterations identified in recent genomic studies of squamous cell lung cancers and their approximate frequencies. The table describes the alterations depicted in the pie chart.
FGFRs
FGFR kinases comprise the most frequently altered tyrosine kinase family in lung SqCC, with alterations found in at least 12% of cases [15]. Prior to the widely available use of next-generation sequencing, multiple groups independently described amplification of chromosome 8p12 in lung SqCC, a region containing the FGFR1 gene [12,13,16,17]. Two reports went on to show that some lung cancer cell lines with 8p12 amplification are sensitive to small-molecule FGFR kinase inhibitors [13,16], and several FGFR kinase inhibitors are now in clinical development for use in lung or other cancer types; however, at this time, no studies have yet been completed to support the use of FGFR kinase inhibitors in a specifically selected population of cancer patients.
Point estimates of the prevalence of FGFR1 amplification, measured by SNP array profiling, array-comparative genomic hybridization or FISH, have ranged from 7 to 20% in lung SqCCs [13,15,16,18]. Encouragingly, there have been reports of response to the FGFR kinase inhibitor BGJ398 (Novartis, Switzerland) in a Phase I study in heavily pretreated individuals with lung SqCC [19]. Several clinical trials of FGFR kinase inhibitors are now accruing subjects with lung SqCC harboring FGFR1 amplification. Most Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories use FISH to detect FGFR1 amplification in accruing clinical trials, but this will likely be replaced over time by multiplexed copy number platforms, such as digital droplet PCR, or copy number estimates from next-generation sequencing. Despite these advances, it remains unclear which patients with FGFR1 amplifications are most likely to respond to anti-FGFR therapy, and preclinical data suggest that additional biomarkers of FGFR pathway activation may be required to aid the selection of subjects most likely to respond to this drug class.
In addition to FGFR1 amplification, mutation of all four FGFR kinases has now been described in lung SqCC by whole-exome and RNA sequencing [15], confirming isolated reports of FGFR mutations described by Sanger sequencing in the past [20]. This is consistent with observations from other cancer types, notably endometrial and bladder cancer, where mutations in FGFR2 and FGFR3 have been shown to be oncogenic and to sensitize cancer cells to FGFR kinase inhibitors [20–23]. Indeed, some mutations observed in lung SqCC occur at identical sites to mutations driving these diseases, namely K660E and K660N in FGFR2 and R248C and S249C in FGFR3, an observation nominating these events as drivers of lung SqCC. The mutations in FGFR2 and FGFR3 are found in at least 5% of lung SqCCs, and initial experimental data support that they predict response to targeted therapeutics, although this requires validation in prospective clinical trials. Upcoming trials of FGFR kinase inhibitors in lung SqCC have been designed to include subjects with either amplifications or mutations of FGFR kinase genes, and the results of these studies are eagerly anticipated.
DDRs
Sanger sequencing of tyrosine kinases in a cohort of 68 individuals with lung SqCC identified recurrent mutations in the DDR2 receptor tyrosine kinase [14]. DDR2 mutations had been previously described in lung adenocarcinoma [24] and were thought to be potentially important, given that DDR2 is a high-affinity target of several small-molecule kinase inhibitors that have demonstrated efficacy against the BCR–ABL oncoprotein, notably imatinib, nilotinib and dasatinib [25]. Preclinical studies suggest that lung cancer cell lines with DDR2 mutations are sensitive to dasatinib and other tyrosine kinase inhibitors and that mutated DDR2 is oncogenic, although the mechanism of its oncogenicity remains unknown. Extended Sanger sequencing studies and whole-exome sequencing performed by the TCGA have confirmed DDR2 mutations are present in 3–4% of lung SqCCs [14].
Two clinical trials of dasatinib have been reported for subjects with advanced NSCLC. In unselected patients, the response rate to dasatinib is inferior to conventional chemotherapy, although some individuals did respond [26–28]. One of these subjects, who had an extended partial response to the combination of dasatinib and erlotinib, was found to harbor an activating DDR2 S768R mutation and no EGFR mutation. A national Phase II clinical trial of dasatinib is accruing at this time for individuals with lung SqCC and DDR2 mutations, as well as inactivating BRAF mutations, given preclinical data suggesting that specific BRAF mutations may be an additional biomarker of response to dasatinib [28]. Identification of novel agents with more selective anti-DDR2 activity is being pursued at this time to develop more optimal therapies for individuals whose lung SqCC tumors harbor DDR2 mutations.
EGFRs
Targeting of EGFRs has been one of the most successful therapeutic strategies in lung adenocarcinoma, with response rates to small-molecule EGFR tyrosine kinase inhibitors of 70–80% in patients with canonical EGFR kinase domain mutations [8]. Mutations in another ERBB family member, ERBB2 (HER2), have also been successfully targeted with small-molecule kinase inhibitors and antibodies in lung adenocarcinoma patients [29].
By contrast, clinical trials of EGFR tyrosine kinase inhibitors in unselected NSCLC patients, including those with lung SqCC, have reported modest response rates to these drugs (<10%), presumably due to a lack of sensitizing alterations in EGFR [30]. However, monoclonal antibody therapy with cetuximab in combination with chemotherapy did demonstrate a significant overall survival advantage as compared with chemotherapy alone for lung SqCC patients with advanced disease [31]. A predictive biomarker for response to cetuximab in lung SqCC patients has not yet been identified, although data describing correlations among immunohistochemical staining patterns for EGFR and response are provocative [32], and are the subject of prospective studies at this time.
In lung SqCC, EGFR and ERBB2 are targets of gene amplification in approximately 7 and 4% of cases, respectively [15]. EGFR mutations are rarely encountered in pure squamous tumors, in which the canonical exon 19 deletion and L858R mutations described in lung adenocarcinoma are absent [33]. Some experts have suggested that detection of one of these EGFR mutations in a lung SqCC sample should prompt reconsideration of the diagnosis, particularly the consideration of a mixed adenosquamous tumor. In agreement with this, the TCGA Network reported only two cases with the noncanonical EGFR L861Q mutation in 178 lung SqCCs, an alteration that is associated with gefitinib response [15]. Large deletions in EGFR, specifically the variant III form, have been described in lung SqCC in individuals from Asia [34], but these reports have not been validated in larger independent cohorts. Based on these observations, it is likely that EGFR-directed kinase inhibitors and monoclonal antibodies have some role in the therapy of lung SqCC, albeit in a much more narrow patient population than lung adenocarcinoma.
PI3K/PKB
Approximately half of individuals with lung SqCC harbor an alteration in a gene in the PI3K/PKB (AKT) pathway, most commonly point mutations in or amplification of PIK3CA, deletions and mutations of PTEN and overexpression of AKT3 [15]. Other genes that regulate this pathway include TSC1, TSC2 and STK11, which are also altered in lung SqCC. The overall frequency of these alterations suggests that targeting the PI3K/AKT pathway with selective inhibitors could be an attractive approach for patients with lung SqCC.
However, there has yet to be a compelling demonstration of efficacy of this class of drugs in a selected or unselected lung cancer patient population. This is likely due to several factors, including a lack of direct dependency of cancer cells on alterations in this pathway and a lack of understanding of additional molecular biomarkers that may predict response to drugs that target this pathway. Furthermore, alterations in the PI3K pathway often co-occur with other candidate oncogenes, suggesting that inhibiting PI3K alone may be insufficient. Given that many genes in this pathway are altered and that many compounds currently in development differ in their selectivity and potency with respect to different isoforms of PI3K, AKT and other pathway members, it will likely take substantial preclinical and clinical investigation to define the role of PI3K and/or AKT inhibitors in the treatment of NSCLC. Additionally, how these agents should be used in genotypically selected populations will need to be defined, either as single agents or in combination with other chemotherapeutics or radiotherapy.
Other kinases
In lung adenocarcinoma, the majority of tumors contain an alteration in a known oncogenic kinase (most commonly EGFR) or in the GTPase KRAS. Strikingly, while either EGFR or KRAS mutations are found in a third to half of patients with lung adenocarcinoma in the USA, mutations in either of these genes are very rare in lung SqCC [3]. As noted above, only EGFR amplification and the L861Q mutation have been described by the TCGA in lung SqCC, and HRAS is the more commonly altered RAS family member in lung SqCC.
In lung adenocarcinoma, kinase alterations such as BRAF V600E, AKT1 E17K and ERBB2 mutations have been identified as targetable alterations in a small percentage of cases. By contrast, in lung SqCC, additional candidate therapeutic targets have been identified by genomic studies and include mutations and amplifications of PDGFRA, mutations in BRAF at sites other than V600 and mutations in JAK2, ABL1, ABL2, MET and ERBB2–4 [15]. Although it is unclear whether any of these alterations represent a targetable dependency in lung SqCC, given the success in targeting alterations in these genes in other cancer types, the study of these more infrequent alterations in lung SqCC is a promising avenue for further research.
Nonkinase targets
It bears mentioning that the most commonly altered genes in lung SqCC are TP53 and CDKN2A/RB1, with nearly all lung SqCCs demonstrating alteration of one or both of the related pathways [15]. While drug discovery efforts targeting these events have been challenging, the ability to selectively target either of these lesions would have a tremendous impact on patients with lung SqCC, as well as other tumor types, most of which commonly alter these pathways.
Previous studies have shown that the SOX2 gene, a lineage-specific transcription factor that regulates squamous cell differentiation, is focally amplified on chromosome 3q in a substantial fraction of lung SqCCs, as well as SqCCs from other tissue sites such as esophagus, cervix, skin and head and neck cancers. Some cancer cell lines harboring focal amplification of SOX2 display SOX2 dependency, suggesting that SOX2 may be therapeutically targetable if it can either be directly targeted or targeted via a synthetic lethal interaction [12]. SOX2 is one of several genes altered in lung SqCC that has a defined role in squamous differentiation, with others including NOTCH1, NOTCH2, TP63, FOXP1 and ASCL4 [15]. These alterations suggest that targeting these molecules as a prodifferentiation therapy could be an attractive therapeutic approach given success with similar strategies in hematologic malignancies.
A second transcription factor pathway comprised of the genes NFE2L2, KEAP1 and CUL3 is altered by amplification or mutation in a third of lung SqCCs [15]. NFE2L2, which encodes the NRF2 protein, drives the transcription of genes involved in the oxidative or chemical stress response [35,36]. NRF2 levels are usually kept at low cellular levels by a physical interaction with KEAP1, which targets NRF2 for degradation by the CUL3 E3 ubiquitin ligase [37,38]. Previous studies have demonstrated that cancer cell lines with activating NFE2L2 or inhibiting KEAP1 mutations are dependent on NRF2, and that cells expressing high levels of NRF2 are resistant to chemotherapeutic agents [39,40]. Ongoing studies are investigating the role of this pathway in the development of lung SqCC, and are aimed at identifying ways in which this pathway may be amenable to targeted inhibition.
Immunotherapy
Despite a history of limited efficacy in lung cancer, immunotherapeutic approaches have generated renewed interest based on recent successes in early-phase clinical trials. Specifically, antibodies directed against negative costimulatory receptors and ligands (CTLA-4, PD-1, PD1-L1) have demonstrated efficacy in subjects with NSCLC, with perhaps even more impressive effects in individuals with lung SqCC than those with lung adenocarcinoma [41–44]. Given the current lack of effective therapeutic options in relapsed/recurrent lung SqCC, there is tremendous interest in expanding these clinical trials and in understanding the predictive biomarkers of response to immunotherapies in lung cancers and other cancer types.
A surprising result from genomic studies of lung SqCC and other cancers is that somatic alteration of the expression and sequence of genes that play a role in the immune response is commonly observed. In lung SqCC, mutation and downregulation of MHC genes and β2 microglobulin have been observed along with varying degrees of expression of genes involved in costimulation, such as PD1L1 and OX40L [15]. Mutation and downregulation of these genes may explain how some tumors are able to evade the immune response and suggest that specific individuals are likely to benefit from different immunotherapy approaches. Computational methods have now been established to identify tumor antigens based on the somatic sequence of a tumor, and it is likely that this information will lead to improved vaccine strategies to accompany the recent successes in targeting regulators of costimulation. Clinical trials are already being designed and executed in which immunotherapeutic approaches are being coupled with targeted kinase inhibitors, and we envision that this type of approach will be of utmost importance to studies in lung cancer populations and beyond, given the need to augment the activity of kinase inhibitors that are almost uniformly associated with acquired resistance when used as a single agent.
Conclusion & future perspective
Prior to the genomic studies reviewed here, little was known about potential targetable alterations in lung SqCCs and there had been no demonstration of efficacy of any targeted agent developed specifically for the treatment of this disease. We now know that lung SqCCs harbor targetable alterations in kinases and other genes, although the enumeration of these events is far from complete. No recurrent gene rearrangements have yet been identified in lung SqCCs, in contrast to lung adenocarcinoma; this is likely due to a paucity of whole-genome and RNA sequencing of these tumors. In addition, given the very high overall somatic mutation rate in lung SqCC, a complete understanding of the alterations that drive this disease will require the genomic profiling of many more tumor/normal pairs. As in lung adenocarcinoma, there are likely clinical and demographic features associated with specific alterations in lung SqCC, and these will also require careful consideration when designing studies.
We are now at a point where we understand the basic landscape of genomic alterations in lung SqCC to the extent that we should be able to identify a substantial proportion of individuals with targetable alterations. These patients should be actively recruited for clinical trials assessing novel targeted agents, particularly given the lack of options in relapsed/recurrent lung SqCC. There are several barriers to identifying these patients, the most significant of which is generating multiplexed assays to identify multiple types of genomic alterations within a tumor specimen. While the process of generating these assays took several years for lung adenocarcinoma because of the need to analyze tumor samples using multiple technologies, it should now be possible to use next-generation sequencing to define relevant translocations, amplifications, deletions and mutations within cancer specimens. The implementation of these assays will require close collaboration among translational scientists, departments of pathology and pharmaceutical companies, and cooperative groups who will help to design and sponsor relevant clinical trials.
It should be the goal of the lung cancer community to offer individuals with lung SqCC at least the same level of personalized care as is now offered to patients with lung adenocarcinoma. The identification of patients with targetable alterations described here, and the use of single drugs to target these lesions, will only be the beginning of the realization of this goal in the care of patients with lung SqCC. We will also need to design relevant combinations of targeted agents, immunotherapies and conventional surgical, chemotherapeutic and radiation approaches in order to design the best possible plan of care for each individual with lung SqCC, using the genomic alterations identified in the tumor as a guide.
Practice Points.
Targeted therapeutics have been successful in treating advanced lung adenocarcinoma and molecular genotyping has now become the standard of care.
Recent advances in lung squamous cell carcinoma (SqCC) suggest that multiple targetable genomic alterations are present in this disease.
The types and frequencies of targetable alterations in lung SqCCs and lung adenocarcinomas are distinct.
FGFR kinases are the dominant altered kinase family in SqCCs and appear targetable with available agents in clinical development based on preclinical and Phase I data.
Molecular genotyping of lung SqCCs and clinical trials of targeted agents are warranted at this time.
Acknowledgments
The authors would like to thank L Sholl of the Department of Pathology at the Brigham and Women’s Hospital for providing the images for Figure 1.
Footnotes
Financial & competing interests disclosure
PS Hammerman is supported by a Young Investigator Award from the Conquer Cancer Foundation. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haura EB, Camidge DR, Reckamp K, et al. Molecular origins of lung cancer: prospects for personalized prevention and therapy. J Thorac Oncol. 2010;5(6 Suppl 3):S207–S213. doi: 10.1097/JTO.0b013e3181e2f682. [DOI] [PubMed] [Google Scholar]
- 3.Pao W. New approaches to targeted therapy in lung cancer. Proc Am Thorac Soc. 2012;9(2):72–73. doi: 10.1513/pats.201112-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer. Correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 8.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small- cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26(15):2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 9.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 11.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 12▪.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41(11):1238–1242. doi: 10.1038/ng.465. Describes SOX2 as a lineage survival gene in squamous cell carcinomas and suggests it may be a therapeutic target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13▪.Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6(6):e20351. doi: 10.1371/journal.pone.0020351. One of several studies implicating FGFR1 amplification as a therapuetic target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14▪▪.Hammerman PS, Sos ML, Ramos A, Xu C. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1(1):78–89. doi: 10.1158/2159-8274.CD-11-0005. DDR2 mutations are being studied in clinical trials as potential therapeutic targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15▪▪.The Cancer Genome Atlas Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. First comprehensive genomic study of lung squamous cell carcinomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16▪▪.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. One of several studies implicating FGFR1 amplification as a therapuetic target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockwood WW, Chari R, Coe BP, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med. 2010;7(7):e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.102. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf J, Lorusso PM, Camidge RD, et al. A Phase I dose escalation study of NVP-BGJ398, a selective pan FGFR inhibitor in genetically preselected advanced solid tumors. Cancer Res. 2012;72(8 Suppl):LB-122. [Google Scholar]
- 20.Dutt A, Salvesen HB, Chen TH, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci USA. 2008;105(25):8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qing J, Du X, Chen Y, et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119(5):1216–1229. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Lee BH, Williams IR, et al. FGFR3 as a therapeutic target of the small molecule inhibitor PKC412 in hematopoietic malignancies. Oncogene. 2005;24(56):8259–8267. doi: 10.1038/sj.onc.1208989. [DOI] [PubMed] [Google Scholar]
- 23.Cappellen D, De Oliveira C, Ricol D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23(1):18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 24.Davies H, Hunter C, Smith R, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65(17):7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 25.Day E, Waters B, Spiegel K, et al. Inhibition of collagen-induced discoidin domain receptor 1 and 2 activation by imatinib, nilotinib and dasatinib. Eur J Pharmacol. 2008;599(1–3):44–53. doi: 10.1016/j.ejphar.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Johnson FM, Bekele BN, Feng L, et al. Phase II study of dasatinib in patients with advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(30):4609–4615. doi: 10.1200/JCO.2010.30.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haura EB, Tanvetyanon T, Chiappori A, et al. Phase I/II study of the Src inhibitor dasatinib in combination with erlotinib in advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(8):1387–1394. doi: 10.1200/JCO.2009.25.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen B, Peng S, Tang X, et al. Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med. 2012;4(136):136ra170. doi: 10.1126/scitranslmed.3003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cappuzzo F, Bemis L, Varella-Garcia M. HER2 mutation and response to trastuzumab therapy in non-small-cell lung cancer. N Engl J Med. 2006;354(24):2619–2621. doi: 10.1056/NEJMc060020. [DOI] [PubMed] [Google Scholar]
- 30.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small- cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 31.Pirker R, Pereira JR, Szczesna A, et al. Cetuximab plus chemotherapy in patients with advanced non-small-cell lung cancer (FLEX). An open-label randomised Phase III trial. Lancet. 2009;373(9674):1525–1531. doi: 10.1016/S0140-6736(09)60569-9. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch FR, Herbst RS, Olsen C, et al. Increased EGFR gene copy number detected by fluorescent in situ hybridization predicts outcome in non-small-cell lung cancer patients treated with cetuximab and chemotherapy. J Clin Oncol. 2008;26(20):3351–3357. doi: 10.1200/JCO.2007.14.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rekhtman N, Paik PK, Arcila ME, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung. Lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18(4):1167–1176. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji H, Zhao X, Yuza Y, et al. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci USA. 2006;103(20):7817–7822. doi: 10.1073/pnas.0510284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- 36.Solis LM, Behrens C, Dong W, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16(14):3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3(10):e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata T, Ohta T, Tong KI, et al. Cancer related mutations in NRF2 impair its recognition by Keap1–Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci USA. 2008;105(36):13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A, Bodas M, Wakabayashi N, Bunz F, Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal. 2010;13(11):1627–1637. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shibata T, Kokubu A, Gotoh M, et al. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135(4):1358–1368. 1368.e1–4. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 41.Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors. Safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lynch TJ, Bondarenko IN, Luft A, et al. Phase II trial of ipilimumab (IPI) and paclitaxel/carboplatin (P/C) in first-line stage IIIb/IV non-small cell lung cancer (NSCLC) J Clin Oncol. 2010;28(15 Suppl):Abstract 7531. [Google Scholar]
- 43.Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366(26):2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 101.National Cancer Institute. Surveillance, Epidemiology and End Results. http://seer.cancer.gov.