Abstract
The maintenance of muscle mass is critical for health and issues associated with the quality of life. Over the last decade, extensive progress has been made with regards to our understanding of the molecules that regulate skeletal muscle mass. Not surprisingly, many of these molecules are intimately involved in the regulation of protein synthesis and protein degradation [e.g. the mammalian target of rapamycin (mTOR), eukaryotic initiation factor 2B (eIF2B), eukaryotic initiation factor 3f (eIF3f) and the forkhead box O (FoxO) transcription factors]. It is also becoming apparent that molecules which sense, or control, the energetic status of the cell play a key role in the regulation of muscle mass [e.g. AMP-activated protein kinase (AMPK) and peroxisome proliferator-activated receptor gamma coactivator-1 α (PGC1α)]. In this review we will attempt to summarize the current knowledge of how these molecules regulate skeletal muscle mass.
Keywords: skeletal muscle, hypertrophy, atrophy, protein metabolism
1. Introduction
Skeletal muscles, which comprise up to 40-50% of the body's mass, are not only the motors that drive locomotion, but they also play a crucial role in whole body metabolism [1, 2]. It has also been well recognized that the maintenance of skeletal muscle mass plays an important role in disease prevention and issues associated with the quality of life [3]. Skeletal muscle mass can change quite rapidly and these changes can be evoked by a variety of stimuli including mechanical loads, nutrients, neural activity, cytokines, growth factors and hormones [4-6]. All of these stimuli induce changes in muscle mass by altering the net balance between protein synthesis and protein degradation. Hence, it should not be surprising that molecules involved in the regulation of protein synthesis (e.g. mTOR, eIF3f, eIF2B) have recently been shown to induce an increase in muscle mass, while molecules that activate protein degradation (e.g. FoxO, atrogin-1) induce a decrease in muscle mass. What might be less apparent is that many of the regulatory events that control protein metabolism are influenced by molecules that sense and / or are involved in the regulation of cellular energetic status (e.g. AMPK and PGC1α), and recent studies have indicated that these molecules also play an important role in the regulation of skeletal muscle mass. In this review, we will provide a brief background with regard to why these molecules have been implicated in the regulation of skeletal muscle mass and summarize recent data which shed light on how these molecules may exert their regulatory effects. We would also like to acknowledge that many other signalling molecules and transcription factors have also been shown to play a role in the regulation of skeletal muscle mass. However, due to limited space, we are not able to discuss all of these factors in the current review. Hence, the reader is referred to the following recent original papers and reviews [7-20].
2. The Role of mTOR in the Regulation of Skeletal Muscle Mass
2.1 mTOR, mTORC1 and Protein Synthesis
In 1965 it was discovered that a microorganism (Streptomyces hygroscopicus) in the soil from the island of Rapa Nui (Easter Island) produced a compound that possessed antibiotic properties [21]. This compound was subsequently given the name rapamycin [22]. After its identification, rapamycin was found to be capable of inhibiting the growth of a variety of eukaryotic organisms [23]. It was later determined that the growth regulatory effects of rapamycin were a result of its ability to inhibit signalling by two closely related serine / threonine kinases in yeast, which were designated the target of rapamycin (TOR) 1 and 2 [24]. A single mammalian ortholog of the yeast TOR genes was discovered [25, 26] and later termed the mammalian target of rapamycin (mTOR) [27]. More recently, it has been shown that mTOR exists in two functionally distinct multi-protein signalling complexes, mTORC1 and mTORC2 [28, 29]. In general, only signalling by mTORC1 is inhibited by rapamycin, and thus the growth regulatory effects of rapamycin are believed to be primarily exerted through the mTORC1 complex [30, 31]. Over the last decade, our knowledge of mTOR has rapidly expanded and it is now widely appreciated that signalling by mTORC1 is involved in the regulation of several anabolic processes including protein synthesis, ribosome biogenesis, and mitochondrial biogenesis, as well as catabolic processes such as autophagy [31-33].
Two of the most studied mTORC1 targets are the eukaryotic initiation factor 4E binding protein (4E-BP1) and the ribosomal S6 kinase (p70S6k1), which both play important roles in the initiation of mRNA translation. For example, eIF4E, which binds to the 7-methyl-guanosine ‘cap’ (found on the 5’-end of all cellular mRNAs), is inhibited from binding to eIF4G by 4E-BP1, thus suppressing cap-dependent translation initiation [34]. The phosphorylation of 4E-BP1 by mTORC1 results in the dissociation of 4E-BP1 from eIF4E which allows eIF4G to bind to eIF4E and this, in-turn, promotes an increase in cap-dependent translation (Figure 1) [34]. Another widely recognized function of mTORC1 involves its ability to control the selective translation of mRNAs that contain a long and highly structured 5’ untranslated region (5’UTR). These types of mRNAs often encode proteins with growth regulatory functions such as myc, HIF1 , cyclin D1, and insulin-like growth factor II (IGF-II) [34-36]. For a comprehensive review of these topics see Ma and Blenis, 2009 [34].
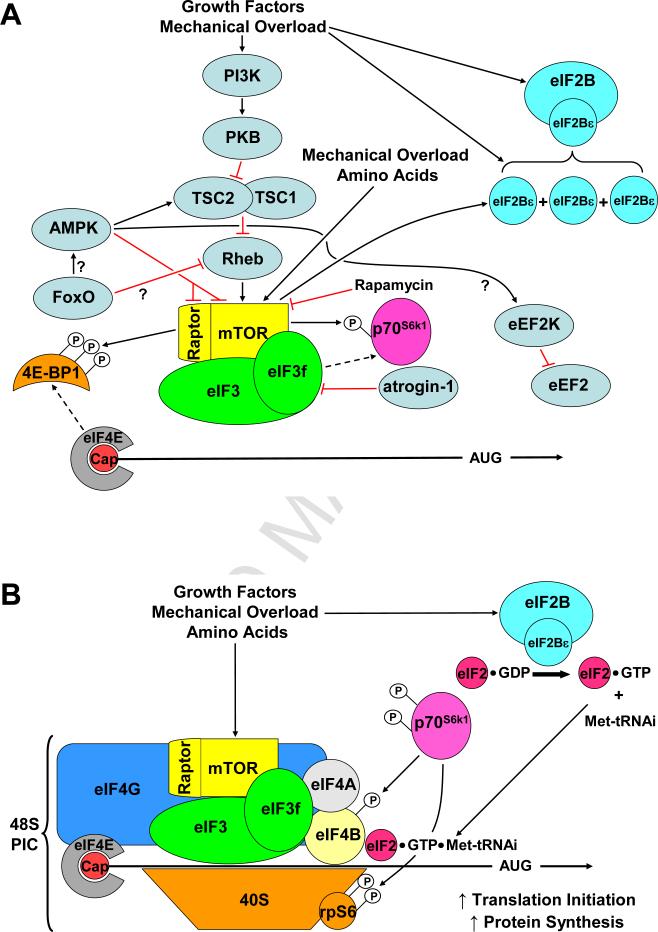
Figure 1. A general overview of some of the factors that regulate translation initiation and protein synthesis in skeletal muscle.
(A) Mammalian target of rapamycin complex 1 (mTORC1) and eukaryotic initiation factor (eIF) 2B activity, and eIF2B subunit abundance, are up-regulated in response to growth factors, nutrients and mechanical overload. mTORC1 may, in part, be responsible for the increase in eIF2B expression. Activated mTORC1 binds to eIF3f causing the activation and release of ribosomal protein S6 kinase 1 (p70S6k1). Atrogin-1 can ubiquitinate eIF3f, targeting it for degradation via the ubiquitin proteasome system leading to decreased mTORC1 signalling. mTORC1 also phosphorylates 4E-binding protein (4E-BP1), relieving it's binding to eIF4E. AMPK-activated protein kinase (AMPK) inhibits mTORC1 activity by activating the GAP activity of the tuberous sclerosis complex (TSC1/2) complex, and by directly phosphorylating Raptor and mTOR. AMPK may also activate eukaryotic elongation factor 2 kinase (eEF2K) leading to inhibition of eEF2 and peptide elongation. It remains to be determined whether Forkhead box containing protein, O subclass (FoxO) transcription factors indirectly lead to AMPK activation and Ras homologue enriched in the brain (Rheb) inhibition. (B) mTORC1-induced phosphorylation of 4E-BP1 and its dissociation from eIF4E allows the recruitment other translation initiation factors (e.g. eIF4G, 4A & 4B, the eIF3 complex, and GTP/Met-tRNAi bound eIF2) and the 40s ribosomal subunit to the 5’-Cap structure of mRNA leading to the formation of the 48S pre-initiation complex (PIC) and increased translation initiation and protein synthesis. Activated p70S6k1 phosphorylates ribosomal protein S6 (rpS6) and eIF4B, and enhances translation initiation efficiency.
In skeletal muscle, signalling by mTORC1 has been shown to be regulated by a variety of different stimuli that control skeletal muscle mass. For example, signalling by mTORC1 is activated in response to hypertrophic stimuli such as increased mechanical loading, feeding and growth factors [37-39]. On the other hand, signalling by mTORC1 is inhibited by atrophic stimuli such as decreased mechanical loading, food deprivation and glucocorticoids. [40-42]. Studies with rapamycin also suggest that signalling through mTORC1 is necessary for the hypertrophic effects of several stimuli. For instance, hypertrophy induced by mechanical loading, IGF-I and clenbuterol has been shown to be significantly, if not completely, blocked by rapamycin [38, 43, 44]. There is also evidence which suggests that the activation of mTORC1 signalling is sufficient to induce hypertrophy. For example, overexpression of constitutively active PKB (c.a.-PKB) activates mTORC1 signalling and induces hypertrophy through a rapamycin-sensitive mechanism [45]. Combined, these types of observations have led many to conclude that the activation of mTORC1 signalling is both necessary and sufficient for the induction of skeletal muscle hypertrophy [45, 46].
The aforementioned studies support the hypothesis that signalling through mTORC1 is sufficient to induce hypertrophy, however, the hypertrophic stimuli employed in these studies also induce signalling through phosphatidyinositol 3-kinase (PI3K) and PKB. This is an important point because signalling through PI3K/PKB can regulate mTOR-independent growth regulatory molecules such as the glycogen synthase kinase 3β (GSK3β), tuberin (TSC2) and the FoxO transcription factors [4, 5, 47]. Thus, based on these original studies, it was not clear if signalling by mTORC1 was sufficient, or simply permissive, for the induction of hypertrophy. To address this issue, overexpression of Rheb was recently used as a means to induce a PI3K/PKB-independent activation of mTORC1 [48]. Rheb was selected for this study because in-vitro studies had demonstrated that purified Rheb can directly activate mTORC1 signalling [49]. Consistent with these studies, it was determined that overexpression of Rheb induced a PI3K/PKB-independent activation of mTORC1 in skeletal muscle. Furthermore, overexpression of Rheb was sufficient to induce an increase in protein synthesis and hypertrophy [48, 50]. Finally, the hypertrophic effect of Rheb was shown to occur through a rapamycin-sensitive mechanism [48]. Taken together, these results suggested that the activation of mTORC1 is indeed sufficient to induce hypertrophy, at least in part by increasing protein synthesis.
Rapamycin is considered to be a highly specific inhibitor of mTORC1, and thus, it has been widely accepted that a rapamycin-sensitive hypertrophic response implies an mTORC1-dependent response [6, 46, 51]. However, like most pharmacological inhibitors, rapamycin can exert non-specific (mTORC1-independent) actions. For instance, rapamycin can bind and sequester the FKBP12 protein, which has been shown to play an important role in the function of the ryanodine receptor and signalling by members of the TGF-β superfamily (e.g. myostatin) [52-54]. Furthermore, systemic administration of rapamycin would be expected to inhibit mTORC1 signalling in all cells throughout the body. Thus, it is not clear if the anti-hypertrophic effects of rapamycin are due to the inhibition of mTORC1 signalling in skeletal muscle cells, other cell types within skeletal muscle tissue (e.g., immune cells), or distant effects in other tissues. However, the recent development of mice with skeletal muscle specific expression of rapamycin-resistant (RR) or rapamycin-resistant kinase dead (RRKD) mutants of mTOR has begun to shed light on this topic [55]. For example, it has been shown that the hypertrophic effects of Rheb are completely blocked by rapamycin in muscles from wild type and RRKD-mTOR mice, but not RR-mTOR mice. Similar observations have also been observed during the regenerative hypertrophy that occurs in response to myotoxin induced injury [55]. In other words, these recent studies indicate that mTOR, in skeletal muscle cells, is the rapamycin-sensitive element that confers hypertrophy, and the kinase activity of mTOR is necessary for this event. In the future, it will be important to determine if these points hold true during other types of rapamycin-sensitive hypertrophic events such as that which occurs in response to increased mechanical loading.
2.2 mTORC1, Autophagy and Protein Degradation
Another potential mechanism by which mTORC1 may control skeletal muscle mass is through the regulation of protein degradation via the process of autophagy (‘self-eating’) [56]. In general, autophagy is a process that involves the membranous engulfing of cellular components, which are then subjected to lysosome-dependent degradation [57]. Conditions which inhibit mTORC1 signalling, such as starvation and rapamycin, result in increased autophagy, while under nutrient rich conditions mTORC1 inhibits autophagy [56]. Although the exact mechanism by which mTORC1 regulates autophagy remains to be fully determined, current evidence suggests that mTORC1 associates with, phosphorylates, and inhibits Atg13 (autophagy gene 13) and ULK1 (UNC-51 like kinase), which are key protein kinases involved in the initial formation of autophagasomes (Figure 2) [33]. Thus, reduced mTORC1 signalling may result in increased autophagy-induced protein degradation in skeletal muscle. Indeed, rapamycin has been shown to increase autophagic flux [58] and lysosomal proteolysis rates [59] in myoblasts and myotubes, respectively. To date, however, evidence that mTORC1 plays a major role in regulating skeletal muscle autophagy is lacking [57]. For example, treatment of C2C12 myotubes with rapamycin induces only relatively small increases in lysosomal proteolysis rates (~10%) [59]. Furthermore, rapamycin or RNAi-mediated mTOR knockdown in vivo are not sufficient to induce the formation of autophagasomes [60]. In contrast, however, starvation and rapamycin have been shown to increase autophagic flux in skeletal muscle in vivo [58]. Thus, the magnitude of mTORC1's role in regulating skeletal muscle autophagy remains to be definitively determined. The paucity of studies in this area of investigation highlights the need for more research into mTORC1 and autophagy in skeletal muscle.
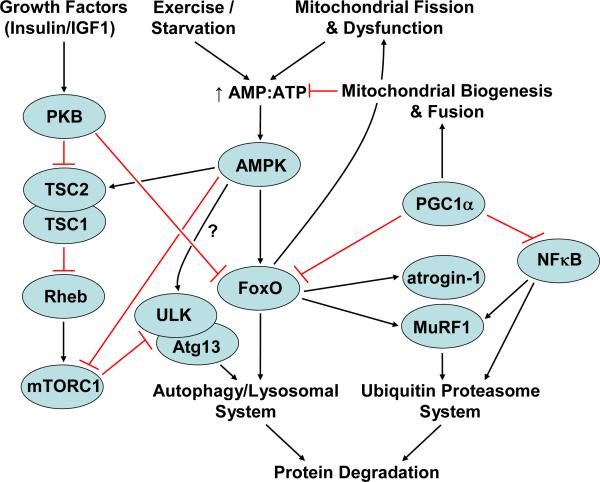
Figure 2. A general overview of some of the signalling molecules involved in the regulation of protein degradation in skeletal muscle.
Activated Forkhead box containing protein, O subclass (FoxO) transcription factors induce the transcription of genes encoding proteins involved in protein degradation via both the autophagy/lysosomal and ubiquitin proteasome pathways. AMP-activated protein kinase (AMPK), which is activated by a decrease in ATP levels due to conditions such as exercise, starvation, mitochondrial fission and/or dysfunction, ultimately leads to the activation of FoxOs. Activated FoxOs induce further mitochondrial fission/dysfunction. It is yet to be determined in skeletal muscle whether AMPK can directly activate autophagy by the phosphorylation of UNC-51 like kinase (ULK). Overexpression of peroxisome proliferator-activated receptor-γ coactivator-1α (PGC1α) inhibits FoxO transcriptional activity, possibly by inducing mitochondrial biogenesis and fusion, restoring ATP levels and thus decreasing AMPK activation. PGC1α overexpression also inhibits nuclear factor kappa B (NFκB) transcriptional activity. Growth factor induced signalling through protein kinase B (PKB) inhibits FoxO transcriptional activity and activates mTORC1 which is an inhibitor of ULK and autophagy gene 13 (Atg13), and thus, an inhibitor of autophagy.
In summary, recent studies strongly indicate that the activation of signalling through mTORC1 is sufficient to induce an increase in muscle mass and this effect is at least partially driven by an increase in protein synthesis. Recent studies have also implicated mTOR in the regulation of ribosome biogenesis, mitochondrial biogenesis and autophagy, but the potential involvement of these mTOR-dependent mechanisms in the regulation of skeletal muscle mass remain to be clearly defined.
3. The eIF3f can Regulate Protein Synthesis and Skeletal Muscle Mass
The eukaryotic initiation factor 3 (eIF3) is a multi-protein complex composed of 12 subunits (eIF3a-eIF3l) [61]. When combined with the 40S ribosomal subunit, eIF3 becomes part of the pre-initiation complex (PIC) which, in-turn, plays a fundamental role in the regulation of translation initiation [61]. The eIF3 complex has also been shown to act as a scaffold that interconnects mTORC1 with p70S6k1 [62]. In this model, inactive p70S6k1 is bound to eIF3, and upon mitogen/growth factor/amino acid stimulation, the mTORC1 complex binds to eIF3-p70S6k1, which in-turn, leads to the phosphorylation / activation of p70S6k1. Active p70S6k1 is then released from eIF3 and can subsequently phosphorylate various downstream substrates that have been implicated in the regulation of protein synthesis, such as the ribosomal protein S6 (rpS6) and eukaryotic initiation factors 4B (eIF4B) (Figure 1) [62]. Hence, the eIF3 complex has the potential to regulate protein synthesis and skeletal muscle mass via both mTOR-dependent (e.g. mTORC1-eIF3-p70S6k1) and mTOR-independent (e.g. PIC formation) mechanisms.
In support of a role for eIF3 in the regulation of skeletal muscle mass, recent studies have demonstrated that overexpression of the eIF3f subunit can induce muscle hypertrophy both in-vitro and in-vivo [63]. Furthermore, eIF3f overexpression promotes an increase in the phosphorylation of mTORC1 substrates such as p70S6k1, rpS6 and 4E-BP1 along with a concomitant increase in the rate of protein synthesis [64]. Conversely, knockdown of eIF3f expression induces myotube atrophy and this is associated with a decrease in the phosphorylation of mTORC1 substrates and protein synthesis [63, 65]. Combined, these studies suggest that simply altering the expression level of eIF3f can result in strong effects on mTORC1 signalling, protein synthesis and muscle mass.
The expression of eIF3f can be regulated by a muscle specific E3 ubquitin ligase called atrogin-1 [63], whose expression increases in response to atrophic stimuli [66-69]. Interestingly, eIF3f expression decreases during various atrophic conditions, and knockdown of atrogin-1 or inhibition of the proteasome system can prevent this decrease [63]. Since the expression level of eIF3f can regulate protein synthesis, it has been proposed that the atrophic effects of atrogin-1 are at least partially due to a decrease in protein synthesis that is driven by the loss of eIF3f [70]. In support of the hypothesis, six highly conserved lysine residues on the C-terminal end of eIF3f have been shown to be necessary for atrogin-1 mediated ubiquitination and degradation, and mutation of these residues (eIF3f K5-10R) confers increased stability to eIF3f protein [64]. Furthermore, under basal conditions, overexpression of the eIF3f K5-10R mutant is capable of inducing a more robust increase in mTORC1 signalling, protein synthesis and hypertrophy than the wild type protein. Finally, the eIF3f K5-10R mutant can attenuate the development of atrophy under both in vitro and in vivo conditions [64, 65].
Additional support for the hypothesis that atrogin-1 serves as a regulator eIF3f levels and protein synthesis has also recently been obtained in a study that examined the atrophic effects of berberine, an inhibitor of mitochondrial respiration [71]. Berberine-induced atrophy is associated with decreased eIF3f expression, mTORC1 signalling and protein synthesis, as well as increased protein degradation. Importantly, knockdown of atrogin-1 prevented the berberine-induced decrease in eIF3f levels, protein synthesis and atrophy [71].
In summary, it appears that the expression of eIF3f plays a fundamental role in the regulation of mTORC1 signalling, protein synthesis and skeletal muscle mass [63]. In addition, recent studies indicate that under atrophic conditions, atrogin-1 can directly regulate mTORC1 signalling and protein synthesis via the ubiquitination and subsequent degradation of eIF3f. Interestingly, the translation of eIF3f is regulated via a rapamycin-sensitive mechanism which raises the possibility that mTORC1 activation could induce eIF3f expression and protein synthesis in a feed-forward manner [72]. Additional studies are required to further examine the interplay between mTORC1, eIF3f and atrogin-1 under different hypertrophic and atrophic conditions, as well as the potential role of other eIF3 subunits in the regulation of skeletal muscle mass.
4. The eIF2B Regulates Global Protein Synthesis and Skeletal Muscle Mass
The eukaryotic initiation factor 2 (eIF2) plays a key role in translation initiation. Specifically, in a GTP dependent manner, eIF2 shuttles the initiator tRNA (Met-tRNAi) to the ribosome during the formation of the 48S PIC (Figure 1B). The GTP on eIF2 is then hydrolyzed to GDP which induces the release of eIF2·GDP, as well as other initiation factors, from the PIC. Once these factors have been released, a functional 80S ribosome is formed by addition of the large 60S ribosomal subunit, at which point translation elongation can commence. In order to participate in another round of translation initiation, the GDP bound to eIF2 must be exchanged for GTP. This reaction is catalyzed by the eIF2B holoenzyme, which is composed of five (α-ε) subunits. The ε subunit serves the catalytic function of eIF2B and its activity can be regulated by a variety of stimuli including hormones, nutrients, and mechanical loading [73-77]. Furthermore, several studies have shown that, in skeletal muscle, the activity of eIF2B correlates well with the changes in protein synthesis that occur in response to mechanical loading, amino acid starvation and sepsis [73, 75, 78]. Thus, by controlling rates of protein synthesis, eIF2B has the potential to play a role in the regulation of skeletal muscle mass during a variety of different conditions.
Unlike mTORC1, which controls the translation of a selective subset of mRNAs, eIF2-eIF2B appears to control the translation initiation of nearly all mRNAs, and thus controls global rates of protein synthesis. However, the activity of eIF2B can also exert some transcript specific regulatory functions. For example, a reduction in eIF2B activity leads to an increase in the translation of a small subset of mRNAs, including the activating transcription factor 4 (ATF4) [79]. ATF4 is a protein that has been reported to be both necessary and sufficient for the induction of atrophy in adult mouse skeletal muscle [80]. Thus, it is possible that inhibition of eIF2B activity could promote atrophy via a mechanism that involves both a decrease in global protein synthesis along with a selective increase in the translation of ATF4, however, this possibility remains to be explored in skeletal muscle.
One of most widely recognized mechanisms that controls eIF2B activity is a change in the abundance of the eIF2Bε subunit. For example, it was demonstrated that overexpression of eIF2Bε is sufficient to induce an increase in eIF2B activity and global protein synthesis, while knockdown of eIF2Bε results in a decrease in both of these end points [81, 82]. Thus, it appears that the abundance of eIF2Bε is a rate limiting factor for global protein synthesis. This is an important point because previous studies have shown that eIF2Bε abundance and eIF2B activity are elevated by hypertrophic stimuli such as mechanical loading [75, 83-86]. Furthermore, it was recently demonstrated that overexpression of eIF2Bε was sufficient to induce hypertrophy invivo [85]. Thus, changes in eIF2Bε and eIF2B activity appear to be a fundamental mechanism through which muscle mass can be regulated.
The activity of eIF2B is regulated not only by the abundance of eIF2Bε, but may also be regulated by changes in the phosphorylation state of eIF2Bε at Ser539 [87], although evidence that this occurs in skeletal muscle is scant. Furthermore, phosphorylation of the α subunit of eIF2 can have a major inhibitory influence on eIF2B activity, however, a detailed discussion of this mechanism is beyond the scope of this review and hence the reader is referred to Wek et al., 2006 [88]. It should also be mentioned that only a limited number of studies in skeletal muscle have been aimed at examining the phosphorylation eIF2α [89, 90]. Thus, further investigation is required into how eIF2-eIF2B activity is regulated in skeletal muscle.
In summary, eIF2B activity plays a fundamental role in the regulation of protein synthesis and recent studies have shown that increased eIF2Bε abundance, which is sufficient to increase eIF2B activity, is also sufficient to induce an increase in muscle mass [85]. In addition, there is evidence which suggests that a decrease in eIF2B activity can lead to a decrease in protein synthesis [81, 82]. Although there are several mechanisms through which eIF2B activity can be regulated, these mechanisms remain rather unexplored in skeletal muscle. Interestingly, eIF2B activity can be regulated by the amount of eIF2Bε subunit and recent reports indicate that the translation of eIF2Bε is regulated via an mTORC1-dependent mechanism [81, 83]. Thus, the effects of mTORC1 on protein synthesis may be due, in part, to a selective increase in the translation of eIF2Bε. In other words, there is still much to learn about eIF2-eIF2B and this will likely be an exciting area of study in the near future.
5. FoxO Transcription Factors and the Regulation of Skeletal Muscle Mass
The FoxO (Forkhead box containing protein, O-subclass) proteins are members of a large family of conserved DNA binding transcription factors that regulate metabolism, cellular proliferation and differentiation, apoptosis, stress tolerance and longevity [91]. The FoxO subclass is comprised of FoxO1 (a.k.a. forkhead in Rhabdomyosarcomas; FKHR), FoxO3 (FKHR-like protein 1; FKHRL1 or FoxO3a), FoxO4 (acute leukemia fusion gene located in chromosome X; AFX), and the most recently described FoxO6 [92]. With the exception of FoxO6 [93], the transcription activity of these proteins is largely controlled by shuttling between the cytoplasm and the nucleus where FoxOs can bind specific DNA sequences [93]. The major determinant of FoxO cellular location is its phosphorylation status and several kinases have been found to phosphorylate FoxOs [93]. The most widely studied kinase is PKB, with PKB-dependent phosphorylation inducing translocation of the FoxOs from the nucleus to the cytoplasm, thereby inhibiting their transcriptional activity [94, 95].
The FoxOs are expressed in skeletal muscle cells [96] and play roles in myotube differentiation [97], myotube fusion [98], and the regulation of carbohydrate and fatty acid metabolism [99-101]. Skeletal muscle FoxO nuclear localization and transcriptional activities are suppressed by IGF-1/PI3K/PKB [102, 103] and stress-activated protein kinases (JNK and p38) [104] signalling, and have recently been shown to be modulated by acetylation [105]. Importantly, FoxOs have also been implicated in the regulation of skeletal muscle mass. For example, FoxO1 expression is up regulated in various models of muscle atrophy [68, 69], and this is associated with increased nuclear levels of FoxO [102, 103]. Moreover, transgenic overexpression of FoxO1 results in reduced muscle mass [106], while knocking down FoxO1 expression promotes an increase muscle mass [107]. Furthermore, overexpression of FoxO3 is sufficient to induce a decrease in muscle mass [59, 102, 108]. These data suggest that changes FoxO activity are somehow involved in regulation of protein degradation and/or proteins synthesis. Indeed, overexpression of FoxO3 can stimulate proteolysis in myotubes [59, 60, 109] and knocking down FoxO1 expression decreases protein degradation in myotubes [110]. The two main processes of cellular protein degradation in skeletal muscle are the ubiquitin proteasome system (UPS) and autophagic/lysosomal protein degradation [111] and recent studies have implicated FoxO transcription factors in the regulation of both these catabolic pathways.
5.1 FoxOs and the Ubiquitin Proteasome System
Many atrophic conditions are associated with an up regulation of the muscle specific E3-ubiquitin ligases atrogin-1 and MuRF1 [66-69]. Poly-ubiquitination by atrogin-1 and MuRF1 targets selected proteins for degradation by the 26S proteasome [111]. UPS-mediated protein degradation is responsible for the degradation of myofibrillar and most soluble proteins [112], and thus is responsible for the majority of protein degradation in vivo [113]. Overexpression of FoxO3 has been shown to increase UPS mediated proteolysis in myotubes [59]. Furthermore, wild type and constitutively active FoxO3 (c.a.-FoxO3) overexpression regulates atrogin-1 and MuRF1 promoter activities and mRNA expression [102, 108, 109], albeit through potentially different mechanisms [114]. Moreover, FoxO3 knock down and expression of dominant negative FoxO3 inhibits increases in atrogin-1 and MuRF1 promoter activities under some [102, 115], but not all atrophic conditions [110]. Thus, there is significant evidence that FoxO3 plays a key role in the activation of UPS-mediated protein degradation in skeletal muscle (Figure 2).
Similar to FoxO3, FoxO1 knock down inhibits increases in atrogin-1 promoter activity [102] and atrogin-1 and MuRF1 expression under certain atrophic conditions [110]. Furthermore, FoxO1 has been shown to bind to, and co-activate, the MuRF1 promoter [116], although this binding may not be required for MuRF1 gene expression [117]. Interestingly, unlike FoxO3, overexpression of wild type or constitutively active FoxO1 is not sufficient to induce an increase in atrogin-1 or MuRF1 expression [103] which suggests possible differences in the mechanisms through which FoxO1 and FoxO3 regulate the expression of these E3 ligases [116].
To date, there has been very little investigation into FoxO4-mediated regulation of atrogin-1, MuRF1 and the UPS. Recently, however, it was shown that TNFα-induced increases in atrogin-1 were mediated by FoxO4 nuclear localization, independent of PKB phosphorylation, FoxO1 and FoxO3 [118]. These data further suggest that different stimuli may activate different members of the FoxO family and thus regulate the UPS via different mechanisms.
5.2 FoxOs and Autophagic/Lysosomal Protein Degradation
In addition to regulating the UPS mediated protein degradation, FoxOs also appear to play a role in the regulation of autophagic/lysosomal protein degradation [59, 60]. For example, recent studies have demonstrated that the expression of several autophagy related genes is up regulated in various models of skeletal muscle atrophy [60, 68, 119]. Moreover, FoxO3 binds to and increases the transcription of several key autophagy genes, including LC3 (microtubule-associated protein1 light chain 3) and Bnip3 (Bcl-2/adenovirus E1B 19-kDa interacting protein 3) [59, 60]. Overexpression of c.a.-FoxO3 also increases the formation of LC3 positive autophagasomes and stimulates lysosomal protein degradation [59, 60]. Indeed, it has recently been suggested that FoxO3 is both necessary and sufficient for the induction of autophagy in skeletal muscle [60]. Thus, several lines of evidence indicate that FoxO3 plays a fundamentally important role in the regulation of protein degradation via autophagy in skeletal muscle (Figure 2).
While there has been much focus on the role of FoxO3 in the induction of autophagy in skeletal muscle, there has been less research on whether FoxO1 plays a similar role. Muscles overexpressing FoxO1 have, however, been reported to have elevated levels of the lysosomal protease cathepsin L [106]. More recently, the expression of cathepsin L has been shown to be a direct target of FoxO1 in skeletal muscle, with fasting-induced expression of cathepsin L being inhibited in FoxO1 knockout mice and by expression of a dominant negative mutant of FoxO1 [120]. Thus, FoxO1 may play a role in lysosomal protein degradation in skeletal muscle, however, the magnitude of this role, whether it plays a role in the formation of autophagasomes and the degree of redundancy between FoxO1 and 3 are yet to be determined. To date, there has been no investigation into the role of FoxO4 in the regulation of skeletal muscle autophagy.
5.3 Potential Regulation of mTORC1 Signalling by FoxOs?
Studies across different metazoans have shown the potential for FoxO-induced inhibition of TORC1 signalling [121]. In mammalian non-muscle cells, FoxO1 activation indirectly activates AMPK, which then phosphorylates TSC2 and activates its GAP activity (see AMPK section below), leading to reduced mTORC1 signalling [122]. FoxO3 may also attenuate mTORC1 signalling by increasing the expression of Bnip3 [60] which interacts with Rheb and reduces Rheb's ability to induce mTORC1 signalling (Figure 1) [123]. It has also been suggested that FoxO3 may also regulate mTORC1 via a TSC1 dependent mechanism [124]. If FoxOs are able to regulate mTORC1 signalling in skeletal muscle cells, they may also play a role in regulating skeletal muscle mass by inhibiting mTORC1-mediated protein synthesis. Indeed, FoxO1 induces proteasome-dependent degradation of some of the components of mTORC1 (mTOR, Raptor, p70S6k1) in differentiating C2C12 myoblasts [125], thus potentially reducing mTORC1 signalling. Interestingly, however, constitutively active and dominant negative FoxO3 constructs were shown to have no effect on mTORC1 signalling (p70S6k1 and 4E-BP1 phosphorylation) in differentiated myotubes [59, 60]. Thus, more research is required to elucidate any potential role for the FoxOs in regulating mTORC1 signalling, and protein synthesis, in skeletal muscle under basal, hypertrophic or atrophic conditions.
In summary, FoxO transcription factors play a key role in the regulation of skeletal muscle mass by coordinating the activation of both UPS and autophagic/lysosomal mediated protein degradation. While considerably more research has focused on FoxO3, further research is required into the differential mechanisms of FoxO1, 3 and 4 activation, and their respective roles in regulating skeletal muscle protein degradation via the UPS and autophagy. Recent data also suggest that FoxOs could potentially down regulate mTORC1 signalling, and thus regulate skeletal muscle mass by decreasing protein synthesis.
6. The Role of AMPK in the Regulation of Skeletal Muscle Mass
5’-AMP-activated protein kinase (AMPK) is a heterotrimeric serine / threonine kinase composed of a catalytic subunit (α) and two regulatory subunits (β and γ) [126]. AMPK serves as an energy sensor and is activated by an elevation in the AMP/ATP ratio [126]. Enhanced binding of AMP to the subunit leads to an increase in the phosphorylation of the Thr172 residue on AMPK and subsequently induces an increase in AMPK activity [126]. In other words, conditions that cause significant cellular energy stress (e.g. exercise, starvation or mitochondrial dysfunction) will promote an increase in AMPK activity.
AMPK has been implicated in the regulation of a variety of different metabolic processes. For example, it has been shown that activation of AMPK inhibits energy consuming anabolic processes such as protein synthesis and stimulates catabolic energy producing processes such as glycolysis, fatty acid oxidation and protein degradation [126]. The ability of AMPK to exert such diverse effects on metabolism suggests that it could play a fundamental role in the regulation of skeletal muscle mass. In support of this possibility, the expression of a kinase dead AMPK α2 subunit induces muscle hypertrophy in-vivo [127]. Furthermore, knockout models of AMPK α1 (AMPK α1-/-) and α1/α2 (AMPKα1-/- α2-/-) have shown that the loss of these genes promotes hypertrophy both in-vitro and in-vivo [128, 129]. Conversely, activation of AMPK with 5-aminoimidazole-4-carboxamide riboside (AICAR) induces atrophy in myotubes [130]. Moreover, in atrophic double p70S6k1/S6k2 knockout myotubes, in which AMPK is activated, the expression of kinase dead AMPK rescues the atrophic phenotype [127]. In other words, several studies have clearly implicated AMPK in the regulation of muscle cell size, and there is evidence which suggests that AMPK exerts its effects by controlling both protein synthesis and protein degradation.
6.1 AMPK and Skeletal Muscle Protein Degradation
Consistent with the studies mentioned above, there is a growing body of evidence which indicates that the activation of AMPK results in signalling events that promote an increase in protein degradation (Figure 2). For example, activation of AMPK with AICAR has been shown to induce an increase in protein degradation and this is associated with an increase in the expression of atrogin-1 and MuRF1 [131-133]. Other AMPK activators such as metformin and 2-deoxyglucose also increase atrogin-1 and MuRF1 expression [131]. Conversely, inhibition of AMPK with Compound C blocks AICAR-induced atrogin-1 and MuRF1 expression [130, 131]. It has also been proposed that AMPK regulates the expression of atrogin-1 and MuRF1 via a FoxO3 dependent mechanism that involves an AMPK dependent increase in total [132], and nuclear localized, FoxO3 [130], as well as increased FoxO3 binding to atrogin-1 and MuRF1 promoters (Figure 2) [108]. The AMPK-induced activation of FoxO3 transcription activity may involve the phosphorylation of FoxO3 on residues that are distinct from those phosphorylated by PKB [134], although evidence in skeletal muscle is lacking. Thus, there is significant evidence that signalling through AMPK is sufficient for the induction of protein degradation via the UPS, and this is at least partially the result of the ability of AMPK to induce a FoxO3-dependent transcription of atrogin-1 and MuRF1.
Another potential mechanism to explain AMPK-induced proteolysis is via the activation of autophagy. In fact, in non-muscle cells, AMPK is required for autophagy-induced proteolysis [135]. Recent studies suggest that AMPK can directly phosphorylate and activate ULK1 (the exact phosphorylation site is yet to be confirmed), leading to the formation of a protein complex (ULK1/Atg13/FIP200) involved in the early formation of autophagasomes (Figure 2) [136, 137]. This mechanism would provide a direct pathway for cellular energy stress to trigger the catabolism of cellular proteins and provide substrates for energy production and survival. While attractive, such a mechanism remains to be established in skeletal muscle cells. Also, as discussed above, given that FoxO3 plays a role in the regulation of skeletal muscle autophagy [59, 60], AMPK may also induce activation of the autophagic/lysosomal pathway via FoxO3 (Figure 2).
Recently, it was shown that mitochondrial remodeling (i.e. mitochondrial fission), induced during atrophic conditions such as fasting, denervation and FoxO3 overexpression, was associated with increased activation of AMPK [108]. Furthermore, expression of the autophagy-related atrogene Bnip3 and mitochondrial fission proteins, DRP1 and Fis1, also increase AMPK activation in cell culture. Moreover, the expression of these proteins is sufficient to induce atrophy in vivo, and this effect can be completely inhibited by expression of a dominant negative AMPK, and knockdown of either AMPK or FoxO3 [108]. Importantly, these observations provided evidence for an amplification loop in which FoxO3 induces mitochondrial fission and perturbs mitochondrial energy metabolism, leading to increased activation of AMPK and possibly further activation of FoxO3-induced activation of autophagic [59] and UPS protein degradation [102]. Hence, it appears that alterations in mitochondrial morphology and mitochondrial function lead to activation of AMPK/FoxO3 and the exacerbation of catabolic processes induced during various atrophic conditions.
Further support that impaired mitochondrial function and AMPK activation may play a role in skeletal muscle atrophy comes from a recent study investigating muscle atrophy in response to administration of anti-diabetic agent, berberine [71]. Since berberine had previously been shown to impair mitochondrial function [138], it was hypothesized that berberine-induced mitochondrial dysfunction would activate AMPK and promote the induction of atrogin-1 expression. Indeed, in the absence of changes in the phosphorylated PKB or PKB phosphorylation of FoxO1 & 3, berberine increased AMPK activation and atrogin-1 expression, and stimulated protein degradation [71]. This study further supports the hypothesis that abnormal mitochondrial function, and the subsequent activation of AMPK, may play a causal role in the induction of skeletal muscle atrophy.
Taken together, the aforementioned studies show a consistent link between AMPK activation, increased atrogin-1 expression and protein degradation, and ultimately muscle atrophy. Moreover, most studies show a role for FoxO3 in the AMPK-induced activation of atrogin-1. Furthermore, recent studies provide important evidence that mitochondrial morphology and/or function may play critical roles in the process of skeletal muscle atrophy, at least in part by activating AMPK and the UPS. In addition, there is growing evidence that AMPK may also regulate autophagic protein degradation pathways. Additional studies are required to determine whether changes in mitochondrial morphology and function play a role in other models of skeletal muscle atrophy (i.e. diabetes, cancer cachexia, chronic kidney disease, sepsis, cardiac failure). Also, further research is needed to determine how AMPK induces FoxO3 transcriptional activity and whether AMPK activation leads to direct and /or indirect activation of autophagy.
6.2 AMPK and the Regulation of Skeletal Muscle Protein Synthesis
AMPK has not only been implicated in the regulation of protein degradation, but there is also evidence that it can regulate protein synthesis. For example, activation of AMPK with AICAR has been shown to induce a decrease in the rate of protein synthesis in myotubes [133] and in skeletal muscle in vivo [139]. Furthermore, inhibition of AMPK activity via knockout of the AMPK α1 and α2 isoforms (AMPKα1-/- α2-/-) results in myotubes with a dramatically elevated rates of protein synthesis [129]. Hence, it appears that active AMPK exerts a negative regulatory influence on the rate of protein synthesis.
The exact mechanisms through which AMPK regulates protein synthesis in skeletal muscle have not been clearly defined; however, numerous studies suggest that the regulation of mTORC1 signalling is involved in this process. This point is based on several studies which have shown that pharmacological activation (AICAR, metformin, berberine) of AMPK induces a decrease in markers of mTORC1 signalling [127, 139, 140]. On the other hand, muscles with diminished AMPK activity (dominant negative AMPK or AMPKα1-/- α2-/-) reveal enhanced mTORC1 signalling [129, 141]. Studies to date suggest that AMPK may regulate mTORC1 signalling via the phosphorylation of TSC2, leading to increased GAP activity toward Rheb [140, 142], direct phosphorylation and inhibition mTOR [143], and / or the phosphorylation and dissociation of the critical mTORC1 protein, Raptor (Figure 1) [129, 144]. Thus, it appears that AMPK can directly and / or indirectly effect mTORC1 signalling, and this likely serves as one mechanism through which AMPK regulates protein synthesis.
Another potential mechanism linking AMPK with the regulation of protein synthesis is based on the ability of AMPK to regulate atrogin-1 expression [131]. As mentioned previously, atrogin-1 can regulate the levels of eIF3f and the levels of eIF3f can regulate the rate of protein synthesis. Thus, it is possible that an increase in AMPK activity may also inhibit protein synthesis, in part, via an atrogin-1-induced decrease in eIF3f levels. Furthermore, AMPK has also been proposed to regulate protein synthesis by inhibiting translation elongation. In non-muscle cells, AMPK has been shown to phosphorylate and activate the eukaryotic elongation factor 2 (eEF2) kinase. Active eEF2 kinase can then phosphorylate and inhibit the activity of eEF2, and consequently, inhibit peptide elongation (Figure 1) [145, 146]. Whether a similar mechanism exists in resting or contracting skeletal muscle, however, remains to be determined [147].
Taken together, the aforementioned studies indicate that there are potentially several mechanisms via which AMPK could regulate skeletal muscle protein synthesis and thus play a role in the regulation of skeletal muscle mass.
6.3 AMPK, mTORC1 Signaling and Compensatory Skeletal Muscle Growth
As mentioned above, AMPK can directly influence mTORC1 signalling. Signalling from AMPK also appears to be able to override the ability of other stimuli to regulate mTORC1. For instance, it has been shown that AICAR-induced activation of AMPK can prevent the activation of mTORC1 signalling that normally occurs in response to agonists such as insulin or high-resistance contractions [148, 149]. Conversely, in myotubes with impaired AMPK activity (AMPK α1-/-), overexpression of c.a.-PKB promotes a greater increase in mTORC1 signalling than that which is observed in wild type myotubes [128]. Interestingly, in AMPKα1-/- α2-/- myotubes, basal mTORC1 signalling is already elevated and c.a.-PKB fails to promote a further increase in mTORC1 signalling [129]. Thus, in the absence of AMPK activity, mTORC1 signalling appears to be maximally activated and cannot be driven higher by c.a.-PKB [129]. These observations have led to the hypothesis that, under basal conditions, AMPK exerts an inhibitory effect on mTORC1 activity, which presumably puts the brakes on growth promoting processes and thus preserves cellular energy.
The idea that AMPK exerts inhibitory effects on growth promoting processes has led many to question whether AMPK activation could also play a role in limiting compensatory changes in skeletal muscle mass in response to stimuli such as increased mechanical loading [150]. In support of this possibility, a strong negative correlation has been shown between AMPK Thr172 phosphorylation and the magnitude of hypertrophy in response to chronic overload [151]. Significant negative correlations have also been found between AMPK Thr172 phosphorylation and markers of mTORC1 signalling [151]. More recently, it was shown that inhibition of AMPK activity (Compound C and AMPK α1-/-) allows muscles to undergo an accelerated hypertrophy in response to mechanical loading and this effect is associated, in part, with a greater increase in mTORC1 signalling [128, 152]. Conversely, activation of AMPK with AICAR reduces the hypertrophic effects of mechanical loading [152]. Thus, a growing body of evidence suggests that AMPK activity can have a potent effect on compensatory changes in skeletal muscle mass.
In summary, cell culture and rodent studies clearly show that AMPK has the potential to regulate skeletal muscle mass by influencing both protein synthesis and/or protein degradation. This is a particularly important point when considering that the AMPK activating compound metformin is a first line therapy for the control of blood glucose in pre-diabetics and type II diabetics. Based on the studies mentioned above it would appear that such AMPK agonists could have negative effects on muscle mass and thus further complicate the diabetic state. However, further research will be needed to assess the potential negative consequences of pharmacological activation of AMPK on skeletal muscle mass.
7. PGC1α and the Regulation of Skeletal Muscle Mass
Peroxisome Proliferator-Activated Receptor-γ Coactivator-1α (PGC1α) was originally identified as a transcriptional co-activator of PPAR-γ induced by cold exposure in brown adipose tissue [153]. It has since been shown to co-activate several nuclear hormone receptors and transcription factors (e.g. ERRα, PPARα, NRF-1, MEF2) which leads to an up regulation of mitochondrial gene expression and an increase in mitochondrial DNA in tissues such as skeletal muscle (for review see [154]). The expression of PGC1α in skeletal muscle is greatly influenced by levels of physical activity with endurance exercise increasing PGC1α expression [155, 156] and physical inactivity leading to decreased expression [157].
PGC1α has been implicated in the regulation of skeletal muscle mass, particularly in conditions of muscle atrophy [158] (Figure 2). For example, PGC1α expression decreases in a multitude of different muscle atrophic models [109, 157-165], although some models have reported increases in PGC1α expression [166-169]. Recently, it was proposed that a decrease in PGC1α mRNA, prior to the induction of denervation-induced muscle atrophy, may contribute to ensuing muscle atrophy. [158]. To date, however, no studies have demonstrated that a decrease in muscle PGC1α is sufficient to induce muscle fiber atrophy. In fact, muscle specific knockout of PGC1α does not exacerbate denervation-induced atrophy, nor increase the expression of atrogin-1 or MuRF1 [170]. Thus, notwithstanding potential compensatory adaptations in these transgenic models, to date, there is little support for the hypothesis that the loss of PGC1α expression per se is sufficient to induce muscle atrophy. Thus, it appears that the decrease in PGC1α expression observed in various models of muscle atrophy may be a secondary event to the atrophic condition itself and/or due to reduced levels of physical activity that are associated with the atrophic condition/disease.
Although reduced PGC1α expression may not be sufficient to induce skeletal muscle atrophy, significant evidence exists which demonstrates that increased expression of PGC1α is sufficient to inhibit increases in protein degradation and protect skeletal muscle mass from various atrophic stimuli. For example, transient overexpression of PGC1α in vivo blocks c.a.-FoxO3-induced muscle fiber atrophy, and transgenic muscle specific PGC1α overexpression protects against denervation- and fasting-induced atrophy, an effect associated with a blunting of the expression of atrogin-1, MuRF1 and cathepsin L [158]. Moreover, PGC1α overexpression also protects aging muscle from sarcopenia and is associated with reduced apoptotic markers and suppression of autophagy and UPS genes [171]. Together, these data suggest that PGC1α overexpression may inhibit protein degradation. Indeed, PGC1α overexpression has been shown to decrease basal, and block c.a.-FoxO3-induced, protein degradation rates in myotubes by inhibiting both the UPS and lysosomal degradation systems, with no effect on rates of protein synthesis [109] The anti-atrophic effects of PGC1α may be due, at least in part, to the suppression of FoxO3 binding to, and activation of genes such as atrogin-1 [109, 158, 172]. NFκB, a transcription factor also implicated in skeletal muscle atrophy [173, 174], has also recently been shown to be profoundly inhibited by PGC1α overexpression in vivo [109] and could explain some of the PGC1α effect. Thus, PGC1α overexpression has the ability to inhibit muscle atrophy and this appears to be mediated, at least in part, via an attenuation of the UPS and autophagic/lysosomal protein degradation pathways (Figure 2).
As mentioned earlier, it has recently been shown that some models of atrophy are associated with disruption of the mitochondrial network (i.e. mitochondrial fission) [108], and an increase in AMPK activation that may lead to increased FoxO3-induced activation of autophagic [59] and proteasomal protein degradation [102]. This suggests that mitochondrial morphology and function may play key roles in the maintenance of muscle mass [172, 175]. Hence, the anti-atrophic effects of PGC1α overexpression may potentially result from its ability to induce mitochondrial biogenesis [176] and promote mitochondrial fusion [177], which would counteract atrophy associated mitochondrial fission, and thus help to maintain cellular ATP levels. In fact, PGC1α overexpression has recently been shown to induce mitochondrial biogenesis, and reduce the activation of AMPK and atrogin-1 expression that is associated with berbine-induced skeletal muscle atrophy [71]. Thus, by inhibiting AMPK activation, PGC1α overexpression has the potential to inhibit protein degradation and muscle atrophy.
In summary, there is a growing body of evidence which demonstrates that PGC1α overexpression can exert a protective effect against skeletal muscle atrophy in rodents. While some progress has been made in understanding the mechanism of this effect, more work is required to further elucidate the signals generated or inhibited by PGC1α overexpression. The anti-atrophic effect of PGC1α suggests that the development of therapeutic compounds which increase muscle PGC1α expression may be beneficial for maintaining skeletal muscle mass in patients with various muscle wasting conditions, particularly in those with limited ability for exercise [178]. In this regard, further work is required to determine the level of PGC1α expression needed to have a therapeutic effect, and the long terms effects of PGC1α overexpression [179].
8. Summary
Recent progress has significantly expanded our understanding of the molecular mechanisms that regulate skeletal muscle protein synthesis and degradation. Despite this, considerably more research is required to fully elucidate the many different mechanisms that potentially regulate these two processes and ultimately determine skeletal muscle mass. Moreover, defining these mechanisms in the myriad of different models used to investigate skeletal muscle hypertrophy and atrophy will be a complex task. This task may be made easier by identifying common elements that might link the various models together. For example, mitochondrial shape and function, cellular energetic status, and AMPK activation, may play a fundamental role in regulating protein synthesis and degradation in many of the different models of atrophy currently under investigation [180]. Successful identification of such common regulatory molecules / pathways will greatly aid our understanding of how different types of stimuli promote changes in skeletal muscle mass. Furthermore, future findings may lead to the development of novel therapeutics for use in a range of clinical conditions associated with muscle atrophy / wasting, metabolic disease and reduced mobility, and thus help to improve quality of life.
Highlights.
We review recent progress on selected potential regulators of skeletal muscle mass.
mTOR, eIF3f, and eIF2Bε play critical roles in the regulation of protein synthesis.
Activated FoxOs regulate protein degradation via the proteasome and autophagy.
AMPK can activate protein degradation and suppress protein synthesis.
PGC1α and mitochondria are implicated in the regulation of muscle mass.
Acknowledgements
This work was supported by NIH grants AR057347 to T.A.H. and F30AG31623 to D.L.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: 4E-BP1, 4E binding protein 1; 5’UTR, 5’ untranslated region; AICAR, 5-aminoimidazole-4-carboxamide riboside; AMP, adenosine monophosphate; AMPK, 5’-AMP-activated protein kinase; AMPK α1-/-, AMPK α1 subunit knockout; AMPKα1-/- α2-/- AMPK α1 and α2 subunit knockout; AFX, acute leukemia fusion gene located in chromosome X; ATF4, activating transcription factor 4; Atg13, autophagy gene 13; ATP, adenosine triphosphate; Bnip3, (Bcl-2/adenovirus E1B 19-kDa interacting protein 3; c.a.-, constitutively active; DNA, deoxyribonucleic acid; DRP1, dynamin-related protein 1; eIF2B, eukaryotic initiation factor 2B; ERRα, estrogen-related receptor alpha; eIF2Bε, eukaryotic initiation factor 2B epsilon subunit; eIF3, eukaryotic initiation factor 3; eIF3f, eukaryotic initiation factor 3 subunit f; eIF4B, eukaryotic initiation factor 4B; eIF4G, eukaryotic initiation factor 4G; FIP200, focal adhesion kinase family interacting protein of 200 kD; Fis1, fission 1 gene; FKBP12, FK506 binding protein 12; FKHR, forkhead in Rhabdomyosarcomas; FKHRL1, FKHR-like protein 1; FoxO, Forkhead box containing protein, O-subclass; GAP, GTP-activating protein; GSK3β, glycogen synthase kinase 3β; GDP, guanosine diphosphate; GTP, guanosine triphosphate; IGF, insulin-like growth factor; JNK, c-Jun NH2-terminal kinase; LC3, microtubule-associated protein1 light chain 3; MEF2, myocyte enhancer factor-2; Met-tRNAi, eukaryotic initiation factor methionyl transfer ribonucleic acid; mRNA, messenger ribonucleic acid; mTOR, mammalian target of rapamycin; mTORC1, mTOR complex 1; mTORC2, mTOR complex 2; MuRF1, muscle ring finger 1; NF κB, nuclear factor kappa B; NRF-1, nuclear respiratory factor 1; p70S6k1, ribosomal S6 kinase 1; PI3K, phosphoinositide-3 kinase; PGC1α, peroxisome proliferator-activated receptor-γ coactivator-1α; PIC, pre-initiation complex; PKB, protein kinase B; PPARα, peroxisome proliferator-activated receptor-alpha; PPAR-γ, peroxisome proliferator-activated receptor- gamma; Rheb, ras homologue enriched in brain; rpS6, ribosomal protein S6; RR-mTOR, rapamycin-resistant mutant of mTOR; RRKD-mTOR, rapamycin-resistant kinase dead mutant of mTOR; TGF-β, transforming growth factor-beta; Thr, threonine; TOR, target of rapamycin; tRNA, transfer ribonucleic acid; TSC1/2, tuberous sclerosis complex; TSC1, hamartin; TSC2, tuberin; ULK1, UNC-51 like kinase; UPS, ubiquitin proteasome system,
References
- 1.Lee RC, Wang Z, Heo M, Ross R, Janssen I, Heymsfield SB. Am J Clin Nutr. 2000;72:796–803. doi: 10.1093/ajcn/72.3.796. [DOI] [PubMed] [Google Scholar]
- 2.Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seguin R, Nelson ME. Am J Prev Med. 2003;25:141–149. doi: 10.1016/s0749-3797(03)00177-6. [DOI] [PubMed] [Google Scholar]
- 4.Sandri M. Physiology. 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- 5.Frost RA, Lang CH. J Appl Physiol. 2007;103:378–387. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- 6.Bodine SC. Med Sci Sports Exerc. 2006;38:1950–1957. doi: 10.1249/01.mss.0000233797.24035.35. [DOI] [PubMed] [Google Scholar]
- 7.Wang XH, Hu J, Du J, Klein JD. Gene Ther. 2007;14:711–720. doi: 10.1038/sj.gt.3302927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raffaello A, Milan G, Masiero E, Carnio S, Lee D, Lanfranchi G, Goldberg AL, Sandri M. J Cell Biol. 2010;191:101–113. doi: 10.1083/jcb.201001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartori R, Milan G, Patron M, Mammucari C, Blaauw B, Abraham R, Sandri M. Am J Physiol Cell Physiol. 2009;296:C1248–1257. doi: 10.1152/ajpcell.00104.2009. [DOI] [PubMed] [Google Scholar]
- 10.Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Mourkioti F, Rosenthal N. J Mol Med. 2008;86:747–759. doi: 10.1007/s00109-008-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy JJ, Esser KA. Curr Opin Clin Nutr Metab Care. 2010 doi: 10.1097/MCO.0b013e32833781b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Russell AP. Clin Exp Pharmacol Physiol. 2010;37:378–384. doi: 10.1111/j.1440-1681.2009.05265.x. [DOI] [PubMed] [Google Scholar]
- 14.Lynch GS, Ryall JG. Physiol. Rev. 2008;88:729–767. doi: 10.1152/physrev.00028.2007. [DOI] [PubMed] [Google Scholar]
- 15.Tisdale MJ. Physiol. Rev. 2009;89:381–410. doi: 10.1152/physrev.00016.2008. [DOI] [PubMed] [Google Scholar]
- 16.Huang Z, Chen X, Chen D. Cell Signal. 2011;23:1441–1446. doi: 10.1016/j.cellsig.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Glass DJ. Curr Top Microbiol Immunol. 2010;346 doi: 10.1007/82_2010_78. [DOI] [PubMed] [Google Scholar]
- 18.Rennie MJ, Wackerhage H, Spangenburg EE, Booth FW. Annu Rev Physiol. 2004;66:799–828. doi: 10.1146/annurev.physiol.66.052102.134444. [DOI] [PubMed] [Google Scholar]
- 19.Spangenburg EE, Booth FW. Cytokine. 2006;34:125–130. doi: 10.1016/j.cyto.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Hornberger TA. Int J Biochem Cell Biol. 2011 doi: 10.1016/j.biocel.2011.05.007. In press: doi:10.1016/j.biocel.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vezina C, Kudelski A, Sehgal SN. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 22.Sehgal SN. Transplant Proc. 2003;35:S7–S14. [Google Scholar]
- 23.Fingar DC, Blenis J. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 24.Heitman J, Movva NR, Hall MN. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 25.Brown EJ, Albers MW, Bum Shin T, Ichikawa K, Keith CT, Lane WS, Schreiber SL. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 26.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 27.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 28.Kim D-H, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Thoreen C, Wang J, Sabatini D, Gray NS. Drug Discov Today Ther Strateg. 2009;6:47–55. doi: 10.1016/j.ddstr.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoncu R, Efeyan A, Sabatini DM. Nat Rev Mol Cell Biol. 2011 doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunlop EA, Tee AR. Cell Signal. 2009;21:827–835. doi: 10.1016/j.cellsig.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 33.Jung CH, Ro S-H, Cao J, Otto NM, Kim D-H. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma XM, Blenis J. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Proud CG. Physiology. 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- 36.Wall M, Poortinga G, Hannan KM, Pearson RB, Hannan RD, McArthur GA. Blood. 2008;112:2305–2317. doi: 10.1182/blood-2007-09-111856. [DOI] [PubMed] [Google Scholar]
- 37.Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. J Physiol. 2009;587:1535–1546. doi: 10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 39.Glover EI, Oates BR, Tang JE, Moore DR, Tarnopolsky MA, Phillips SM. Am J Physiol Regul Integr Comp Physiol. 2008;295:R604–610. doi: 10.1152/ajpregu.00097.2008. [DOI] [PubMed] [Google Scholar]
- 40.McGhee NK, Jefferson LS, Kimball SR. J Nutr. 2009;139:828–834. doi: 10.3945/jn.108.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You JS, Park MN, Song W, Lee YS. Appl Physiol Nutr Metab. 2010;35:310–318. doi: 10.1139/H10-022. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. J. Biol. Chem. 2006;281:39128–39134. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- 43.Kline WO, Panaro FJ, Yang H, Bodine SC. J Appl Physiol. 2007;102:740–747. doi: 10.1152/japplphysiol.00873.2006. [DOI] [PubMed] [Google Scholar]
- 44.Hornberger TA, McLoughlin TJ, Leszczynski JK, Armstrong DD, Jameson RR, Bowen PE, Hwang ES, Hou H, Moustafa ME, Carlson BA, Hatfield DL, Diamond AM, Esser KA. J Nutr. 2003;133:3091–3097. doi: 10.1093/jn/133.10.3091. [DOI] [PubMed] [Google Scholar]
- 45.Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- 46.Nader GA. Nat Med. 2007;13:1016–1018. doi: 10.1038/nm0907-1016. [DOI] [PubMed] [Google Scholar]
- 47.Hamilton DL, MacKenzie MG, Baar KR. In: Muscle Plasticity - Advances in Biochemical and Physiological Research. Magalhaes J, Ascensao A, editors. The Research Signpost/ Transworld Research Network; Kerala: 2009. pp. 45–93. [Google Scholar]
- 48.Goodman CA, Miu MH, Frey JW, Mabrey DM, Lincoln HC, Ge Y, Chen J, Hornberger TA. Mol. Biol. Cell. 2010;21:3258–3268. doi: 10.1091/mbc.E10-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato T, Nakashima A, Guo L, Tamanoi F. J. Biol. Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Faseb J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies SP, Reddy H, Caivano M, Cohen P. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang T, Donahoe PK. Front Biosci. 2004;9:619–631. doi: 10.2741/1095. [DOI] [PubMed] [Google Scholar]
- 53.Avila G, Lee EH, Perez CF, Allen PD, Dirksen RT. J Biol Chem. 2003;278:22600–22608. doi: 10.1074/jbc.M205866200. [DOI] [PubMed] [Google Scholar]
- 54.Osman B, Doller A, Akool E-S, Holdener M, Hintermann E, Pfeilschifter J, Eberhardt W. Cell Signal. 2009;21:1806–1817. doi: 10.1016/j.cellsig.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 55.Ge Y, Wu A-L, Warnes C, Liu J, Zhang C, Kawasome H, Terada N, Boppart MD, Schoenherr CJ, Chen J. Am J Physiol Cell Physiol. 2009;297:C1434–1444. doi: 10.1152/ajpcell.00248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DCO, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Physiol. Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 57.Sandri M. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 58.Ju JS, Miller SE, Jackson E, Cadwell K, Piwnica-Worms D, Weihl CC. Autophagy. 2009;5:511–519. doi: 10.4161/auto.5.4.7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Hinnebusch AG. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Holz MK, Ballif BA, Gygi SP, Blenis J. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 63.Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA, Segura CT, Leibovitch SA. Embo J. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Csibi A, Leibovitch MP, Cornille K, Tintignac LA, Leibovitch SA. J Biol Chem. 2009;284:4413–4421. doi: 10.1074/jbc.M807641200. [DOI] [PubMed] [Google Scholar]
- 65.Csibi A, Cornille K, Leibovitch M-P, Poupon A, Tintignac LA, Sanchez AMJ, Leibovitch SA. PLoS ONE. 2010;5:e8994. doi: 10.1371/journal.pone.0008994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Proc Natl Acad Sci U S A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 68.Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Faseb J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 69.Sacheck JM, Hyatt J-PK, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 70.Csibi A, Tintignac LA, Leibovitch MP, Leibovitch SA. Cell Cycle. 2008;7:1698–1701. doi: 10.4161/cc.7.12.6090. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, Liu D, Cao P, Lecker S, Hu Z. Diabetes. 2010;59:1879–1889. doi: 10.2337/db10-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. RNA. 2008;14:1730–1736. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimball SR, Horetsky RL, Jefferson LS. J Biol Chem. 1998;273:30945–30953. doi: 10.1074/jbc.273.47.30945. [DOI] [PubMed] [Google Scholar]
- 74.Gilligan M, Welsh GI, Flynn A, Bujalska I, Diggle TA, Denton RM, Proud CG, Docherty K. J Biol Chem. 1996;271:2121–2125. doi: 10.1074/jbc.271.4.2121. [DOI] [PubMed] [Google Scholar]
- 75.Kostyak JC, Kimball SR, Jefferson LS, Farrell PA. J Appl Physiol. 2001;91:79–84. doi: 10.1152/jappl.2001.91.1.79. [DOI] [PubMed] [Google Scholar]
- 76.Bush JA, Kimball SR, O'Connor PMJ, Suryawan A, Orellana RA, Nguyen HV, Jefferson LS, Davis TA. Endocrinology. 2003;144:1273–1283. doi: 10.1210/en.2002-220983. [DOI] [PubMed] [Google Scholar]
- 77.Welsh GI, Miyamoto S, Price NT, Safer B, Proud CG. J Biol Chem. 1996;271:11410–11413. doi: 10.1074/jbc.271.19.11410. [DOI] [PubMed] [Google Scholar]
- 78.Tuckow AP, Vary TC, Kimball SR, Jefferson LS. Am J Physiol Endocrinol Metab. 2010;299:E241–248. doi: 10.1152/ajpendo.00151.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vattem KM, Wek RC. Proc Natl Acad Sci U S A. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ebert SM, Monteys AM, Fox DK, Bongers KS, Shields BE, Malmberg SE, Davidson BL, Suneja M, Adams CM. Mol Endocrinol. 2010;24:790–799. doi: 10.1210/me.2009-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kubica N, Crispino JL, Gallagher JW, Kimball SR, Jefferson LS. Int J Biochem Cell Biol. 2008;40:2522–2533. doi: 10.1016/j.biocel.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gallagher JW, Kubica N, Kimball SR, Jefferson LS. Cancer Res. 2008;68:8752–8760. doi: 10.1158/0008-5472.CAN-08-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. J Biol Chem. 2005;280:7570–7580. doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- 84.Fluckey JD, Knox M, Smith L, Dupont-Versteegden EE, Gaddy D, Tesch PA, Peterson CA. Am J Physiol. 2006;290:E1205–E1211. doi: 10.1152/ajpendo.00593.2005. [DOI] [PubMed] [Google Scholar]
- 85.Mayhew DL, Hornberger TA, Lincoln HC, Bamman MM. J Physiol. 2011;589:3023–3037. doi: 10.1113/jphysiol.2010.202432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kubica N, Kimball SR, Jefferson LS, Farrell PA. J Appl Physiol. 2004;96:679–687. doi: 10.1152/japplphysiol.00962.2003. [DOI] [PubMed] [Google Scholar]
- 87.Hardt SE, Tomita H, Katus HA, Sadoshima J. Circ Res. 2004;94:926–935. doi: 10.1161/01.RES.0000124977.59827.80. [DOI] [PubMed] [Google Scholar]
- 88.Wek RC, Jiang HY, Anthony TG. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 89.Russell ST, Siren PMA, Siren MJ, Tisdale MJ. Exp Cell Res. 2010;316:286–295. doi: 10.1016/j.yexcr.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 90.Vary TC, Deiter G, Kimball SR. Am J Physiol. 2002;283:E1032–E1039. doi: 10.1152/ajpendo.00171.2002. [DOI] [PubMed] [Google Scholar]
- 91.Accili D, Arden KC. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 92.Huang H, Tindall DJ. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 93.van der Heide LP, Jacobs FMJ, Burbach JPH, Hoekman MFM, Smidt MP. Biochem. J. 2005;391:623–629. doi: 10.1042/BJ20050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Van Der Heide LP, Hoekman MFM, Smidt MP. Biochem. J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 96.Furuyama T, Yamashita H, Kitayama K, Higami Y, Shimokawa I, Mori N. Microsc Res Tech. 2002;59:331–334. doi: 10.1002/jemt.10213. [DOI] [PubMed] [Google Scholar]
- 97.Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. J Cell Biol. 2003;162:535–541. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bois PRJ, Grosveld GC. Embo J. 2003;22:1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Furuyama T, Kitayama K, Yamashita H, Mori N. Biochem. J. 2003;375:365–371. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bastie CC, Nahlé Z, McLoughlin T, Esser K, Zhang W, Unterman T, Abumrad NA. J Biol Chem. 2005;280:14222–14229. doi: 10.1074/jbc.M413625200. [DOI] [PubMed] [Google Scholar]
- 101.Kamei Y, Mizukami J, Miura S, Suzuki M, Takahashi N, Kawada T, Taniguchi T, Ezaki O. FEBS Lett. 2003;536:232–236. doi: 10.1016/s0014-5793(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 102.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 104.Clavel S, Siffroi-Fernandez S, Coldefy AS, Boulukos K, Pisani DF, Derijard B. Mol. Cell. Biol. 2010;30:470–480. doi: 10.1128/MCB.00666-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Senf SM, Sandesara PB, Reed SA, Judge AR. American Journal of Physiology - Cell Physiology. 2011 doi: 10.1152/ajpcell.00255.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 107.Liu CM, Yang Z, Liu CW, Wang R, Tien P, Dale R, Sun LQ. Cancer Gene Ther. 2007;14:945–952. doi: 10.1038/sj.cgt.7701091. [DOI] [PubMed] [Google Scholar]
- 108.Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M. Embo J. 2010;29:1774–1785. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brault JJ, Jespersen JG, Goldberg AL. J Biol Chem. 2010;285:19460–19471. doi: 10.1074/jbc.M110.113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith IJ, Alamdari N, O'Neal P, Gonnella P, Aversa Z, Hasselgren P-O. Int J Biochem Cell Biol. 2010;42:701–711. doi: 10.1016/j.biocel.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mitch WE, Goldberg AL. N Engl J Med. 1996;335:1897–1905. doi: 10.1056/NEJM199612193352507. [DOI] [PubMed] [Google Scholar]
- 112.Solomon V, Goldberg AL. J Biol Chem. 1996;271:26690–26697. doi: 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- 113.Lecker SH, Goldberg AL, Mitch WE. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 114.Senf SM, Dodd SL, Judge AR. Am J Physiol Cell Physiol. 2010;298:C38–45. doi: 10.1152/ajpcell.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Senf SM, Dodd SL, McClung JM, Judge AR. FASEB J. 2008;22:3836–3845. doi: 10.1096/fj.08-110163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. Am J Physiol Endocrinol Metab. 2008;295:E785–E797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McLoughlin TJ, Smith SM, DeLong AD, Wang H, Unterman TG, Esser KA. Am J Physiol Cell Physiol. 2009;297:C548–555. doi: 10.1152/ajpcell.00502.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moylan JS, Smith JD, Chambers MA, McLoughlin TJ, Reid MB. Am J Physiol Cell Physiol. 2008;295:C986–993. doi: 10.1152/ajpcell.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jagoe RT, Lecker SH, Gomes M, Goldberg AL. FASEB J. 2002;16:1697–1712. doi: 10.1096/fj.02-0312com. [DOI] [PubMed] [Google Scholar]
- 120.Yamazaki Y, Kamei Y, Sugita S, Akaike F, Kanai S, Miura S, Hirata Y, Troen BR, Kitamura T, Nishino I, Suganami T, Ezaki O, Ogawa Y. Biochem J. 2011;427:171–178. doi: 10.1042/BJ20091346. [DOI] [PubMed] [Google Scholar]
- 121.Hay N. Biochim Biophys Acta. 2011 In Press, doi:10.1016/j.bbamcr.2011.03.013. [Google Scholar]
- 122.Chen C-C, Jeon S-M, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, Park Y, Hay N. Developmental Cell. 2010;18:592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Y, Wang Y, Kim E, Beemiller P, Wang C-Y, Swanson J, You M, Guan K-L. J. Biol. Chem. 2007;282:35803–35813. doi: 10.1074/jbc.M705231200. [DOI] [PubMed] [Google Scholar]
- 124.Khatri S, Yepiskoposyan H, Gallo CA, Tandon P, Plas DR. J Biol Chem. 2010;285:15960–15965. doi: 10.1074/jbc.M110.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu A-L, Kim J-H, Zhang C, Unterman TG, Chen J. Endocrinology. 2008;149:1407–1414. doi: 10.1210/en.2007-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Steinberg GR, Kemp BE. Physiol. Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 127.Aguilar V, Alliouachene S, Sotiropoulos A, Sobering A, Athea Y, Djouadi F, Miraux S, Thiaudiere E, Foretz M, Viollet B, Diolez P, Bastin J, Benit P, Rustin P, Carling D, Sandri M, Ventura-Clapier R, Pende M. Cell Metab. 2007;5:476–487. doi: 10.1016/j.cmet.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 128.Mounier R, Lantier L, Leclerc J, Sotiropoulos A, Pende M, Daegelen D, Sakamoto K, Foretz M, Viollet B. FASEB J. 2009;23:2264–2273. doi: 10.1096/fj.08-119057. [DOI] [PubMed] [Google Scholar]
- 129.Lantier L, Mounier R, Leclerc J, Pende M, Foretz M, Viollet B. Faseb J. 2010;24:3555–3561. doi: 10.1096/fj.10-155994. [DOI] [PubMed] [Google Scholar]
- 130.Tong JF, Yan X, Zhu MJ, Du M. J Cell Biochem. 2009;108:458–468. doi: 10.1002/jcb.22272. [DOI] [PubMed] [Google Scholar]
- 131.Krawiec BJ, Nystrom GJ, Frost RA, Jefferson LS, Lang CH. Am J Physiol Endocrinol Metab. 2007;292:E1555–1567. doi: 10.1152/ajpendo.00622.2006. [DOI] [PubMed] [Google Scholar]
- 132.Nakashima K, Yakabe Y. Biosci Biotechnol Biochem. 2007;71:1650–1656. doi: 10.1271/bbb.70057. [DOI] [PubMed] [Google Scholar]
- 133.Nystrom GJ, Lang CH. Int J Clin Exp Med. 2008;1:50–63. [PMC free article] [PubMed] [Google Scholar]
- 134.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. J. Biol. Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 135.Meley D, Bauvy C, Houben-Weerts JHPM, Dubbelhuis PF, Helmond MTJ, Codogno P, Meijer AJ. J Biol Chem. 2006;281:34870–34879. doi: 10.1074/jbc.M605488200. [DOI] [PubMed] [Google Scholar]
- 136.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim J, Kundu M, Viollet B, Guan K-L. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Turner N, Li J-Y, Gosby A, To SWC, Cheng Z, Miyoshi H, Taketo MM, Cooney GJ, Kraegen EW, James DE, Hu L-H, Li J, Ye J-M. Diabetes. 2008;57:1414–1418. doi: 10.2337/db07-1552. [DOI] [PubMed] [Google Scholar]
- 139.Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. J. Biol. Chem. 2002;277:23977–23980. doi: 10.1074/jbc.C200171200. [DOI] [PubMed] [Google Scholar]
- 140.Williamson DL, Bolster DR, Kimball SR, Jefferson LS. Am J Physiol Endocrinol Metab. 2006;291:E80–89. doi: 10.1152/ajpendo.00566.2005. [DOI] [PubMed] [Google Scholar]
- 141.Ju J-S, Gitcho MA, Casmaer CA, Patil PB, Han D-G, Spencer SA, Fisher JS. Am J Physiol Cell Physiol. 2007;292:C564–C572. doi: 10.1152/ajpcell.00269.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Inoki K, Li Y, Xu T, Guan K-L. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cheng SWY, Fryer LGD, Carling D, Shepherd PR. J Biol Chem. 2004;279:15719–15722. doi: 10.1074/jbc.C300534200. [DOI] [PubMed] [Google Scholar]
- 144.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Horman S, Browne GJ, Krause U, Patel JV, Vertommen D, Bertrand L, Lavoinne A, Hue L, Proud CG, Rider MH. Curr Biol. 2002;12:1419–1423. doi: 10.1016/s0960-9822(02)01077-1. [DOI] [PubMed] [Google Scholar]
- 146.Browne GJ, Finn SG, Proud CG. J Biol Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 147.Rose AJ, Richter EA. J Appl Physiol. 2009;106:1702–1711. doi: 10.1152/japplphysiol.91375.2008. [DOI] [PubMed] [Google Scholar]
- 148.Deshmukh AS, Treebak JT, Long YC, Viollet B, Wojtaszewski JFP, Zierath JR. Mol Endocrinol. 2008;22:1105–1112. doi: 10.1210/me.2007-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Thomson DM, Fick CA, Gordon SE. J Appl Physiol. 2008;104:625–632. doi: 10.1152/japplphysiol.00915.2007. [DOI] [PubMed] [Google Scholar]
- 150.McGee SL, Mustard KJ, Hardie DG, Baar K. J Physiol. 2008;586:1731–1741. doi: 10.1113/jphysiol.2007.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thomson DM, Gordon SE. J Physiol. 2006;574:291–305. doi: 10.1113/jphysiol.2006.107490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gordon SE, Lake JA, Westerkamp CM, Thomson DM. Exerc Sport Sci Rev. 2008;36:179–186. doi: 10.1097/JES.0b013e3181877e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 154.Scarpulla RC. Physiol. Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 155.Pilegaard H, Saltin B, Neufer PD. J Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino J-P, Deriaz O. Diabetes. 2003;52:2874–2881. doi: 10.2337/diabetes.52.12.2874. [DOI] [PubMed] [Google Scholar]
- 157.Aoi W, Naito Y, Mizushima K, Takanami Y, Kawai Y, Ichikawa H, Yoshikawa T. Am J Physiol Endocrinol Metab. 2010;298:E799–806. doi: 10.1152/ajpendo.00448.2009. [DOI] [PubMed] [Google Scholar]
- 158.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. Proc Natl Acad Sci USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Roberts-Wilson TK, Reddy RN, Bailey JL, Zheng B, Ordas R, Gooch JL, Price SR. Biochim Biophys Acta. 2010;1803:960–967. doi: 10.1016/j.bbamcr.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wagatsuma A, Kotake N, Mabuchi K, Yamada S. J Physiol Biochem in press. 2011 doi: 10.1007/s13105-011-0083-5. [DOI] [PubMed] [Google Scholar]
- 161.Crunkhorn S, Dearie F, Mantzoros C, Gami H, da Silva WS, Espinoza D, Faucette R, Barry K, Bianco AC, Patti ME. J Biol Chem. 2007;282:15439–15450. doi: 10.1074/jbc.M611214200. [DOI] [PubMed] [Google Scholar]
- 162.Sáinz N, Rodríguez A, Catalán V, Becerril S, Ramírez B, Gómez-Ambrosi J, Frühbeck G. PLoS ONE. 2009;4:e6808. doi: 10.1371/journal.pone.0006808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Baker DJ, Betik AC, Krause DJ, Hepple RT. J Gerontol A Biol Sci Med Sci. 2006;61:675–684. doi: 10.1093/gerona/61.7.675. [DOI] [PubMed] [Google Scholar]
- 164.Ling C, Poulsen P, Carlsson E, RidderstrÃ¥le M, Almgren P, J.r., Wojtaszewski, Beck-Nielsen H, Groop L, Vaag A. J Clin Invest. 2004;114:1518–1526. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Adhihetty PJ, O'Leary MFN, Chabi B, Wicks KL, Hood DA. J Appl Physiol. 2007;102:1143–1151. doi: 10.1152/japplphysiol.00768.2006. [DOI] [PubMed] [Google Scholar]
- 166.Fuster G, Busquets S, Ametller E, Olivan M, Almendro V, de Oliveira CC, Figueras M, Lopez-Soriano FJ, Argiles JM. Cancer Res. 2007;67:6512–6519. doi: 10.1158/0008-5472.CAN-07-0231. [DOI] [PubMed] [Google Scholar]
- 167.Dobrowolny G, Aucello M, Rizzuto E, Beccafico S, Mammucari C, Bonconpagni S, Belia S, Wannenes F, Nicoletti C, Del Prete Z, Rosenthal N, Molinaro M, Protasi F, Fanò G, Sandri M, Musarò A. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 168.Wagatsuma A, Kotake N, Kawachi T, Shiozuka M, Yamada S, Matsuda R. Mol Cell Biochem. 2011;350:1–11. doi: 10.1007/s11010-010-0677-1. [DOI] [PubMed] [Google Scholar]
- 169.Abruzzo PM, Di Tullio S, Marchionni C, Belia S, Fan G, Zampieri S, Carraro U, Kern H, Sgarbi G, Lenaz G, Marini M. Free Radic Res. 2010;44:563–576. doi: 10.3109/10715761003692487. [DOI] [PubMed] [Google Scholar]
- 170.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 171.Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Proc Natl Acad Sci U S A. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 172.Hanai JI, Cao P, Tanksale P, Imamura S, Koshimizu E, Zhao J, Kishi S, Yamashita M, Phillips PS, Sukhatme VP, Lecker SH. J Clin Invest. 2007;117:3940–3951. doi: 10.1172/JCI32741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cai D, Frantz JD, Tawa NE, Jr., Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, Shoelson SE. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 174.Hasselgren P-O. Journal of Cellular Physiology. 2007;213:679–689. doi: 10.1002/jcp.21190. [DOI] [PubMed] [Google Scholar]
- 175.Wenz T, Diaz F, Spiegelman BM, Moraes CT. Cell Metab. 2008;8:249–256. doi: 10.1016/j.cmet.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 176.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 177.Soriano FX, Liesa M, Bach D, Chan DC, Palacin M, Zorzano A. Diabetes. 2006;55:1783–1791. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 178.Arnold AS, Egger A, Handschin C. Gerontology. 2011;57:37–43. doi: 10.1159/000281883. [DOI] [PubMed] [Google Scholar]
- 179.Miura S, Tomitsuka E, Kamei Y, Yamazaki T, Kai Y, Tamura M, Kita K, Nishino I, Ezaki O. Am J Pathol. 2006;169:1129–1139. doi: 10.2353/ajpath.2006.060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Romanello V, Sandri M. Curr Hypertens Rep. 2010;12:433–439. doi: 10.1007/s11906-010-0157-8. [DOI] [PubMed] [Google Scholar]