In gram-negative bacteria, secretion of macromolecules across the two membranes takes place using diverse macromolecular transport assemblies, from simple one-component systems to complex multicomponent machineries. The type IV secretion system (T4SS) is one of the five major secretion systems that are capable of exporting virulence factors across the membranes of gram-negative bacteria. T4SSs are defined as macromolecular transfer systems, the components of which are homologous in sequence and probably in structure to those of conjugative transfer systems of naturally occurring plasmids (35).
Several important plant and human pathogens have evolved secretion machineries ancestrally related to conjugation systems for the purpose of delivering virulence effectors (proteins or protein-DNA complexes) to eukaryotic-cell targets. Such pathogens include extracellular organisms such as Agrobacterium tumefaciens, the causative agent of crown gall disease, widely used to modify crops genetically; Bordetella pertussis, the agent responsible for whooping cough in children; and Helicobacter pylori, responsible for gastric ulcers and stomach cancer (3, 6, 9, 52). More recently, intracellular bacterial pathogens, such as Brucella suis, the causative agent of brucellosis, and Legionella pneumoniae, the causative agent of Legionnaires' disease, were shown to require such systems for virulence (5, 32). As more bacterial genomes are being sequenced, the list of putative T4SSs is increasing rapidly, suggesting that macromolecular transport by such systems is widespread in nature.
This review summarizes recent progress in determining the molecular scaffolds of the structural components of T4SSs. We do not expand on the structural biology of T4SS effectors such as pertussis toxin (44) or the recently determined catalytic core of the relaxase (13).
PROTEIN COMPONENTS AND ASSEMBLY OF THE TYPE IV SECRETION APPARATUS
The VirB-VirD system of A. tumefaciens, or Ti-DNA transfer system, has served as a prototype for T4SSs (Fig. 1A). In A. tumefaciens, the T4SS is responsible for secreting the virulence factors that lead to the formation of crown galls in infected plants (52). The A. tumefaciens T4SS is composed of 11 proteins encoded by the virB operon and at least 1 protein, VirD4, encoded by the virD operon. Some T4SS members contain a complete set of proteins similar to the A. tumefaciens VirBD proteins, while others do not (Fig. 1A). T4SSs can be classified according to the nature of the effector molecules secreted by the machinery (reviewed in detail in reference 8).
FIG. 1.
(A) Gene organization and nomenclature of T4SSs. The proteins are grouped according to their putative functions: ATPases in green, core components in blue, pilus structures in orange, lytic transglycosylases in yellow, and proteins with no assigned function in magenta. Individual proteins within a functional group are distinguished by different shadings. (B) Model of the type IV secretion machinery. The association of energy-generating transporters with the machinery needs to be defined, as well as the translocation of substrates from the cytoplasm through the core complex. Surface structures (pili) may initiate contact with the host cell, but the substrate translocation pathway is unknown. This assembly model reflects our present knowledge on protein-protein interaction linkage of T4SS components.
The proteins of the type IV secretion machinery can be grouped according to their functions (mostly putative) and/or cellular locations (3, 8). The first group comprises three cytoplasm- or inner-membrane-associated ATPases: VirB4, VirB11, and VirD4. These proteins exhibit the highest sequence conservation among VirBD components. The VirB4 homologues are large inner membrane proteins. A. tumefaciens VirB4 is a homodimer protein containing ATPase activity and may energize transport (10, 11, 42). Sequence analyses of VirB4 proteins predict four putative transmembrane sequences in the middle of the proteins (7). Multiple sequence alignment of the VirB4 proteins indicates at least four notable conserved motifs, including the nucleotide binding sites (29). A recent study has shown that soluble variants of VirB4 homologues, such as TrbE of plasmid RP4 and TrwK of plasmid R388, do not hydrolyze ATP or GTP and behave as monomers in solution (29).
The VirB11 proteins are among the most extensively studied constituents of the T4SS machinery (22, 23, 30, 33). Sequence analysis of VirB11 proteins reveals the presence of highly conserved hydrophilic domains and typical Walker A and B boxes (23). VirB11 proteins are related in sequence to the PulE class of ATPases, which power the type II secretion system (28). VirB11 proteins form hexameric assemblies that exhibit a nucleotide-hydrolyzing activity, which is stimulated by lipids (23). VirB11 proteins are likely located to the inner membrane but may also be found in the cytoplasm (22, 23).
VirD4 (or TraG-like) proteins act as “coupling” proteins. In bacterial conjugation systems, VirD4 proteins are thought to mediate interactions between the DNA-processing (relaxosome) and mating pair formation (T4SS) systems (25, 40, 41). They also function as essential components of T4SSs of several bacterial pathogens such as H. pylori. VirD4 proteins form oligomers (mostly hexamers) and bind DNA in a non-sequence-specific manner (including HP0524, the H. pylori VirD4 homologue). None exhibits the postulated nucleoside triphosphatase (NTPase) activity, but all bind ADP, the product of ATP hydrolysis (41).
The second group comprises proteins forming core complexes in the periplasm and/or membrane, such as VirB6, VirB7, VirB8, VirB9, and VirB10. Several lines of evidence indicate that these proteins interact with each other. At present no structure information is available for any member of this group. VirB6 homologues are relatively poorly conserved, with no strictly conserved residues or motif. VirB7 proteins are small (about 55 residues), except for the H. pylori homologue, called CagT (370 residues). VirB8 homologues are proteins of about 250 residues with one putative transmembrane domain in the N-terminal domain (NTD). Except for several patches of significant conserved residues, the overall sequence conservation of the protein family is not very high. VirB9 homologues are proteins of approximately 300 residues with moderate levels of sequence conservation. VirB10 homologues are proteins of about 400 residues, except for the H. pylori homologue, HP0527 (1,819 residues composed of repetitive sequence motifs). Only the hydrophobic C-terminal region of 150 residues is well conserved. VirB8, -9, and -10 are believed to form the core of the transfer machinery, possibly forming a pore spanning the two membranes (4, 12, 14, 15, 21).
The third group, comprising VirB2 and VirB5, contains components of the pilus or the cell surface structure. The proposed role of the pilus in conjugation is to establish physical contact between the donor and recipient cells. The contact is initiated when the donor cell attaches the tip of the pilus to the recipient cell. A depolymerization step is thought to pull the donor and recipient together, thus allowing the cell envelopes to engage in intimate contact (2). The VirB2 protein is the major component of the pilus. Of particular interest is the finding that TrbC and VirB2 (or T pilin), encoded by plasmids IncP (RP4) and Ti (A. tumefaciens), respectively, are cyclic peptides (20, 24). The VirB5 protein is known as a minor component of the pilus structure (39). VirB5 proteins are essential for T4SS virulence. The T4SSs of B. pertussis and H. pylori encode the VirB2 proteins PtlA and HP0546, respectively, but no VirB5 homologues. While there is no evidence for a pilus in B. pertussis, a recent study provides evidence for a filamentous surface organelle which is part of the H. pylori T4SS (31).
The protein interaction network contributing to T4SS assembly has been studied extensively (12, 21, 47). A study using high-resolution two-hybrid libraries provided details of the protein-protein interactions among VirB proteins from A. tumefaciens (47). Interactions among VirB11, VirB8, VirB9, and VirB10, and among VirB4, VirB8, and VirB10 provide evidence for T4SS interaction pathways linking the cytoplasmic components of the machinery to the core VirB8-VirB9-VirB10 complex. The NTD of VirB11 appears to harbor the regions interacting with other VirB proteins. In another study using a biochemical approach, protein-protein interactions in the membranes of A. tumefaciens were analyzed after extraction with the mild detergent dodecyl-β-d-maltoside followed by separation under native conditions (21). This work revealed two classes of protein complexes containing VirB7. The first class consists of the T-pilus major component VirB2, the minor component VirB5, and an associated protein, VirB7, in the low-molecular-weight portion of the gel (about 100 kDa). The second class contains the putative translocation complex core components VirB8, VirB9, VirB10, and VirB7 in the high-molecular-weight portion of the gel (about 232 kDa). This work provides evidence for a VirB7-mediated link between pilus constituents and the core VirB8-VirB9-VirB10 complex. TraG/VirD4 of the RP4 T4SS has been shown to interact with the corresponding relaxase TraI, and the VirD4 homologue of the R388 conjugative T4SS has been shown to interact with TrwE/VirB10 (26, 40). Thus, VirD4 couples the relaxosome to the T4SS for conjugative transfer of plasmid DNA.
MOLECULAR SCAFFOLDS AND STRUCTURAL INSIGHTS INTO TYPE IV COMPONENTS
VirB11 proteins. (i) Crystal structure of HP0525.
HP0525 is the structural prototype for VirB11 proteins. Three representative states of the full-length HP0525 protein (330 amino acids) have been structurally characterized to date: the HP0525-ATPγS (protein-substrate) complex, the HP0525-ADP (protein-product) complex, and the apoprotein (37, 50). The monomer structure contains two domains that are formed by the two contiguous parts of the amino acid sequence (Fig. 2A). Each domain is composed of an extended central β sheet containing six β strands in the NTD and seven β strands in the C-terminal domain (CTD). Topologically, the CTD adopts the RecA fold (45), while the NTD is a fold unique to HP0525. The ADP molecule is bound at the interface between the two domains.
FIG. 2.
Molecular features of HP0525 (PDB code 1g6o). (A) Ribbon presentation of HP0525 bound to ADP. The N and C termini are indicated (N-ter and C-ter, respectively). The two domains are colored differently: NTDs in gold and CTDs in magenta. The ADP molecule is in cyan. (B) Ribbon diagram showing the two-ring feature of the HP0525 hexamer. The color scheme is the same as that in panel A. The proposed membrane-associated state of the HP0525 hexamer is modeled. (C) HP0525 hexamer viewed down the small hole of the chamber formed by the CTDs. The six subunits are color coded differently. (D) Surface of the HP0525 hexamer, color coded according to charge: blue for the most positive regions and red for the most negative regions, with interpolations in between. (Left) View along the sixfold axis membrane pole. (Center) Cutaway of the HP0525 hexamer to highlight the inside channel. The orientation corresponds to that shown in panel B. (Right) Surface of HP0525 viewed down the small hole of the chamber. This side of the hexamer, with a highly negative charge, would face the cytoplasm. The orientation corresponds to that shown in panel C.
The six HP0525 subunits assemble in an intertwined propeller shape whereby residues in both domains of individual subunits participate in the subunit-subunit interface (Fig. 2). The overall shape of the hexamer is that of a six-clawed grapple mounted on a hexameric ring. Indeed, as shown in Fig. 2B, it is apparent that the NTD and CTD form two rings, which together form a dome-like chamber open on one side and closed on the other. The HP0525 hexamer has overall dimensions of 100 Å in diameter and 50 Å in height. The internal chamber has a diameter of 50 Å and a depth of about 30 Å (50). The outside surfaces of the hexameric N-terminal ring and the part of the CTD ring juxtaposed to it are relatively hydrophobic (Fig. 2D), suggesting that these parts may be inserted into the membrane.
(ii) VirB11 ATPases as dynamic hexameric assemblies.
The structure of apo-HP0525 at 3 Å resolution reveals an asymmetric hexameric assembly that is significantly different from the HP0525-ADP complex structure (37). While the CTD rings retain their “six-clawed grapple” shape, the NTDs undergo rigid-body rotations about the linker region between the NTD and CTD and away from the center of the chamber. The magnitude of the conformational change is different in each subunit. Sedimentation equilibrium experiments confirm that nucleotide-dependent conformational changes occur (37). Thus, VirB11 proteins are dynamic molecular assemblies, and the cycling of the subunits through a series of conformational changes induced by ATP binding and hydrolysis, followed by the release of ADP, may be important for function. Structure-based mutagenesis also suggests that the NTDs play a crucial role (37). Indeed, an R18A HP0525 variant (R18 is located in the N-terminal helix αA, far away from the nucleotide-binding site) exhibits increased ATPase activity but decreased stability of the hexamer. It is also biologically inactive.
Based on the structural evidence, together with structure-based functional studies, a four-step cycling mechanism for the VirB11 ATPases was described, as follows. (i) The nucleotide-free VirB11 form exists as an asymmetric hexamer possessing conformational flexibility in the NTDs while the CTDs are responsible for maintaining a pseudohexameric scaffold. (ii) The binding of three ATP molecules locks three subunits into a rigid conformation. (iii) Hydrolysis of these three ATPs to ADPs, with concomitant binding of ATP to the remaining three nucleotide-free subunits, results in a rigid, symmetric hexamer. (iv) The molecule retains this rigid hexameric form until all ATP molecules are hydrolyzed, at which point VirB11 ATPases can return to their nucleotide-free form. The alternate ATP binding mechanism was proposed in light of the alternate occupancy of ATP-γS within the HP0525-ATP-γS complex structure.
Another interesting observation is that the only significant structural similarity HP0525 has with other proteins in the Protein Data Bank (PDB) is that with the p97 AAA ATPase (51), a protein involved in homotypic membrane fusion and organelle biogenesis. This finding suggests perhaps a functional homology between VirB11 ATPases and p97 proteins. Although membrane fusion events have never been observed in bacteria utilizing type IV secretion machineries, it is known that during mating and conjugation, the pilus, which may have served for attachment and early recognition events between mating pairs, disappears and the outer membranes of the mating partners come into close juxtaposition (36). Could T4SSs be ancestral fusion systems? This would at this point in time be a very speculative proposition. However, VirB11 ATPases could use the mechanical force that they appear to be able to exert through ATP-driven conformational changes to recruit, assemble, or disassemble type IV secretion protein components, making them available for insertion into the nascent secretion apparatus and/or facilitating substrate translocation across the membrane. A high-resolution yeast two-hybrid study with A. tumefaciens identified several protein partners for VirB11 within VirB protein components of the type IV secretion apparatus (see above). Interestingly, the NTDs of VirB11, not the CTDs, harbor the interacting sites, which is consistent with our observation that the NTDs are the most likely effectors of VirB11 ATPase activities.
VirD4 proteins. (i) Crystal structure of TrwB.
TrwB, the VirD4 protein from the R388 plasmid, is a protein of 507 residues and binds single-stranded DNA (ssDNA) and double-stranded DNA nonspecifically and independently of nucleoside triphosphate (NTP) binding (27). The crystal structure of a soluble fragment lacking the 70 residues of the N-terminal transmembrane part (TrwBΔN70) unveiled the molecular architecture of TrwB (17, 18). Three representative states of TrwBΔN70 are available to date: protein-substrate, protein-product, and apoprotein. The TrwBΔN70 monomer has an orange slice shape with approximate dimensions of 90 by 45 by 40 Å (Fig. 3A). It consists of two domains: a cytosol-oriented all-α-helical domain (AAD) and a membrane-proximal nucleotide-binding domain (NBD). The NBD is composed of a central, twisted, nine-stranded, mixed β-pleated sheet, which is flanked by four and seven helices on either side. On top of the NBD, the smaller AAD (residues Gly184 to Gly297) is inserted between β4 and αL of the NBD (18). This AAD contains seven helices. Topologically, the NBD reveals a high structural similarity to the equivalent domain of RecA and other RecA-like core-encompassing enzymes. Although helicase motifs cannot be found in the sequences of VirD4-like proteins, several helicases/ATPase-like proteins also display an AAD besides the core NBD. However, the TrwB AAD appears to bear significant structural similarity only to NTD1 of the site-specific recombinase XerD of the λ-integrase family (46), and thus it has been proposed to contribute to DNA binding (18).
FIG. 3.
Molecular features of TrwB (PDB code 1e9r). (A) Ribbon presentation of TrwBΔN70 bound to ADP. The N and C termini are indicated (N-ter and C-ter, respectively). Magenta, AAD; gold, NBD. A bound ADP molecule is shown as a cyan stick model to position the nucleotide-binding site. (B) TrwBΔN70 hexamer viewed from the side. The color-coding scheme is the same as that in panel A. (C) TrwBΔN70 hexamer viewed down the small hole at the base of the hexamer formed by the AADs. Each monomer is color coded differently. (D) Surface of the TrwBΔN70 hexamer, color coded as described in the legend to Fig. 2D. (Left) View along the sixfold axis membrane pole. (Center) Cutaway of the TrwBΔN70 hexamer to highlight the inside channel. The orientation corresponds to that shown in panel B. (Right) Surface of the TrwBΔN70 hexamer viewed down the hole in the cytoplasmic side. The orientation corresponds to that shown in panel C.
Six TrwBΔN70 subunits associate tightly to form an almost orange-shaped hexamer, somewhat flattened at both poles, with overall dimensions of 110 Å in diameter and 90 Å in height (Fig. 3B, C, and D). TrwB is very similar to the F1-ATPase α3β3 heterohexamer (1), except for being a homohexamer. In addition, TrwB shares with F1-ATPase the property of being a membrane-associated protein. A central channel runs from the cytosolic pole (formed by the AADs) to the membrane pole (formed by the NBDs), ending at the putative transmembrane pore. Thus, the central channel may connect cytoplasm and periplasm. The channel entrance is plugged by a crown of asparagine residues and restricted to a diameter of ∼8 Å. This is the narrowest part of the channel, which is ∼20 Å wide all along and ends up at its membrane side with an opening of ∼22 Å, although a narrower section may occur at the modeled transmembrane domain.
(ii) VirD4 coupling proteins as gatekeepers in conjugation.
VirD4 coupling proteins are present in all conjugative systems (25). How would a coupling protein function? Their location and interactions with other proteins strongly suggest that these proteins do indeed act as coupling factors between the relaxosome and the T4SS machinery. Direct interaction has been observed in vitro between the relaxase TraI (a VirD2 homologue) and TraG (a VirD4 homologue) of plasmid RP4 (40). Very recently, direct interaction has been observed between VirD4 and VirB10 homologues (16, 26). The ability of VirD4 proteins to bind DNA and the possibility that they may be able to harness the energy derived from NTP binding and hydrolysis (perhaps aided by a cofactor) suggest that they may play a direct role in DNA transport and help thread the DNA toward a transmembrane pore and/or drive the DNA through it.
The strong structural similarity of TrwB to DNA ring helicases suggests that the ssDNA might pass through the central channel of the particle (19, 43). Based on this observation, a working model was proposed: ATP binding and hydrolysis could induce a molecular switch mechanism affecting the channel or triggering domain rearrangements and thereby promote DNA binding and displacement through the channel. A main drawback of this model is that the hole (∼8 Å) at the cytoplasmic side is too narrow for ssDNA to pass through. Also, ATP binding does not appear to be sufficient to trigger conformational changes (18). Conformational changes may, however, be induced by interaction with relaxosome components or other cellular proteins.
VirB5 proteins.
VirB5 proteins are minor components of the pilus in both the A. tumefaciens and pKM101 plasmid T4SSs (38, 39). In Agrobacterium, VirB5 cofractionates with extracellular VirB2, the major pilus component, and also with VirB7, an outer membrane protein (21, 34). Despite the fact that VirB5 proteins are essential for type IV secretion, little is known about the functions that these proteins fulfill in these machineries. Sequences of about 15 VirB5 proteins are currently available in the data bank. These proteins are characteristically about 220 residues long and acidic (theoretical pI, 4 to 5), and they contain N-terminal signal peptide sequences targeting the protein to the periplasmic space.
(i) Crystal structure of TraC.
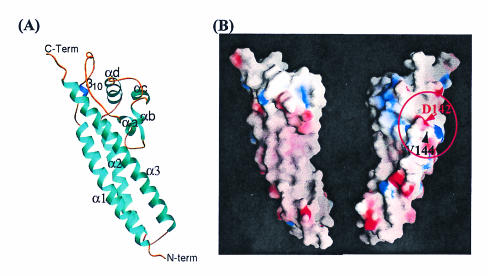
The VirB5 orthologue of the Escherichia coli conjugative plasmid pKM101, TraC, is a protein of 237 residues. It is also extracellular and is associated with the conjugative pilus, which is predicted to contain the VirB2 orthologue, TraM, as a major component (39, 48). The crystal structure of TraC unveiled VirB5 as a single domain protein with a mostly α-helical, elongated structure (Fig. 4) (49). Three long α-helices form the backbone of the structure. This backbone supports a loose α-helical appendage formed by four short helices. Based on the surface electrostatic-potential calculation, TraC presents to the solvent a mostly polar surface.
FIG. 4.
Structure of TraC (PDB code 1r8i) and mapping of functional residues onto the TraC surface. (A) Ribbon diagram of the structure of the TraC monomer. α-Helices, the 310 helix, and loops are colored cyan, blue, and amber, respectively. The three major helices are labeled as α1 to α3, while the short helices between α1 and α2 are labeled as αa to αd. (B) Electrostatic-potential surface of TraC. (Left) TraC is in the same orientation as in panel A. (Right) The surface has been rotated 180° from the view on the left. The red circle delineates the region found to differentially affect the functions of TraC. It contains residues D142 and V144.
(ii) A role for VirB5 proteins.
Our recent efforts to assign TraC a function based on its structure provided functional evidence suggesting that VirB5 proteins mediate some of the pilus functions, such as phage attachment and cell adhesion. In that study, a surface region likely to be involved in protein-protein interaction was identified (Fig. 4B). Two TraC variants in this region that exhibit remarkable properties, D142E and V144W, were found: they are incorporated into the pilus, and yet they affect conjugative transfer dramatically, either decreasing it (D142E) or increasing it (V144W) (49). The D142E variant is also defective in supporting both PRD1 and IKe phage infection; the V144W mutant is defective in promoting infection by PRD1, while infection by IKe proceeds successfully. Thus, this study showed that TraC proteins are able to locate to the pilus yet are unable either to carry out all pilus functions (D142E) or to carry out selective functions (V144W). A simple and straightforward interpretation for such behavior and for other properties of VirB5 proteins reported in the literature is that TraC/VirB5 may be involved in adhesion and that it is also indispensable for pilus biogenesis. However, direct evidence has not been produced, and thus the role of VirB5 proteins in adhesion remains unclear. The findings that TraC variants (V144E, V144W, and V144D) were able to distinguish between two bacteriophages, IKe and PRD1, which have been shown, respectively, to adhere to the tip of the pilus and to bind at the base of the pilus, suggest that TraC may be the receptor for both phages and may decorate the pilus at various points, including the tip, where it can promote attachment of IKe, and the base, where it can promote attachment of PRD1.
CONCLUDING REMARKS
We anticipate that many challenging aspects of T4SSs will be addressed over the next several years. Once a structure of at least one representative homologue of each VirB protein family is determined, insights into the function of each of these proteins will be gained. But will they be sufficient for understanding how T4SSs work? Probably not. The structures of VirB11, VirD4, and VirB5 homologues have provided considerable insights into the way these proteins may work. But, disappointingly, they have provided little information as to what they actually do within the type IV secretion machinery. Further progress will be contingent on generating views of the numerous protein-protein interaction complexes that these proteins form within T4SSs, thus allowing us to reconstitute the structure of the entire system. We anticipate that this will be achieved by use of combinations of crystallographic and electron microscopic analyses.
Acknowledgments
This work was funded by Wellcome Trust grant 065932.
We thank Erich Lanka for valuable comments and suggestions.
REFERENCES
- 1.Abrahams, J. P., A. G. Leslie, R. Lutter, and J. E. Walker. 1994. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370:621-628. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M., G. Morelli, and S. Schwuchow. 1978. Cell-cell interactions in conjugating Escherichia coli: role of F pili and fate of mating aggregates. J. Bacteriol. 135:1053-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1366. [DOI] [PubMed] [Google Scholar]
- 4.Beaupré, C. E., J. Bohne, E. M. Dale, and A. N. Binns. 1997. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 179:78-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boschiroli, M. L., S. Ouahrani-Bettache, V. Foulongne, S. Michaux-Charachon, G. Bourg, A. Allardet-Servent, C. Cazevieille, J. P. Lavigne, J. P. Liautard, M. Ramuz, and D. O'Callaghan. 2002. Type IV secretion and Brucella virulence. Vet. Microbiol. 90:341-348. [DOI] [PubMed] [Google Scholar]
- 6.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 7.Cao, T. B., and M. H. Saier, Jr. 2001. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology 147:3201-3214. [DOI] [PubMed] [Google Scholar]
- 8.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 10.Dang, T. A., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, T. A., X. R. Zhou, B. Graf, and P. J. Christie. 1999. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on the assembly and function of the T-DNA transporter. Mol. Microbiol. 32:1239-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, A., and Y. H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta, S., C. Larkin, and J. F. Schildbach. 2003. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure 11:1369-1379. [DOI] [PubMed] [Google Scholar]
- 14.Ding, Z., Z. Zhao, S. J. Jakubowski, A. Krishnamohan, W. Margolin, and P. J. Christie. 2002. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 184:5572-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finberg, K. E., T. R. Muth, S. P. Young, J. B. Maken, S. M. Heitritter, A. N. Binns, and L. M. Banta. 1995. Interactions of VirB9, -10, and -11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations. J. Bacteriol. 177:4881-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmour, M. W., J. E. Gunton, T. D. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 17.Gomis-Ruth, F. X., and M. Coll. 2001. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 33:839-843. [DOI] [PubMed] [Google Scholar]
- 18.Gomis-Ruth, F. X., G. Moncalian, F. de la Cruz, and M. Coll. 2002. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277:7556-7566. [DOI] [PubMed] [Google Scholar]
- 19.Guasch, A., J. Pous, B. Ibarra, F. X. Gomis-Ruth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa, and M. Coll. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 315:663-676. [DOI] [PubMed] [Google Scholar]
- 20.Kalkum, M., R. Eisenbrandt, R. Lurz, and E. Lanka. 2002. Tying rings for sex. Trends Microbiol. 10:382-387. [DOI] [PubMed] [Google Scholar]
- 21.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krause, S., M. Barcena, W. Pansegrau, R. Lurz, J. M. Carazo, and E. Lanka. 2000. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc. Natl. Acad. Sci. USA 97:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krause, S., W. Pansegrau, R. Lurz, F. de la Cruz, and E. Lanka. 2000. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J. Bacteriol. 182:2761-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, E. M., R. Eisenbrandt, M. Kalkum, E. Lanka, and C. I. Kado. 2002. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 184:327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 26.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moncalian, G., E. Cabezon, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goni, and F. de La Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 28.Motallebi-Veshareh, M., D. Balzer, E. Lanka, G. Jagura-Burdzy, and C. M. Thomas. 1992. Conjugative transfer functions of broad-host-range plasmid RK2 are coregulated with vegetative replication. Mol. Microbiol. 6:907-920. [DOI] [PubMed] [Google Scholar]
- 29.Rabel, C., A. M. Grahn, R. Lurz, and E. Lanka. 2003. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 185:1045-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivas, S., S. Bolland, E. Cabezon, F. M. Goni, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272:25583-25590. [DOI] [PubMed] [Google Scholar]
- 31.Rohde, M., J. Puls, R. Buhrdorf, W. Fischer, and R. Haas. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 49:219-234. [DOI] [PubMed] [Google Scholar]
- 32.Roy, C. R., and L. G. Tilney. 2002. The road less traveled: transport of Legionella to the endoplasmic reticulum. J. Cell Biol. 158:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sagulenko, E., V. Sagulenko, J. Chen, and P. J. Christie. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 183:5813-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmond, G. P. C. 1994. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32:181-200. [Google Scholar]
- 36.Samuels, A. L., E. Lanka, and J. E. Davies. 2000. Conjugative junctions in RP4-mediated mating of Escherichia coli. J. Bacteriol. 182:2709-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savvides, S. N., H. J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt-Eisenlohr, H., N. Domke, and C. Baron. 1999. TraC of IncN plasmid pKM101 associates with membranes and extracellular high molecular weight structures in Escherichia coli. J. Bacteriol. 181:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schröder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185:4371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirasu, K., Z. Koukolikova-Nicola, B. Hohn, and C. I. Kado. 1994. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol. Microbiol. 11:581-588. [DOI] [PubMed] [Google Scholar]
- 43.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage φ29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein, P. E., A. Boodhoo, G. D. Armstrong, S. A. Cockle, M. H. Klein, and R. J. Read. 1994. The crystal structure of pertussis toxin. Structure 2:45-57. [DOI] [PubMed] [Google Scholar]
- 45.Story, R. M., I. T. Weber, and T. A. Steitz. 1992. The structure of the E. coli recA protein monomer and polymer. Nature 355:318-325. [DOI] [PubMed] [Google Scholar]
- 46.Subramanya, H. S., L. K. Arciszewska, R. A. Baker, L. E. Bird, D. J. Sherratt, and D. B. Wigley. 1997. Crystal structure of the site-specific recombinase, XerD. EMBO J. 16:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winans, S. C., and G. C. Walker. 1985. Conjugal transfer system of the N incompatibility plasmid pKM101. J. Bacteriol. 161:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeo, H.-J., Q. Yuan, M. R. Beck, C. Baron, and G. Waksman. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc. Natl. Acad. Sci. USA 100:15947-15952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeo, H. J., S. N. Savvides, A. B. Herr, E. Lanka, and G. Waksman. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol. Cell 6:1461-1472. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, X., A. Shaw, P. A. Bates, R. H. Newman, B. Gowen, E. Orlova, M. A. Gorman, H. Kondo, P. Dokurno, J. Lally, G. Leonard, H. Meyer, M. van Heel, and P. S. Freemont. 2000. Structure of the AAA ATPase p97. Mol. Cell 6:1473-1484. [DOI] [PubMed] [Google Scholar]
- 52.Zupan, J., T. R. Muth, O. Draper, and P. C. Zambryski. 2000. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 23:11-28. [DOI] [PubMed] [Google Scholar]