Abstract
In the model of the A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) regulatory cascade in Streptomyces griseus, A-factor binds ArpA, the A-factor receptor protein, that has bound to the adpA promoter and dissociates it from the DNA, thus inducing the transcription of adpA. AdpA switches on the transcription of a number of genes required for secondary metabolism and morphological differentiation, forming an AdpA regulon. Consistent with this model, arpA null mutants produced streptomycin and a yellow pigment in larger amounts and formed aerial hyphae from an earlier growth stage than the wild-type strain. On the other hand, mutant MK2, expressing a mutant ArpA (Trp119Ala), neither produced secondary metabolites nor formed aerial hyphae, because this A-factor-insensitive mutant ArpA always bound to and repressed the adpA promoter due to the amino acid replacement of Trp-119 with Ala. Introduction of adpA under the control of a foreign promoter into mutant MK2 restored all of the phenotypes that we could observe, which suggests that the only significant target of ArpA is adpA. In contrast to other γ-butyrolactone regulatory systems, disruption of arpA had no effect on A-factor production, indicating that ArpA does not regulate A-factor biosynthesis. Instead, A-factor production was found to be repressed by AdpA in a two-step regulatory feedback loop.
The transcriptional factors ArpA and AdpA which we have long studied are key members of the A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) regulatory cascade controlling the initiation of secondary metabolism and morphological differentiation in Streptomyces griseus (2, 5-7). A-factor acts as a switch for these processes, and an A-factor-deficient mutant neither produces any of the secondary metabolites nor forms aerial mycelium. During the early stages of growth, when the concentration of A-factor is still low, the A-factor-specific receptor, ArpA, binds to the promoter region of adpA and represses its transcription. When the concentration of A-factor reaches a critical level at or near the decision point, A-factor binds to the DNA-bound ArpA and releases it from the DNA, thus activating the transcription of adpA. The decision point is at the middle of the exponential growth phase (5), and the transcription of adpA therefore starts just before or at the decision point. A number of genes required for secondary metabolism and morphological development are then switched on by the transcriptional activator, AdpA. These include strR, encoding the pathway-specific transcriptional activator for streptomycin biosynthetic genes (19); adsA, encoding an extracytoplasmic function sigma factor essential for aerial mycelium formation (31); amfR, encoding a transcriptional factor for the amf operon essential for aerial mycelium formation (33); sgmA, encoding a zinc-containing metalloendopeptidase important for aerial mycelium formation (9); and ssgA, encoding a small acidic protein essential for septum formation in aerial hyphae (32). We have also identified several proteases as targets of AdpA (our unpublished data).
The above model for the early steps of the A-factor regulatory cascade has been formulated by the following observations. S. griseus mutant HH1 is unable to form aerial hyphae or streptomycin because it cannot synthesize A-factor. Suppressor mutants KM7 and HO1, which form aerial hyphae and produce streptomycin even in the absence of A-factor, lack the A-factor receptor activity (15, 23). Consistent with the repressor-like behavior of ArpA in vivo, in vitro analysis showed that it binds a 22-bp palindrome with the sequence 5′-GG(T/C)CGGT(A/T)(T/C)G(T/G)-3′ as one half of the palindrome, and upon binding A-factor it immediately dissociates from the DNA (22). At this stage of the research, we postulated that ArpA bound specific genes, named Xs, and repressed their transcription in an early stage of growth. Apart from these studies, we purified an A-factor-dependent protein (AdpA) that can bind the upstream activation sequence of strR and identified adpA as one of the gene Xs, since ArpA bound the promoter of adpA (19). Disruption of adpA caused S. griseus to be unable to form aerial hyphae or produce secondary metabolites such as streptomycin and a yellow pigment. The yellow pigment has been determined to be a phenoxazinone derivative and it was named grixazone (our unpublished data).
Because the arpA mutants examined so far were derived from mutant HH1, which contains large deletions (14), including afsA, which is essential for A-factor biosynthesis (1), , we generated an arpA null mutation, starting with S. griseus IFO13350, which has the wild-type genetic background. One of the purposes of constructing this mutant was to examine A-factor production, since orthologous γ-butyrolactone receptor mutations, including BarA, which serves as a receptor for virginiae butanolides in Streptomyces virginiae (11, 16, 20), FarA for IM-2 in Streptomyces lavendulae (12, 13, 30), and ScbR for SCB1 in Streptomyces coelicolor A3(2) (26), affect the biosynthesis of the respective γ-butyrolactones in some unknown way. We also generated an arpA mutant (MK2) that contained a site-directed mutation at position 119 of ArpA; the tryptophan residue at this position is essential for A-factor binding (25). The Trp residue corresponding to Trp119 of ArpA is conserved in all the γ-butyrolactone receptors so far reported and is involved in the formation of a ligand-binding pocket, as determined by X-ray crystallography of CprB (17), an ArpA homologue in S. coelicolor A3(2). Site-directed mutagenesis of ArpA has shown that the Trp119Ala mutant lacks the A-factor binding activity but not the DNA-binding activity (25). Characterization of the arpA null mutant and MK2 clearly showed the function of ArpA as a repressor of morphogenesis and secondary metabolite production.
Concerning this model of the A-factor regulatory cascade, the question of whether ArpA targets additional genes other than adpA was raised. To answer this question, we expressed adpA under a foreign promoter in mutant MK2. If ArpA has additional targets, expression of adpA under a constitutive promoter would not be enough to restore the phenotypes of MK2, because the additional target gene(s), which might also be essential for morphogenesis and secondary metabolite production, must be repressed by the mutant Trp119Ala ArpA. In this short communication we discuss that the target of ArpA must be adpA for all the phenotypes that we can observe and that, unlike in other γ-butyrolactone regulatory systems, the null arpA mutation does not affect A-factor biosynthesis. To fully understand the regulatory mechanism by which various γ-butyrolactones switch on the expression of the genes important for secondary metabolism and morphogenesis in various Streptomyces spp., it is important to determine the detailed phenotypes resulting from mutations of arpA and adpA in the A-factor regulatory cascade and to determine clear differences between the A-factor regulatory system in S. griseus and other γ-butyrolactone regulatory systems in various Streptomyces strains.
Phenotypes of an arpA null mutant.
The S. griseus mutant KM7, encoding an ArpA protein that is defective for A-factor binding, developed normally and produced streptomycin and grixazone in the absence of A-factor. This suggested that the A-factor receptor protein (ArpA) served as a repressor for these phenotypes (15). Consistent with this idea, mutant HO1, containing a mutant ArpA (Pro115Ser) with A-factor-binding ability but without DNA-binding ability, showed the same phenotype as mutant KM7 (23). Both KM7 and HO1 were derived from S. griseus mutant HH1, which had large deletions, including afsA, probably encoding an enzyme for the biosynthesis of A-factor (1). We therefore generated a null arpA mutant by in-frame deletion in the wild-type genetic background, starting with S. griseus IFO13350, as follows. The 4.4-kb SalI fragment containing the whole arpA gene was cloned previously in pUC19 (pARP-Sal) (21). By Southern hybridization and colony hybridization, the 2.8-kb BamHI fragment containing the region downstream of arpA was cloned in pUC19 (pARP-Bam). The 0.3-kb BamHI-EcoRI fragment (corresponding to the sequence upstream from the Gln-4 codon of arpA) was amplified with primers 5′-GGGTTCGGTTCGCCGGGACC-3′ and 5′-CGgaattcCTGCTTCGCCATTTCCTGCC-3′ (the lowercase letters indicate an EcoRI site), and the absence of PCR errors was confirmed by nucleotide sequencing. The 1.3-kb BamHI-SalI fragment containing arpA in pARP-Sal was replaced with this amplified fragment, and the resultant 3.4-kb SalI-EcoRI fragment was ligated with the 2.8-kb EcoRI-BamHI fragment from pARP-Bam in pUC19. A 1.9-kb BamHI-HindIII fragment carrying the kanamycin resistance gene from Tn5 was inserted in the multicloning site.
The plasmid prepared from Escherichia coli JM110 was introduced by transformation into S. griseus IFO13350, and kanamycin-resistant transformants were isolated. Kanamycin-sensitive colonies, derived after a second crossover in one of the transformants, were candidates for arpA-disrupted strains. Southern hybridization with the 4.4-kb SalI fragment in pARP-Sal and the kanamycin resistance gene as 32P-labeled probes against the chromosomal DNA digested with EcoRI plus SalI identified true arpA disruptants (ΔarpA). Mutant ΔarpA strains constructed in this way contained a deletion from Ala-5 to Gly-245 of ArpA (Fig. 1A).
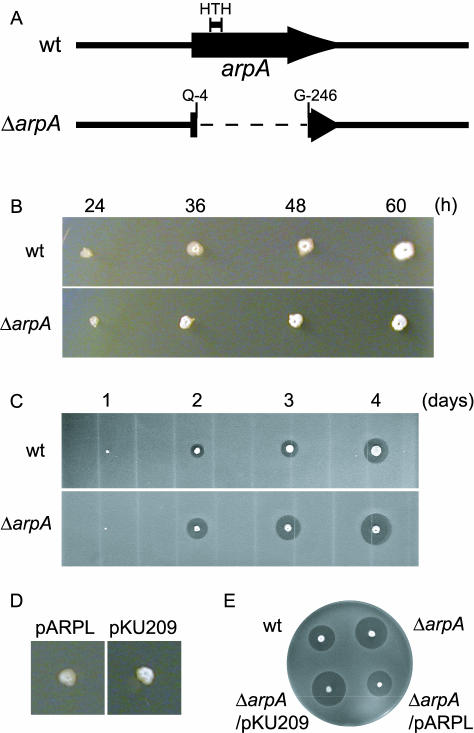
FIG. 1.
Phenotypes of an arpA null mutant, the ΔarpA mutant, of S. griseus. (A) Schematic representation of the structures of arpA on the chromosome of S. griseus IFO13350 (wild type, wt) and an arpA deletion mutant (ΔarpA). ArpA contains a helix-turn-helix (HTH) DNA-binding motif near the N terminus. The ΔarpA mutant contains an in-frame deletion from Ala-5 to Gly-245. (B) Effects of the ΔarpA mutation on morphological differentiation. Both strains were inoculated with a toothpick and grown on YMPD agar medium (19) at 28°C for the indicated periods. (C) Effects of the ΔarpA mutation on streptomycin production. Strains were inoculated with a toothpick and grown on Bennett agar medium (10) without glucose at 28°C for the indicated periods. Spores of Bacillus subtilis were then overlaid, and the plates were incubated at 37°C for 1 day. Streptomycin production was detected by the formation of a growth inhibition zone. (D) Morphological differentiation of the ΔarpA mutant harboring pARPL at the normal time. The ΔarpA mutant harboring pKU209, grown for 36 h in the same way as in B, prematurely forms aerial mycelium, whereas the mutant harboring pARPL remains as substrate hyphae. (E) Streptomycin production by the ΔarpA mutant harboring pARPL in the normal yield at the normal time. Strains were grown for 3 days, and streptomycin production was assayed in the same way as in C.
The ΔarpA mutant produced A-factor with the same kinetics and in the same amount as the wild-type strain (see below). However, it formed aerial mycelium and spores earlier than the wild type; earlier sporulation of the mutant slightly reduced the diameter of its colony on agar medium (Fig. 1B). The ΔarpA mutant produced streptomycin earlier than the wild type and accumulated it in a larger amount (Fig. 1C). Consistent with this, adpA was transcribed more actively at all time points in the ΔarpA mutant than in the wild-type strain (Fig. 2A). Introduction of pARPL containing the whole arpA gene on low-copy-number plasmid pKU209 into the ΔarpA mutant repressed the early aerial mycelium formation at 36 h (Fig. 1D) and the overproduction of streptomycin (Fig. 1E). These phenotypes of the ΔarpA mutant were the same as those of mutants KM7 and HO1, showing that elimination of the repressor function of ArpA led to uncontrolled switch-on of the A-factor regulatory cascade irrespective of the presence of afsA.
FIG. 2.
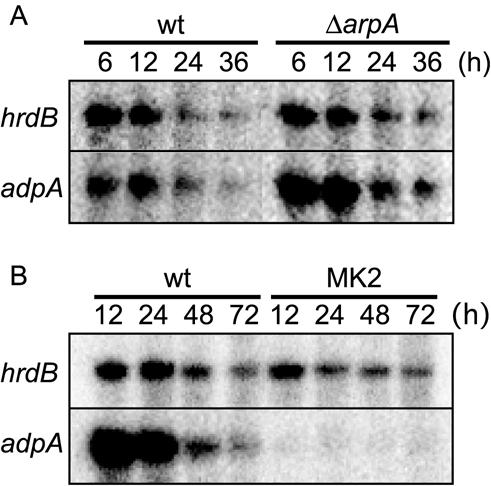
Transcription of adpA in ΔarpA (A) and MK2 (Trp119Ala) (B) mutants. RNA was prepared from mycelium grown in YMPD liquid medium (A) and on YMPD agar medium (B). We used the liquid medium for RNA preparation in A because we could hardly prepare enough RNA from the mycelium grown on cellophane on the surface of agar medium for S1 mapping at 6 h. S1 mapping was performed with the 32P-labeled probe corresponding to nucleotide positions −274 to + 150 with respect to the transcriptional start point of adpA, as described previously (19). hrdB, encoding a principal σ factor of RNA polymerase, was used to check the purity and amounts of RNA used.
Phenotypes of a mutant containing the Trp119Ala mutation.
Site-directed mutagenesis of ArpA showed that Trp-119 is essential for A-factor binding but nonessential for DNA binding. The Trp119Ala mutant is an A-factor-insensitive mutant that binds to its target DNA normally in both the presence and absence of A-factor (25). If the in vitro A-factor-insensitive feature of the Trp119Ala mutant is also the case in vivo, S. griseus mutant MK2 containing the Trp119Ala mutation would fail to develop aerial hyphae and to produce secondary metabolites because of constant binding to and hence repression of the target promoters.
We introduced the mutated arpA gene containing the Trp119Ala mutation into S. griseus. For generation of the S. griseus mutant, we used the mutated arpA gene containing the Trp119Ala mutation and introduced it into S. griseus IFO13350, essentially in the same way as for generating the ΔarpA mutant. A single crossover by homologous recombination yielded mutant MK1 and the second crossover yielded mutant MK2 (Fig. 3A). As expected, mutant MK2 remained bald (Fig. 3B, left) and failed to produce streptomycin (Fig. 3C, left) or grixazone (Fig. 3D, left). Neither addition of A-factor nor introduction of pADP10L containing the entire adpA gene and its promoter on low-copy-number plasmid pKU209 (19) corrected these defects of mutant MK2, whereas pADP20L, containing the adpA sequence under the control of the E. coli lac promoter on pKU209 (19), restored the full wild-type phenotypes (Fig. 3B, C, and D, right). The spores of mutant MK2 harboring pADP20L were the same in shape, length, and diameter as those of the wild-type strain (Fig. 3E).
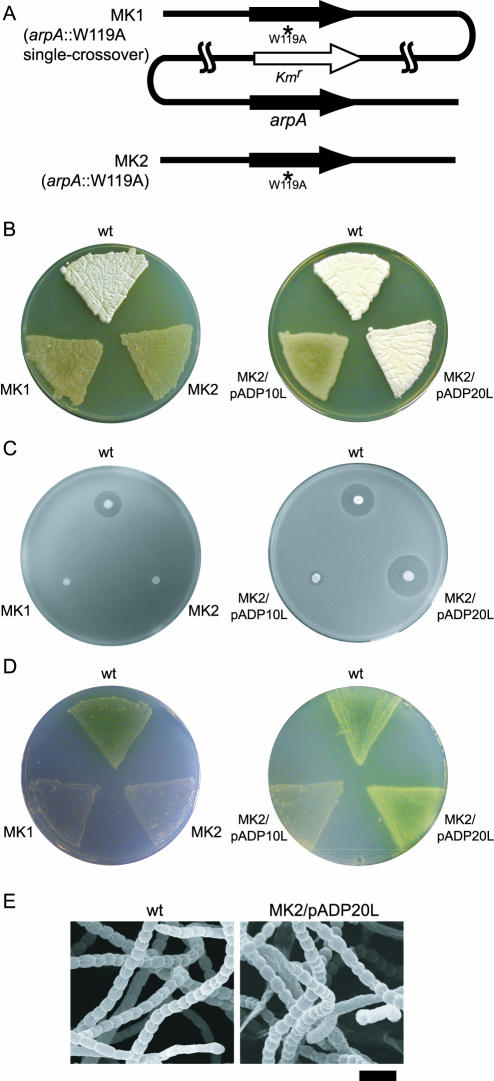
FIG. 3.
Phenotypes of mutants MK1 and MK2. (A) Schematic representation of the arpA region on the chromosomes of mutants MK1 and MK2. Mutant MK1 contains both the intact arpA and the mutated arpA (Trp119Ala) in addition to the kanamycin resistance gene. (B) Effects of the arpA W119A mutation on morphological differentiation. S. griseus IFO13350 (wild type, wt), MK1, and MK2 were grown on YMPD agar medium at 28°C for 3 days. Neither MK1 nor MK2 formed aerial hyphae or spores. Introduction of pADP20L containing the adpA-coding sequence under the control of the E. coli lac promoter resulted in restoration of sporulation in MK2, whereas pADP10L, containing the intact adpA, did not restore the defect in aerial hypha formation of MK2. (C) Effects of the Trp119Ala mutation on streptomycin production. Strains were grown on Bennett agar medium without glucose at 28°C for 3 days, as described in the legend toFig. 1. (D) Effects of the Trp119Ala mutation on grixazone production. Strains were grown on phosphate-depleted standard minimal medium at 28°C for 3 days, as described previously (31). Grixazone is produced only on phosphate-depleted medium. (E) Scanning electron micrographs of spores formed by S. griseus IFO13350 and mutant MK2 harboring pADP20L. Both strains were grown on R2YE agar medium (10) at 28°C for 3 days. Bar, 2 μm.
The E. coli lac promoter is known to be expressed in Streptomyces spp. (19, 29), and we confirmed constant transcription of the lac promoter by S1 mapping (data not shown). In mutant MK2, no transcription of adpA was detected by S1 nuclease mapping (Fig. 2B). The failure of pADP10L to complement the defects therefore resulted from the constant repression of the adpA promoter by the Trp119Ala form of ArpA. Since the presence of AdpA corrects all the defects arising from the constant binding of ArpA (Trp119Ala) to the target promoters, the adpA promoter is the only critical target of ArpA.
Consistent with this idea, we failed to isolate other significant targets for ArpA during repeated attempts by a gel mobility shift-PCR method (8) and Southern hybridization. The gel mobility shift-PCR method was used successfully for isolation of a number of DNA fragments to which AdpA binds (8, 31). Although we isolated two additional ArpA-binding sites (ABS2 and ABS3) other than the adpA promoter by this method, both were found to have no biological functions. ABS2, with the sequence 5′-CGATACCTGCCGTTCGGTATTT-3′, was located between two divergent genes, each encoding a protein with no similarity to known proteins, and ABS3, with the sequence 5′-CAGAACCGGGCGGTCGGTCGGC-3′, was located between convergent genes, each encoding a product with no similarity to known proteins. Replacement of ABS2 on the chromosome with a mutant site unable to bind ArpA had no effects on the transcription of the two divergent genes or on detectable phenotypes. Concerning ABS3, we examined the presence of promoters near or at ABS3 by S1 mapping but found no promoter activity. We suppose that these two ArpA-binding sites, showing similarity to the ArpA-binding consensus sequence 5′-(A/C)C(A/G)(T/A)ACCG(A/G)CCGG(T/C)CGGT(A/T)(T/C)G(T/G)-3′, have no significant biological functions. Repeated Southern hybridization with the 22-bp ArpA-binding consensus sequence as a 32P-labeled probe and the chromosomal DNA of S. griseus digested with various restriction enzymes as targets also gave ABS3.
Mutant MK1, obtained by a single crossover, contained both the intact arpA gene and the mutated arpA (Trp119Ala) gene (Fig. 3A). Because this mutant showed the same phenotypes as mutant MK2, Trp119Ala is a dominant-negative mutation.
A-factor production by arpA and adpA mutants.
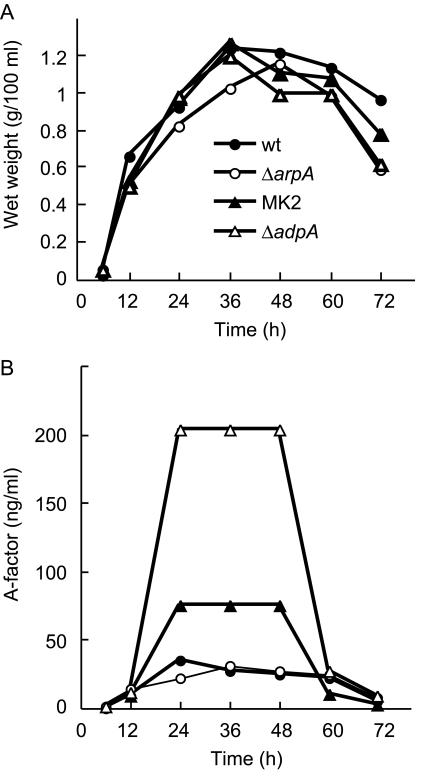
Disruption of the arpA homologues barA in S. virginiae (16) and scbR in S. coelicolor A3(2) (26) resulted in complete loss of production of the respective γ-butyrolactones. In addition, a farA mutant of S. lavendulae produced a greatly reduced amount of the γ-butyrolactone (13). The S. griseus ΔarpA mutant, however, produced the same amount of A-factor in the same amount of time, as determined by the streptomycin cosynthesis method (4) (Fig. 4), which is a vivid contrast to the above γ-butyrolactone regulatory systems. Mutant MK2 (Trp119Ala) unexpectedly produced A-factor in a larger amount than the wild type and ΔarpA mutant (Fig. 4). Furthermore, the ΔadpA mutant produced a much larger amount of A-factor. Since AdpA is absent in the ΔadpA mutant and the AdpA activity in mutant MK2 probably remains to a very small extent, we assume that AdpA acts as a repressor of A-factor production in some unknown way. The ΔarpA mutant, in which AdpA must be present in a larger amount than in the wild-type strain, produced almost the same amount of A-factor as the wild type. We assume that the amount of AdpA in the wild-type strain is sufficient for full repression of A-factor production.
FIG. 4.
A-factor production by the wild-type (wt) S. griseus strain and ΔarpA, MK2, and ΔadpA mutants. (A) Growth was monitored by measuring the wet weight of mycelium grown in YMPD liquid medium. (B) The A-factor concentration was measured by the streptomycin cosynthesis method, as described previously (4). The values are the averages of results for two independent samples.
Similarity and differences between the A-factor regulatory cascade and other γ-butyrolactone regulatory systems.
γ-Butyrolactone regulatory systems have been reported in several Streptomyces spp. There are four such systems in which a receptor and its cognate γ-butyrolactone are known; these are ArpA and A-factor in S. griseus, BarA and virginiae butanolides in S. virginiae (20), FarA and IM-2 in S. lavendulae (30), and ScbR and SCB1 in S. coelicolor A3(2) (26). The phenotypes of MK1 and MK2, in combination with adpA, enable us to compare the function of the A-factor receptor with that of other receptors. Of the four receptors, only arpA acts as a switch for aerial mycelium formation, and the others exert no detectable effects on morphogenesis. Concerning secondary metabolism, arpA acts as a repressor of the production of streptomycin, grixazone, and other secondary metabolites. Similarly, barA acts as a repressor of virginiamycin production in S. virginiae (16), and farA acts as a repressor of production of a blue pigment and a nucleosidic antibiotic in S. lavendulae (13). In S. coelicolor A3(2), however, an ScbR-SCB1 complex is required for actinorhodin production; SCB1 induces actinorhodin production, and an scbR disruptant does not produce actinorhodin (26, 27).
A vivid contrast between the A-factor regulatory system and the other γ-butyrolactone regulatory systems is that arpA appears not to affect A-factor production, but barA, farA, and scbR are required for production of the respective γ-butyrolactones (13, 16, 26). In S. griseus, A-factor production is controlled directly or indirectly by adpA, which we suppose is the unique target of ArpA. A-factor at a critical concentration dissociates ArpA from the adpA promoter and induces the production of AdpA, which in turn serves as a negative regulator of A-factor production, thus preventing overproduction of A-factor. We speculate that, in the case of BarA (11), FarA (12), and ScbR (26), these receptors control the production of the respective γ-butyrolactones as a positive factor by a different mechanism, autorepression of their own promoters. In contrast, ArpA does not bind its own promoter (data not shown).
In S. griseus, adpA serves as a global transcriptional activator gene for a number of genes required for both secondary metabolism and morphological differentiation. Because of this linear cascade, starting with A-factor to ArpA to AdpA and then to a number of genes in S. griseus, both secondary metabolism and morphological differentiation are switched on by A-factor. The targets of AdpA include the pathway-specific transcriptional activator (strR) in the streptomycin biosynthetic gene cluster (19). In the regulation of antibiotic production by other γ-butyrolactone regulatory systems, a receptor (TylP) itself appears to bind to and control the promoter of the pathway-specific transcriptional activator (tylS) in the tylosin biosynthetic gene cluster in Streptomyces fradiae (24). Similarly, a γ-butyrolactone receptor (SpbR) also appears to control the pathway-specific regulator (papR1) in the pristinamycin biosynthetic gene cluster in Streptomyces pristinaespiralis (3). Furthermore, in some gene clusters for antibiotic biosynthesis, γ-butyrolactone receptor homologues were found; MmfR for methylenomycin biosynthesis in S. coelicolor A3(2) (accession no. AJ276673), JadR2 for jadomycin B biosynthesis in Streptomyces venezuelae (34), and AAM78022.1 for neocarzinostatin biosynthesis in Streptomyces carzinostaticus subsp. neocarzinostaticus (accession no. AY117439).
BarA is also probably encoded in the virginiamycin biosynthetic gene cluster. This means that BarA and these probable receptors directly control the expression of the individual gene clusters. We assume that the presence of AdpA between ArpA and the pathway-specific regulators, including StrR in the linear A-factor regulatory cascade, and the diverse regulatory functions of AdpA make A-factor the key master switch for production of most, if not all, secondary metabolites and aerial mycelium formation in S. griseus. In other Streptomyces spp., the strict signal relay from ArpA to AdpA may be lost; in S. coelicolor A3(2), for example, a probable adpA orthologue (bldH) that is involved both in morphological differentiation and in secondary metabolism is not under the control of γ-butyrolactones (18, 28).
Acknowledgments
J. Kato was supported by the Japan Society for the Promotion of Science. This work was supported by the Asahi Glass Foundation and by the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan (BDP-03-VI-2-7).
REFERENCES
- 1.Ando, N., N. Matsumori, S. Sakuda, T. Beppu, and S. Horinouchi. 1997. Involvement of afsA in A-factor biosynthesis as a key enzyme. J. Antibiot. (Tokyo) 50:847-852. [DOI] [PubMed] [Google Scholar]
- 2.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 3.Folcher, M., H. Gaillard, L. T. Nguyen, K. T. Nguyen, P. Lacroix, N. Bamas-Jacques, M. Rinkel, and C. J. Thompson. 2001. Pleiotropic functions of a Streptomyces pristinaespiralis autoregulator receptor in development, antibiotic biosynthesis, and expression of a superoxide dismutase. J. Biol. Chem. 276:44297-44306. [DOI] [PubMed] [Google Scholar]
- 4.Hara, O., and T. Beppu. 1982. Mutants blocked in streptomycin production in Streptomyces griseus—the role of A-factor. J. Antibiot. (Tokyo) 35:349-358. [DOI] [PubMed] [Google Scholar]
- 5.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 6.Horinouchi, S., and T. Beppu. 1992. Autoregulatory factors and communication in actinomycetes. Annu. Rev. Microbiol. 46:377-398. [DOI] [PubMed] [Google Scholar]
- 7.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 8.Horinouchi, S., H. Onaka, H. Yamazaki, S. Kameyama, and Y. Ohnishi. 2000. Isolation of DNA fragments bound by transcriptional factors, AdpA and ArpA, in the A-factor regulatory cascade. Actinomycetologica 14:11-16. [Google Scholar]
- 9.Kato, J., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieser, H., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 11.Kinoshita, H., H. Ipposhi, S. Okamoto, H. Nakano, T. Nihira, and Y. Yamada. 1997. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J. Bacteriol. 179:6986-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitani, S., H. Kinoshita, T. Nihira, and Y. Yamada. 1999. In vitro analysis of the butyrolactone autoregulator receptor protein (FarA) of Streptomyces lavendulae FRI-5 reveals that FarA acts as a DNA-binding transcriptional regulator that controls its own synthesis. J. Bacteriol. 181:5081-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitani, S., Y. Yamada, and T. Nihira. 2001. Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J. Bacteriol. 183:4357-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lezhava, A., D. Kameoka, H. Sugino, K. Goshi, H. Shinkawa, O. Nimi, S. Horinouchi, T. Beppu, and H. Kinashi. 1997. Chromosomal deletions in Streptomyces griseus that remove the afsA locus. Mol. Gen. Genet. 253:478-483. [DOI] [PubMed] [Google Scholar]
- 15.Miyake, K., T. Kuzuyama, S. Horinouchi, and T. Beppu. 1990. The A-factor-binding protein of Streptomyces griseus negatively controls streptomycin production and sporulation. J. Bacteriol. 172:3003-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakano, H., E. Takehara, T. Nihira, and Y. Yamada. 1998. Gene replacement analysis of the Streptomyces virginiae barA gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J. Bacteriol. 180:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natsume, R., Y. Ohnishi, T. Senda, and S. Horinouchi. 2004. Crystal structure of a γ-butyrolactone autoregulator receptor protein in Streptomyces coelicolor A3(2). J. Mol. Biol. 336:409-419. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen, K. T., J. Tenor, H. Stettler, L. T. Nguyen, L. D. Nguyen, and C. J. Thompson. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185:7291-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto, S., K. Nakamura, T. Nihira, and Y. Yamada. 1995. Virginiae butanolide binding protein from Streptomyces virginiae. Evidence that VbrA is not the virginiae butanolide binding protein and reidentification of the true binding protein. J. Biol. Chem. 270:12319-12326. [DOI] [PubMed] [Google Scholar]
- 21.Onaka, H., N. Ando, T. Nihira, Y. Yamada, T. Beppu, and S. Horinouchi. 1995. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J. Bacteriol. 177:6083-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onaka, H., and S. Horinouchi. 1997. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol. Microbiol. 24:991-1000. [DOI] [PubMed] [Google Scholar]
- 23.Onaka, H., M. Sugiyama, and S. Horinouchi. 1997. A mutation at proline-115 in the A-factor receptor protein of Streptomyces griseus abolishes DNA-binding ability but not ligand-binding ability. J. Bacteriol. 179:2748-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stratigopoulos, G., A. R. Gandecha, and E. Cundliffe. 2002. Regulation of tylosin production and morphological differentiation in Streptomyces fradiae by TylP, a deduced γ-butyrolactone receptor. Mol. Microbiol. 45:735-744. [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama, M., H. Onaka, T. Nakagawa, and S. Horinouchi. 1998. Site-directed mutagenesis of the A-factor receptor protein: Val-41 important for DNA-binding and Trp-119 important for ligand-binding. Gene 222:133-144. [DOI] [PubMed] [Google Scholar]
- 26.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the γ-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 27.Takano, E., T. Nihira, Y. Hara, J. J. Jones, C. J. Gershater, Y. Yamada, and M. Bibb. 2000. Purification and structural determination of SCB1, a γ-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275:11010-11016. [DOI] [PubMed] [Google Scholar]
- 28.Takano, E., M. Tao, F. Long, M. J. Bibb, L. Wang, W. Li, M. J. Buttner, M. J. Bibb, Z. X. Deng, and K. F. Chater. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50:475-486. [DOI] [PubMed] [Google Scholar]
- 29.Ueda, K., T. Umeyama, T. Beppu, and S. Horinouchi. 1996. The aerial mycelium-defective phenotype of Streptomyces griseus resulting from A-factor deficiency is suppressed by a Ser/Thr kinase of S. coelicolor A3(2). Gene 169:91-95. [DOI] [PubMed] [Google Scholar]
- 30.Waki, M., T. Nihira, and Y. Yamada. 1997. Cloning and characterization of the gene (farA) encoding the receptor for an extracellular regulatory factor (IM-2) from Streptomyces sp. strain FRI-5. J. Bacteriol. 179:5131-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 34.Yang, K., L. Han, and L. C. Vining. 1995. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a receptor gene, jadR2. J. Bacteriol. 177:6111-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]