Abstract
Transcription of the bphA1A2A3A4C1B genes, which are responsible for the conversion of biphenyl and polychlorinated biphenyl to the meta-cleavage products in Rhodococcus sp. strain RHA1, was examined. The bphA1 promoter (PbphA1) was identified and was shown to promote transcription induction by biphenyl and ethylbenzene. An 8.8-kb HindIII fragment that promotes transcription induction of PbphA1 in Rhodococcus erythropolis IAM1399 was isolated from the region downstream of bphB by using a reporter plasmid containing PbphA1. Analysis of the nucleotide sequence of this fragment revealed a set of putative two-component regulatory system genes, which were designated bphS and bphT. Deletion analysis of the 8.8-kb HindIII fragment indicated that bphT is responsible for the basal activation of PbphA1 and that both bphS and bphT are required for the elevated basal activation of and transcriptional induction by biphenyl of PbphA1. These results support the notion that bphS and bphT encode a sensor kinase and a response regulator, respectively, of a two-component regulatory system. The bphS and bphT genes promote transcriptional induction by a variety of aromatic compounds, including biphenyl, benzene, alkylbenzenes, and chlorinated benzenes. A promoter activity assay and reverse transcription (RT)-PCR analysis revealed a weak constitutive promoter in the adjacent region upstream of bphS. RT-PCR analysis indicated that there is induced transcription of bphA1 through bphT, in which PbphA1 is thought to take part. An insertionally inactivated bphS mutant, SDR1, did not grow on biphenyl. Growth was restored by introduction of an intact bphS gene into SDR1. These results indicate that at least bphS is indispensably responsible for the growth of RHA1 on biphenyl.
Polychlorinated biphenyls (PCBs) are man-made compounds that have been widely used for industrial purposes due to their exceptional stability. As a consequence, PCBs have caused widespread contamination in the environment. With the goal of remediating contaminated environments, microorganisms that can degrade PCBs have been isolated, and their degradative genes have been characterized (2, 6, 13, 21). These microorganisms cometabolize PCBs through the biphenyl metabolic pathway.
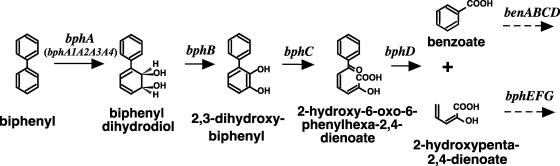
In the aerobic biphenyl metabolic pathway, biphenyl is transformed to benzoate and 2-hydroxypenta-2,4-dienoate by sequential actions of a multicomponent biphenyl dioxygenase (BphA encoded by bphA1A2A3A4) (Fig. 1), a dihydrodiol dehydrogenase (BphB encoded by bphB), a 2,3-dihydroxybiphenyl dioxygenase (BphC encoded by bphC), and a 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (BphD encoded by bphD). 2-Hydroxypenta-2,4-dienoate is further metabolized to pyruvate and acetyl coenzyme A by 2-hydroxypenta-2,4-dienoate hydratase (BphE encoded by bphE), 4-hydroxy-2-oxovalerate aldolase (BphF encoded by bphF), and acetaldehyde dehydrogenase (BphG encoded by bphG). These bph gene-encoded enzymes are usually induced by biphenyl and are involved in the cometabolism of PCBs.
FIG. 1.
Proposed pathway for aerobic bacterial degradation of biphenyl in Rhodococcus sp. strain RHA1. The gene(s) responsible for each enzyme step is indicated above an arrow and is described in the text.
We isolated a gram-positive biphenyl and PCB degrader, Rhodococcus sp. strain RHA1, which has a great capacity to degrade highly chlorinated PCBs (31). In contrast to gram-negative PCB degraders, which have a single set of biphenyl-PCB degradation genes clustered in a single locus (11, 12), RHA1 possesses diverse biphenyl-PCB degradation genes that encode multiple isozymes for each metabolic step and are distributed among multiple clusters (14, 28, 38). Most of these genes are located on linear plasmids, designated pRHL1 and pRHL2 (32). The bphA1A2A3A4C1B genes involved in the upper degradation pathway are located on pRHL1, and all of the other bph genes except bphGF1E1 and bphC5 are located on pRHL2. bphGF1E1 and bphC5 are on the chromosome. bphA1 and bphC1 were found to be transcriptionally induced by biphenyl (14, 28). Gene disruption and enzyme activity analyses suggested that at least the bphA1 and bphC1 genes play major roles in biphenyl-PCB metabolism (10, 20).
Recently, several transcriptional regulatory systems for biphenyl and PCB degradation pathway genes have been described. In Ralstonia eutropha A5, it has been suggested that the bph gene cluster, bphEFGA1A2A3BCD, forms an operon transcribed from a σ70 promoter, which is negatively regulated by a bphS gene-encoded repressor (23). Transcription of the bph gene cluster bphEGF(orf4)A1A2A3BCD(orf1)A4 in Pseudomonas sp. strain KKS102 has been found to be regulated by BphS (24). In Pseudomonas pseudoalcaligenes KF707, it has been found that the ORF0 protein is involved in the regulation of a lower-pathway bph gene operon containing bphX0X1X2X3D (37). In Burkholderia sp. strain LB400, the ORF0 protein was found to be involved in the regulation of transcription from a promoter upstream of bphA1 (3). All these regulators of the biphenyl-PCB degradation pathway in gram-negative bacteria belong to the GntR family of transcriptional regulators. On the other hand, it has been suggested that in gram-positive bacteria, a two-component regulatory system encoded by bpdST is involved in biphenyl-PCB metabolism only in Rhodococcus sp. strain M5 (15). The biphenyl degradation genes of RHA1 are not as similar to those of M5, and the gene organization is distinct from that of M5. In addition, the genes are distributed on linear plasmids. Thus, the regulatory system for biphenyl-PCB metabolism in gram-positive bacteria has attracted much interest.
In the present study we focused on regulation of the bphA1A2A3A4C1B genes, which are known to be expressed and to be responsible for biphenyl and PCB degradation in the gram-positive PCB degrader strain RHA1. Here we describe cloning of the two-component regulatory system for bph gene transcription in RHA1 and present evidence that this system is involved in biphenyl metabolism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Rhodococcus sp. strain RHA1 was grown in Luria-Bertani (LB) medium (10 g of Bacto Tryptone [Difco] per liter, 5 g of yeast extract per liter, 5 g of NaCl per liter) and W minimal medium (20) containing one of the following carbon sources: 0.2% biphenyl, 0.2% sodium benzoate, 0.2% sodium succinate, ethylbenzene, toluene, benzene, or ortho-xylene. Ethylbenzene, toluene, benzene, and ortho-xylene were supplied in the vapor phase. The host strains, Rhodococcus erythropolis IAM1399 (= ATCC 15963) and Escherichia coli JM109 (recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′[traD36 proAB+ lacIq lacZΔM15]) were grown in LB medium. The plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or origin |
|---|---|---|
| Strains | ||
| Rhodococcus sp. strain RHA1 | PCB degrader, BPH+ | 30 |
| Rhodococcus sp. strain SDR1 | bphS mutant of strain RHA1, BPH− | This study |
| R. erythropolis IAM1399 (= ATCC 15963) | Wild type, BPH− | IAM culture collectionb |
| Plasmids | ||
| pBluescript II KS+ | Cloning vector, Apr | Stratagene |
| pG1013F | pBluescript II KS+ with 8.8-kb HindIII fragment of RHA1 carrying bphST | This study |
| pBSL | pBluescript II KS+ with new SpeI site in place of KpnI site | This study |
| pBAB14 | pBSL with 1.4-kb ApaI-BglII fragment of RHA1 carrying bphT | This study |
| pBAB62 | 4.8-kb ApaI fragment carrying bphS inserted into pBAB14 | This study |
| pBAB62ΔSacI | 1.0-kb SacI fragment carrying bphT deleted from pBAB62, leaving bphS | This study |
| pBAS52 | SacI site of pBAB62ΔSacI converted to a SpeI site | This study |
| pET21(+) | Expression vector, Apr | Novagen |
| pETT14 | pET21(+) with SpeI fragment of pBAB14 carrying bphT | This study |
| pETST62 | pET21(+) with SpeI fragment of pBAB62 carrying bphST | This study |
| pKLA1 | Promoter probe vector containing luciferase structural gene, luxAB from V. harveyi, Kmr (neo) | 38 |
| pKLAF1 | pKLA1 with 1.4-kb XhoI-BamHI fragment of RHA1 carrying promoter region of bphA1 | This study |
| pKA851 | pKLAF1 with 8.8-kb HindIII fragment of RHA1 carrying bphST | This study |
| pKAD1-12 | pKLAF1 with deletion derivatives of 8.8-kb HindIII fragment of RHA1 | This study |
| pKLAS1 | pKLAF1 with 5.4-kb SacI fragment of RHA1 carrying bphS | This study |
| pKLAST1 | pKLAF1 with 6.2-kb ApaI-BglII fragment of RHA1 carrying bphST | This study |
| pKLASF | pKLA1 with 0.9-kb PstI-ClaI fragment of RHA1 carrying promoter region of bphS; direction of bphS is identical to that of luxAB reporter gene | This study |
| pKLASR | pKLA1 with 0.9-kb PstI-ClaI fragment of RHA1 carrying promoter region of bphS; direction of bphS is opposite that of luxAB reporter gene | This study |
| pUC-KmD | Gene disruption plasmid, Kmr (aphII) | 29 |
| pUKSD | bphS disruption plasmid pUC-KmD with 1.3-kb ClaI fragment carrying bphS internal fragment of RHA1 | This study |
| pFAJ2574 | Rhodococcus-E. coli shuttle vector, Cmr | 5 |
| pFJS1 | pFAJ2574 with 5.2-kb SpeI fragment of pBAS52 carring bphS | This study |
BPH+, growth on biphenyl; BPH−, no growth on biphenyl.
IAM, Institute of Applied Microbiology.
DNA manipulations and analysis.
All the DNA techniques used, including gene cloning, nucleotide sequencing, electrotransformation (electroporation), and computer analysis of DNA sequences, have been described previously (19, 20, 38).
RNA slot blot analysis.
RHA1 total RNA was prepared as described previously (38). Two micrograms of total RNA was blotted onto a nylon membrane (Hybond N; Amersham International plc, Buckinghamshire, United Kingdom) by using a slot blot apparatus (Bio-Rad, Richmond, Calif.). Probes were labeled as described in the digoxigenin system manual (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). The conditions used for prehybridization, hybridization, washes, and detection were the conditions recommended in the instructions supplied by Boehringer Mannheim for the digoxigenin kit.
Primer extension.
The 5′ end of bphA1 mRNA was mapped by using oligonucleotide primer PEXA1 (5′-TACGAGTTCAGCGATGTCCG-3′, corresponding to nucleotides 215 to 235 relative to the identified transcriptional start site, P1). The primer was end labeled by using T4 polynucleotide kinase (Nippon Gene Co., Tokyo, Japan) with [γ-32P]ATP (Amersham). Two picomoles of the primer was incubated with 20 to 50 μg of RNA in 10 μl of hybridization buffer (10 mM Tris-HCl [pH 8.3], 1 mM EDTA, 0.15 M KCl) at 65°C for 90 min and was allowed to cool. After addition of 30 μl of concentrated reverse transcriptase buffer (containing each deoxynucleoside triphosphate at a concentration of 0.33 mM, 20 mM Tris-HCl [pH 8.3], 10 mM MgCl2, 100 μg of actinomycin D per ml, and 5 mM dithiothreitol) and 20 U of avian myeloblastosis virus reverse transcriptase (Takara Shuzo Co., Ltd., Kyoto, Japan), the mixture was incubated at 42°C for 60 min. The DNA was recovered by ethanol precipitation, after which it was dissolved in 5 μl of formamide loading buffer, and an aliquot was analyzed in an 8% sequencing gel.
RT-PCR.
Reverse transcription (RT)-PCR was performed by using a BcaBEST RNA PCR kit (Takara) for the usual conditions or a ReverTra Dash kit (TOYOBO Co., Ltd., Osaka, Japan) for intensive conditions, as described in the manufacturer's protocol. RT-PCR with ReverTra Dash generates more PCR product and provides superior sensitivity. One to two micrograms of total RNA was reverse transcribed with random primers and PCR amplified with each primer set for 30 cycles at an annealing temperature of 55°C. RNA samples were concurrently analyzed in PCR mixtures without reverse transcriptase to verify the absence of contaminating genomic DNA. PCR mixtures were analyzed on a 2% agarose gel. The primers used for each intergenic region are listed in Table 2.
TABLE 2.
Primers used for RT-PCR
| Primer | Sequence | Region amplified |
|---|---|---|
| 1-F | 5′-TGGATCTATCGATGACGTTCC-3′ | bphS internal |
| 1-R | 5′-GAGGAGAACGAGCTGATTGG-3′ | bphS internal |
| 2-F | 5′-CGGCGGAAACGACCTGTAAG-3′ | bphB-bphS |
| 2-R | 5′-AGCACCCTCACGGTGTCGATTTCC-3′ | bphB-bphS |
| 3-F | 5′-TCGGACAATCCCGATTACC-3′ | bphA1-bphA2 |
| 3-R | 5′-AGAACTGCTCGATCTCGTGC-3′ | bphA1-bphA2 |
| 4-F | 5′-GACCAGAGCACAATTCTCTCC-3′ | bphA2-bphA4 |
| 4-R | 5′-ATAGCCTTCCGAACGCAG-3′ | bphA2-bphA4 |
| 5-F | 5′-GAGTTGCGCGAACTGAATCG-3′ | bphA4-bphC1 |
| 5-R | 5′-ACATCCGCAACCTCGAAGC-3′ | bphA4-bphC1 |
| 6-F | 5′-GTGCGCTACGACAAGATCAGC-3′ | bphC1-bphB |
| 6-R | 5′-GCCGAAGTCGTTGGCAAGC-3′ | bphC1-bphB |
| 7-F | 5′-AGACACCCTGCGGGCCGTGG-3′ | bphS-bphT |
| 7-R | 5′-CGAGGCCGTTGCAGTCGGACAGT-3′ | bphS-bphT |
| 8-F | 5′-GAGGTGCCGATCAGACGATG-3′ | bphT internal |
| 8-R | 5′-CGGAGGCGATCAGCTTCATT-3′ | bphT internal |
Construction of a promoter probe vector.
A promoter probe vector was constructed by using a Rhodococcus-E. coli shuttle vector, pK4 (9), and luxAB luciferase structural genes from Vibrio harveyi, and it was designated pKLA1 (see Fig. 3A). pKLA1 has a unique SalI site for cloning of a DNA fragment containing a promoter region. The promoter fragments were blunt ended and inserted into the blunt-ended SalI site of pKLA1.
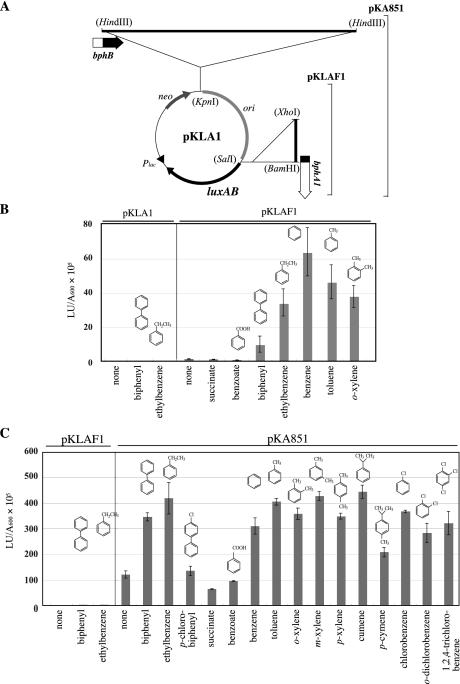
FIG. 3.
Transcriptional activation of bphA1 promoter. (A) Physical maps of pKLAF1 and pKA851. pKLAF1 was constructed by inserting the 1.4-kb XhoI-BamHI bphA1 promoter fragment into a promoter probe vector, pKLA1. The 8.8-kb HindIII fragment containing the region downstream of bphB was cloned in the KpnI site of pKLAF1 to construct pKA851. The restriction sites in parentheses are the sites that were used to generate fragments and were lost by blunt-end formation prior to ligation. (B) Luciferase activity of Rhodococcus sp. strain RHA1 harboring pKLAF1. Cells were grown in 0.2× LB medium in the absence or presence of the substrates. The chemical structures of the substrates are shown. The data are means ± standard deviations from at least three determinations. The luciferase activities of RHA1 cells harboring a promoter probe vector, pKLA1, were all less than 0.1 × 105 light units (LU) per A600 unit (see Materials and Methods). (C) Luciferase activity of R. erythropolis IAM1399 harboring pKLAF1 or pKA851. Cells were grown in 0.2× LB medium in the absence or presence of substrates. The data are means ± standard deviations from at least three determinations.
Luciferase assay.
Recombinant plasmids of pKLA1 were introduced into RHA1 or IAM1399 cells by electroporation. Transformant cells grown on LB medium containing kanamycin (50 μg/ml) were washed with 50 mM sodium phosphate buffer (pH 7.0) and suspended in 10 ml of 0.2× LB medium (2 g of Bacto Tryptone per liter, 1 g of yeast extract per liter, 5 g of NaCl per liter) containing kanamycin (50 μg/ml) at an A600 of 1.0. Each cell suspension was incubated at 30°C for 5 h in the absence or presence of an inducer compound. Solid compounds were each supplied as a powder at a final concentration of 0.2%, and volatile compounds were supplied in vapor. The cell suspension was then diluted 1:10 in lux buffer (25), and a 100-μl aliquot was mixed with 390 μl of lux buffer. After addition of 10 μl of 0.1% (vol/vol) 1-decanal in lux buffer to the resulting 490 μl of diluted cell suspension, the luciferase activity was measured with a luminometer (Lumitester K-100; Kikkoman, Noda, Japan). The total light generated during the initial 15 s was recorded, and the activity was expressed in light units per milliliter of culture per unit of A600.
Detection of gene products.
An ApaI-BglII fragment of pG1013F carrying bphT was inserted between ApaI and BamHI sites of pBSL, yielding plasmid pBAB14. Then the 4.8-kb ApaI fragment of pG1013F carrying the 5′ part of bphS was cloned into the ApaI site of pBAB14. The resulting plasmid, pBAB62, contained the whole bphST region. The SpeI fragments of pBAB14 and pBAB62 carrying bphT and bphST, respectively, were blunt ended and cloned into the blunt-ended BamHI site of pET21(+). The resulting plasmids, pETT14 and pETST62, respectively, were introduced into E. coli BL21. Transformants were grown in LB medium containing ampicillin (50 μg/ml) at 37°C for 2 h and then for 5 h in the presence of isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mM. The cells were washed with 0.5 ml of sodium phosphate buffer (pH 7.0) and resuspended in 0.1 ml of lysis buffer consisting of 100 mM Tris-HCl (pH 6.8), 10% (vol/vol) glycerol, 5% sodium dodecyl sulfate (SDS), and 1 mM 2-mercaptoethanol. Aliquots were boiled for 10 min prior to electrophoresis on an SDS-7% polyacrylamide gel. SDS-polyacrylamide gel electrophoresis (PAGE) was performed as described previously (19).
In vitro expression.
In vitro expression of a cloned gene was carried out with the E. coli S30 extract system (Promega, Madison, Wis.) used according to the manufacturer's protocol. The polypeptides were labeled with [35S]methionine (Amersham). The labeled proteins were separated and detected by SDS-7% PAGE and autoradiography, respectively.
Deletion analysis.
Restriction fragments of pG1013F (see Fig. 6), which were extracted and blunt ended by using T4 DNA polymerase, were cloned into the blunt-ended KpnI site of pKLAF1. Transcription of the bphST genes was started from the kanamycin resistance gene promoter of the pKLAF1 vector. The blunt-ended SpeI fragment of pBAB62 containing bphST was inserted into the blunt-ended KpnI site of pKLAF1, yielding plasmid pKLAST1. To construct the frameshift derivatives pKAD11 and pKAD12, pG1013 was linearized with XmaI and Sse8387I, respectively, and religated after filling in of the ends. Each HindIII fragment containing the bphST genes of pG1013 derivatives was blunt ended and inserted into the blunt-ended KpnI site of pKLAF1. To obtain the bphS plasmid pFJS1, pBAB62ΔSacI was constructed by deleting the SacI fragment from pBAB62, which generated an ApaI-ApaI-SacI insert containing bphS in pBAB62ΔSacI. An SpeI linker was inserted into the blunt-ended SacI site of pBAB62ΔSacI to obtain pBAS52. pFJS1 was constructed by inserting an SpeI fragment containing the bphS insert of pBAS52 into the XbaI site of pFAJ2574, which is a chloramphenicol-resistant E. coli-Rhodococcus shuttle vector.
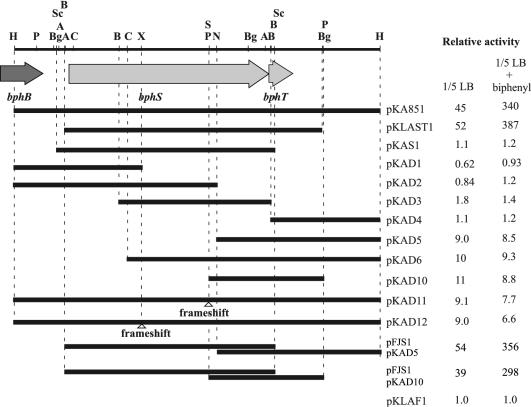
FIG. 6.
Deletion analysis of the 8.8-kb HindIII DNA fragment containing the bphST genes. The segments represented by solid bars were inserted into the KpnI site of pKLAF1. The IAM1399 cells harboring the plasmids were grown in 0.2× LB medium (1/5 LB) in the presence or absence of biphenyl and were subjected to the luciferase assay. The relative luciferase activities conferred by each construct in comparison to the activities of the cells harboring pKLAF1 were estimated and are indicated on the right. pKLAF1 is a reporter plasmid that does not contain any bphST segment. The open triangles indicate the positions of frameshift mutations generated by filling in of restriction fragment termini by T4 polymerase. Restriction enzyme site abbreviations: A, ApaI; B, BamHI; Bg, BglII; C, ClaI; H, HindIII; N, NotI; P, PstI; S, Sse8387I; Sc, SacI; X, XmaI.
Gene disruption and complementation.
To disrupt the bphS gene, a 1.3-kb ClaI fragment containing the internal region of bphS was blunt ended with T4 DNA polymerase and inserted into the SmaI site of pUC-KmD (29), which is a kanamycin-resistant vector and cannot replicate in Rhodococcus strains. The resulting plasmid, pUKSD, was introduced into RHA1 cells by electroporation. A single crossover between bphS and pUKSD sequences was expected to generate tandemly duplicated bphS sequences, leaving a vector containing a kanamycin resistance gene (aphII) between them. Transformants were selected on LB agar plates containing kanamycin (50 μg/ml) and were subjected to a Southern hybridization analysis in order to examine insertion of pUKSD into the bphS gene in RHA1 by single crossover. The insertion mutant obtained was designated SDR1.
To perform bphS gene complementation in SDR1, pFJS1 was introduced into SDR1 by electroporation. A transformant was isolated on an LB agar plate containing chloramphenicol (20 μg/ml). SDR1/pFJS1 and SDR1/pFAJ2574 cells grown in LB medium were washed and resuspended in W minimal medium containing 0.2% biphenyl. The optical density at 600 nm was adjusted to 0.2, and the cell suspension was incubated at 30°C with shaking to examine the growth on biphenyl.
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the DDBJ, EMBL, and GenBank databases under accession no. AB107790.
RESULTS
Expression of the bphA1 promoter in RHA1.
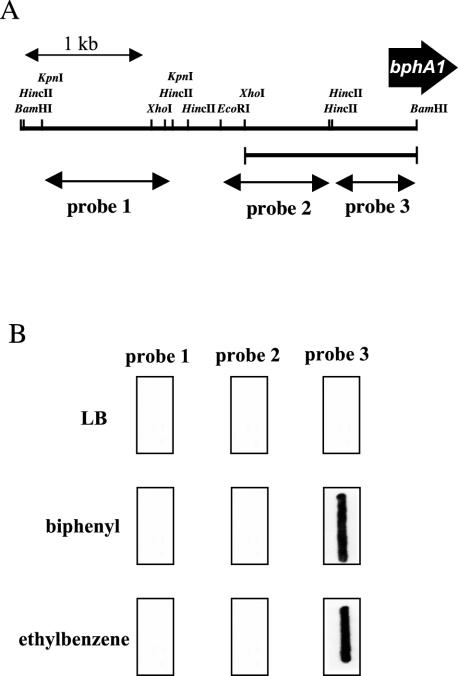
To locate the promoter region of the bphA1 gene in RHA1, transcription of the region upstream of the bphA1 gene was examined by RNA slot blot hybridization analysis by using three separate probes for the bphA1 upstream region (Fig. 2A). Total RNA was extracted from the RHA1 cells grown on LB medium, biphenyl, or ethylbenzene and blotted on a membrane. Neither of the distal upstream probes (probes 1 and 2) hybridized to the total RNA from the cells grown on LB medium, biphenyl, or ethylbenzene (Fig. 2B). The proximal upstream probe, probe 3, hybridized to the RNAs from both biphenyl- and ethylbenzene-grown RHA1 cells. It did not hybridize to RNA from the cells grown on LB medium. These results suggest that the biphenyl- and ethylbenzene-inducible bphA1 promoter (PbphA1) is located in the 1.4-kb XhoI-BamHI fragment of the bphA1 upstream region, which includes probe 3 and its adjacent upstream region.
FIG. 2.
Characterization of the bphA1 promoter. (A) DNA probes used for RNA slot blot hybridization. A 1.1-kb KpnI fragment (probe 1), a 0.9-kb EcoRI-HincII fragment (probe 2), and a 0.7-kb HincII-BamHI fragment (probe 3) of the bphA1 upstream region were used to localize the promoter region of bphA1. The position of the 1.4-kb XhoI-BamHI fragment containing the bphA1 promoter is indicated by a line below the physical map. (B) RNA slot blot hybridization analysis of bphA1 transcripts in Rhodococcus sp. strain RHA1. Two micrograms of total RNA from RHA1 cells grown in LB medium or on a substrate as a sole source of carbon in W minimal medium was blotted onto a nylon membrane and hybridized with digoxigenin-labeled probes 1, 2, and 3.
To instantly examine the expression of PbphA1, the 1.4-kb XhoI-BamHI fragment was inserted into the SalI site of a promoter probe vector, pKLA1 (38) (Fig. 3A). The SalI site in pKLA1 precedes a reporter gene that encodes a LuxAB luciferase of V. harveyi. The resulting plasmid was designated pKLAF1 (Fig. 3A). RHA1 cells harboring pKLAF1 were grown on 0.2× LB medium in the absence or presence of biphenyl, ethylbenzene, toluene, benzene, ortho-xylene, succinate, or benzoate and were subjected to the luciferase assay (see Materials and Methods). Transcription from PbphA1 was activated by biphenyl, ethylbenzene, toluene, benzene, and ortho-xylene (Fig. 3B). Transcription from PbphA1 was induced by neither succinate nor benzoate. The luciferase activity conferred by pKLAF1 in the cells grown in 0.2× LB medium was, however, 85-fold higher than the luciferase activity of pKLA1, suggesting that PbphA1 has basal promoter activity in addition to inducible activity.
Isolation of a fragment activating transcription from PbphA1.
Regulatory genes located adjacent to the corresponding degradation enzyme genes have been reported frequently (15, 16, 33). The 8.8-kb HindIII fragment containing the region downstream of bphB was cloned in the KpnI site of pKLAF1 to obtain pKA851 (Fig. 3A). When pKA851 was introduced into a rhodococcal host strain devoid of bph genes, R. erythropolis IAM1399, induction of PbphA1 was observed with a broad range of aromatic compounds, including biphenyl, ethylbenzene, benzene, toluene, xylenes, cumene, p-cymene, and chlorinated benzenes. Induction was not observed with benzoate, succinate, and p-chlorobiphenyl (Fig. 3C). These results suggest that this 8.8-kb HindIII fragment contains a regulatory gene(s) that promotes induced activation of PbphA1 by a variety of aromatic compounds. In the absence of aromatic compounds, the cells carrying pKA851 exhibited low luciferase activity which was higher than the luciferase activity of the cells carrying pKLAF1, suggesting that the basal activation of PbphA1 is promoted by the regulatory gene(s) in the 8.8-kb HindIII fragment.
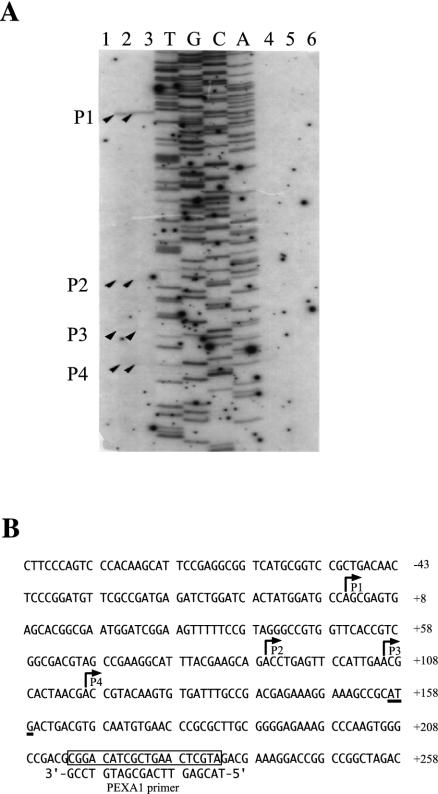
To determine the transcription initiation site of the bphA1 gene, we performed primer extension analysis. Total RNAs were prepared from IAM1399 cells harboring pKA851 grown in LB medium, on biphenyl, or on ethylbenzene. The primer extension products were observed with RNA from the cells grown on biphenyl or ethylbenzene but not with RNA from the cells grown in LB medium. A single major product, designated P1 (Fig. 4), was found, and this product represented transcription starting 156 bp upstream from the ATG initiation codon. Three minor products, P2 to P4 (Fig. 4), were also detected, and these products had transcription starts 66, 50, and 39 bp upstream from the initiation codon, respectively. Total RNA from the IAM1399 cells harboring pKLAF1 gave no product. Possible promoter consensus sequences of E. coli, Bacillus, and Streptomyces coelicolor were not identified at the appropriate positions for the transcription start sites (P1 to P4). The P2 to P4 minor products might have been generated by 5′ end processing of the major P1 transcript.
FIG. 4.
Transcription start site of bphA1. (A) Primer extension analysis of bphA1. Total RNA was isolated from IAM1399 cells harboring pKA851 (lanes 1, 2, and 3) or pKLAF1 (lanes 4, 5, and 6) grown on LB medium (lanes 1 and 4), biphenyl (lanes 2 and 5), or ethylbenzene (lanes 3 and 6). Lanes T, G, C, and A, sequencing ladder for pKLAF1 obtained by using the PEXA1 primer. The primer extension products are indicated by arrowheads and are designated P1, P2, P3, and P4. (B) Nucleotide sequence of the upstream region of bphA1. The deduced transcriptional start sites for P1, P2, P3, and P4 are indicated by bent arrows. The open box indicates the position of the PEXA1 primer, whose sequence is indicated under the box. The start codon of bphA1 is underlined. Nucleotide numbers were assigned by using the transcriptional start site of P1 (position 1) as the reference point.
Sequence analysis of the 8.8-kb HindIII fragment.
The nucleotide sequence of the 8.8-kb HindIII fragment was determined. Eight open reading frames (ORFs) were found in the 8,801-bp HindIII fragment in addition to the carboxyl-terminal portion of bphB. The deduced amino acid sequences encoded by ORF 1 (1,315 to 6,108 bp) and ORF 3 (6,108 to 6,734 bp) showed significant identity with the sequences encoded by putative two-component regulatory genes, bpdS (55%) and bpdT (63%), respectively, of a rhodococcal PCB degrader, Rhodococcus sp. strain M5 (15). ORF 5 (8,296 to 6,761 bp) showed identity with benzoate coenzyme A ligase of Thauera aromatica (21%). The remaining ORFs overlapped these ORFs, and the sequences which they encoded exhibited no homology with known proteins. Thus, ORF 1 and ORF 3 were designated bphS and bphT, respectively. The bphS gene, which starts 627 bp downstream from the bphB stop codon, was 4,794 bp long, and its TGA termination codon overlapped the ATG start codon of bphT, which was 627 bp long. The bphS and bphT genes encoded polypeptides containing 1,598 and 209 amino acids, whose molecular masses were calculated to be 173.5 and 22.9 kDa, respectively.
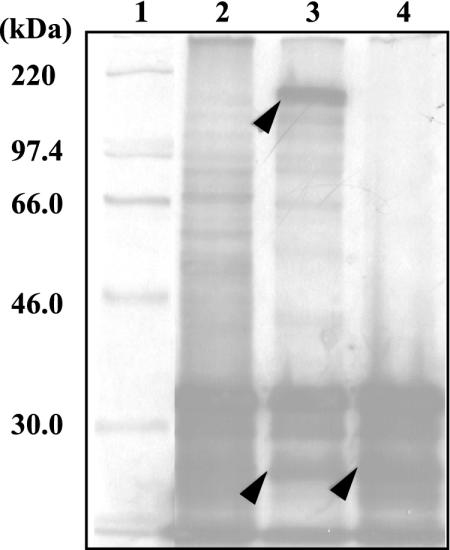
To identify the gene products, the whole-cell lysates of E. coli cells containing pBAB62 and pBAB14, which had inserts of the bphST and bphT genes in the vector pBSL, respectively, were prepared and subjected to SDS-PAGE as described in Materials and Methods. No induced proteins corresponding to the bphS and bphT gene products were observed. We also examined expression of the bphS and bphT genes under control of the T7 promoter of the pET21 vector in E. coli BL21, but the results were negative. We then employed an in vitro transcription-translation assay system. Plasmids pBAB62 and pBAB14 were used as templates for the E. coli S30 extract system (Fig. 5). Two polypeptide bands, at 175 and 25 kDa, were observed with pBAB62, and these molecular masses were in good agreement with those deduced from amino acid sequences of BphS (173.5 kDa) and BphT (22.9 kDa). The 25-kDa polypeptide band was also observed with pBAB14.
FIG. 5.
In vitro expression of the bphS and bphT genes of Rhodococcus sp. strain RHA1. The bphST and bphT genes in pBAB62 and pBAB14, respectively, were expressed in the E. coli S30 extract system containing [35S]methionine. Labeled polypeptides were separated by SDS-PAGE. The expected bphS and bphT products are indicated by arrowheads. Lane 1, 14C-labeled molecular weight marker (Pharmacia); lanes 2, 3, and 4, gene products of pBSL (vector control), pBAB62, and pBAB14, respectively.
Deletion analysis of the 8.8-kb HindIII fragment.
To investigate the functional involvement of the products of bphST in the activation of PbphA1, various subclones of the 8.8-kb HindIII fragment shown in Fig. 6 were constructed by inserting each fragment into the KpnI site of pKLAF1. The resultant plasmids were transformed into IAM1399 by electroporation, and the luciferase activity of each transformant was measured in the presence and absence of biphenyl (Fig. 6). In addition to pKA851 containing the entire 8.8-kb HindIII fragment, pKLAST1 containing just the bphS and bphT genes showed basal activation and biphenyl-induced activation of PbphA1 compared with the reporter plasmid, pKLAF1. Clones not containing the whole bphT gene (pKAS1, pKAD1, pKAD2, pKAD3, and pKAD4) showed no activation of PbphA1 even in the presence of biphenyl. One of these clones, pKAS1, contained the intact bphS gene, suggesting that the bphS gene product has no direct effect on PbphA1. Clones containing only the entire bphT gene (pKAD5, pKAD6, and pKAD10) showed basal activation of PbphA1. The clones containing a frameshift mutation of bphS and an intact bphT gene (pKAD11 and pKAD12) also exhibited basal activation of PbphA1. The basal activation of PbphA1 observed with pKAD5 or pKAD10 was elevated by introduction of pFJS1 carrying an intact bphS gene. These results suggest that bphT has fundamental PbphA1 activation activity, which is elevated by bphS, and that both bphS and bphT are essential for the induced activation by biphenyl. Thus, BphS and BphT seem to be a sensor kinase and a response regulator, respectively, of a two-component regulatory system governing the induced transcription activation of PbphA1.
Constitutive and induced transcription of bphS.
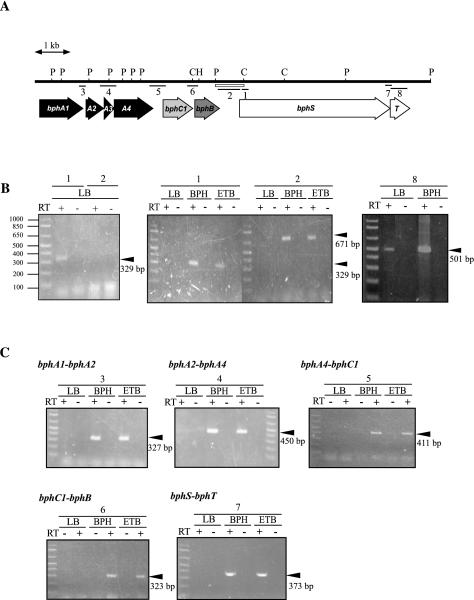
To promote inducible activation of degradation genes, the bphST genes need to be transcribed constitutively in the absence of an inducer. The total RNA from RHA1 cells grown on LB medium was subjected to RT-PCR analysis by using the primer set designed to amplify the internal segment of bphS. No amplification product was observed after RT-PCR performed under the usual conditions (Fig. 7B, center panel). An amplification product of the expected size (329 bp) was obtained when RT-PCR was performed under the intensive conditions (Fig. 7B, left panel), which generated more PCR product and provided higher sensitivity than the usual conditions. The primer set designed to amplify the internal segment of bphT also gave an amplification product corresponding to the expected size (501 bp) under the intensive conditions (Fig. 7B, right panel). The primer set designed to amplify the intergenic segment between bphB and bphS (671 bp) gave no product even under the intensive conditions (Fig. 7B, left panel). Then the promoter activity of the adjacent region upstream of bphS was examined. The PstI-ClaI fragment containing the region upstream of bphS (Fig. 7A) was inserted into the SalI site of pKLA1 to form pKLASF, which was introduced into RHA1. Compared with the activity observed with cells containing pKLA1, about 15-fold-greater luciferase activity was observed in RHA1 cells containing pKLASF even in the absence of biphenyl and ethylbenzene. When the fragment upstream of bphS in pKLASF was reversed to create pKLASR, no increase in luciferase activity was observed in RHA1 cells carrying pKLASR. These results indicate that the bphST genes are transcribed constitutively from a promoter in the adjacent region upstream of bphS in RHA1.
FIG. 7.
Transcription of the bphS gene in RHA1. (A) Physical map of the region from bphA1 to bphT. The open box below the map represents the fragment used to construct reporter plasmids pKLASF and pKLASR. The lines below the map indicate intergenic or internal segments of bph genes that were expected to be amplified by RT-PCR. The numbers below the lines indicate the PCR primer sets shown in Table 2. Restriction enzyme site abbreviations: P, PstI; C, ClaI; H, HindIII. (B) bphS and bphT gene transcripts in RHA1. Total RNAs from RHA1 cells grown on LB medium, on biphenyl (BPH), or on ethylbenzene (ETB) were reverse transcribed under the intensive conditions (left and right panels) or the usual conditions (center panel). The reverse transcripts were subjected to PCR amplification by using the primer sets indicated by the numbers above the panels, which correspond to the numbers in panel A and Table 2. RNA samples were concurrently analyzed in PCR mixtures with (+) and without (−) reverse transcriptase (RT) to verify the absence of total DNA. The position and size of each PCR product are indicated by an arrowhead on the right. (C) Intergenic RT-PCR products of the bphA1A2A3A4C1BST genes in RHA1. Total RNAs from RHA1 were reverse transcribed under the usual conditions, amplified, and analyzed as described above. An arrowhead indicates the position and size of each PCR product.
When RT-PCR analysis of the internal region of bphS was performed with RNA from the cells grown on biphenyl or ethylbenzene, an intense PCR product was observed; in contrast, the cells grown in LB medium gave no product (Fig. 7B, center panel). In this analysis, the primers for an intergenic segment between bphB and bphS also gave an intense product. These results suggest that transcription occurs from another upstream promoter, which is thought to be located within or upstream from bphB. As no terminator sequence was identified by nucleotide sequence analysis between bphB and bphS, it is possible to deduce that there was induced transcription of bphS from PbphA1. Thus, transcription of genes from bphA1 to bphT was examined. The total RNAs prepared from RHA1 cells were subjected to RT-PCR analysis by using the primer sets expected to amplify intergenic segments 2 to 7, as shown in Fig. 7A. All the primer sets gave the corresponding amplification products of the expected sizes with the RNA prepared from the cells grown on biphenyl or ethylbenzene (Fig. 7C). None of the amplification products were detected with RNA from the cells grown in LB medium, indicating that transcription of all the bphA1A2A3A4C1BST genes was induced simultaneously by biphenyl and ethylbenzene.
Disruption of the bphS gene in RHA1.
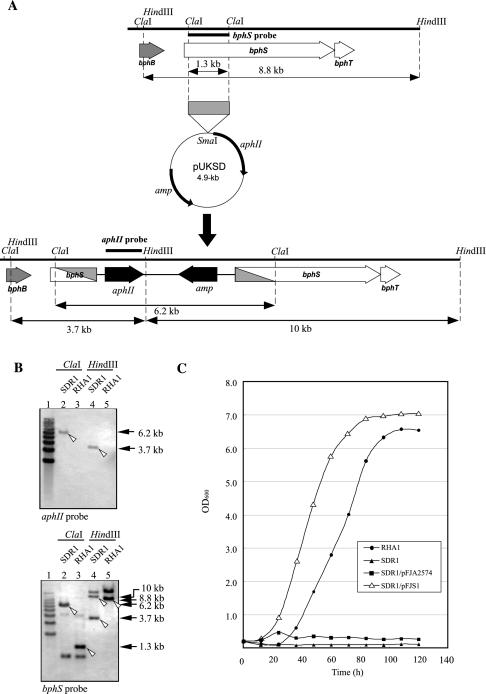
To determine whether the bphS gene is really responsible for biphenyl catabolism in RHA1, the bphS gene was inactivated with a disruption plasmid, pUKSD, by homologous recombination, as shown in Fig. 8A. Southern hybridization analysis was performed with aphII and bphS probes to confirm the expected arrangement of disrupted bphS sequences (Fig. 8B). In a kanamycin-resistant transformant, SDR1, both the aphII and bphS probes hybridized to a 6.2-kb ClaI fragment, which was 4.9 kb larger than the 1.3-kb RHA1 bphS fragment. The aphII probe hybridized to a 3.7-kb HindIII fragment containing aphII. The bphS probe hybridized to 3.7- and 10-kb HindIII fragments of SDR1, as pUKSD contains a HindIII site. The sum of the sizes of the hybridized HindIII fragments was 13.7 kb, and this value was 4.9 kb larger than the size of the RHA1 bphS fragment (8.8 kb). These results indicate that the entire 4.9-kb pUKSD segment was integrated into the bphS gene, as expected. Apart from the expected signals for bphS fragments, we found additional signals which suggested the existence of a bphS homologue in RHA1. SDR1, however, did not grow on biphenyl as a sole carbon source, indicating that the bphS gene alone is essential for growth of RHA1 on biphenyl (Fig. 8C).
FIG. 8.
bphS gene disruption in RHA1. (A) Strategy for bphS gene disruption. Disruption was accomplished by a single crossover between the native bphS gene (top) and the 1.3-kb internal fragment of bphS in pUKSD (middle). The possible region of recombination in the disruption derivative (bottom) is shaded diagonally. The positions of the bphS and aphII probes used in Southern hybridization are indicated by thick lines above bphS (top) and aphII (bottom). The sizes of ClaI and HindIII fragments containing the bphS sequence are indicated below the ORF maps (top and bottom). (B) Southern hybridization analysis of bphS disruption mutant strain SDR1 performed with the aphII (upper panel) and bphS (lower panel) probes. Lane 1, 1-kb ladder marker; lane 2, SDR1 total DNA digested with ClaI; lane 3, RHA1 total DNA digested with ClaI; lane 4, SDR1 total DNA digested with HindIII; lane 5, RHA1 total DNA digested with HindIII. The position and size of each signal derived from the bona fide bphS sequence are indicated by an open arrowhead and an arrow on the right, respectively. (C) Growth on biphenyl of SDR1 and bphS gene-containing SDR1. RHA1, SDR1, SDR1 carrying the vector pFAJ2574, and SDR1 carrying the bphS plasmid pFJS1 were grown in W minimal medium containing 0.2% biphenyl. Growth was measured by determining the optical density at 600 nm (OD600). The data are averages based on triplicate experiments.
To complement the bphS gene defect of SDR1, pFJS1 containing an intact bphS gene was constructed and introduced into SDR1. SDR1 carrying pFJS1 grew on biphenyl better than RHA1 grew on biphenyl (Fig. 8C). Thus, introduction of an intact bphS gene fully restored the growth of SDR1 on biphenyl. The excessive expression of bphS in a multicopy plasmid seemed to cause higher expression of bphA1A2A3C1B, resulting in better growth on biphenyl. These results suggest that the bphS gene alone is responsible for the growth of RHA1 on biphenyl.
DISCUSSION
We found in the present study that the bphS and bphT genes are likely to encode a two-component regulatory system responsible for transcriptional induction of the bphA1 promoter, PbphA1, by aromatic compounds, including biphenyl, based on the following results. (i) BphS and BphT exhibited identity and shared conserved residues with previously characterized sensor kinases and response regulators of two-component regulatory systems. (ii) bphS and bphT promoted transcriptional induction of PbphA1 in a host strain, R. erythropolis IAM1399. (iii) Both the bphS and bphT products were required for the transcriptional induction of PbphA1 in IAM1399. (iv) bphT alone was responsible for basal transcription of PbphA1 in IAM1399. (v) In the absence of bphT, bphS did not activate transcription of PbphA1 at all. (vi) In the presence of bphT, bphS was responsible for elevated basal transcription of PbphA1 and was required for the induced activation of PbphA1 transcription. In Pseudomonas sp. strain Y2, similar basal activation of a responsible promoter of styAB in the absence of an inducer by the two-component regulatory system encoded by stySR was reported previously (36). This activation may be due to both basal activation by the styR product and elevation by the styS product, as observed with RHA1 bphST.
Because these results were obtained with R. erythropolis IAM1399, they do not necessarily indicate that bphS and bphT are indispensably involved in the induction of PbphA1 by biphenyl in RHA1 and in the growth of RHA1 on biphenyl. Thus, we disrupted the bphS gene and complemented the bphS mutation. The results obtained in this study indicated that at least bphS is indispensably responsible for PbphA1 induction by biphenyl and the growth of RHA1 on biphenyl. A Southern hybridization experiment to confirm disruption of the bphS gene indicated that there is a bphS homolog in RHA1. This bphS homolog seems not to be involved in induction of the bphA1A2A3A4C1B genes by biphenyl, as bphS gene disruption was found to result in a stringent growth deficiency on biphenyl. Although all attempts to disrupt the bphT gene failed, the following results suggest that bphT plays at least a partial role in PbphA1 induction by biphenyl. (i) bphT was required for PbphA1 induction by biphenyl in IAM1399. (ii) bphT was expressed transcriptionally in RHA1. (iii) bphT was located next to bphS in a manner implying translational coupling and in the adjacent region downstream of bphA1A2A3A4C1B.
Several two-component regulatory systems involved in the degradation of aromatic compounds have been described previously. These systems include BpdST of PCB-degrading Rhodococcus sp. strain M5 (15), TodST of the toluene degraders Pseudomonas putida F1 (16) and P. putida DOT-T1 (22), TutCB and TutC1B1 of the anaerobic toluene degradation pathway of T. aromatica T1 (4), StySR of the stylene degradation pathway of Pseudomonas sp. strain Y2 (36) and Pseudomonas fluorescens ST (17), TdiSR of the anaerobic toluene degradation pathway of Azoarcus sp. strain T (1) and T. aromatica K172 (18), and TmoST of a toluene degrader, Pseudomonas mendocina KR1 (27). Except for BpdST of M5, these two-component systems are responsible for the degradation of monocyclic aromatic compounds in gram-negative bacteria. With a focus on the transcriptional regulatory systems for biphenyl and PCB degradation pathways, several systems have been described. The systems in gram-negative bacteria, including R. eutropha A5 (23), Pseudomonas sp. strain KKS102 (24), P. pseudoalcaligenes KF707 (37), and Burkholderia sp. strain LB400 (3), belong to the GntR family. In contrast, the systems in gram-positive bacteria, including strain RHA1 and Rhodococcus sp. strain M5, are two-component systems. These results suggest that there was independent evolution of regulatory genes in gram-positive and gram-negative bacteria.
Involvement of a two-component regulatory system in induction of the biphenyl degradation pathway of a rhodococcal PCB degrader has been suggested previously for Rhodococcus sp. strain M5, as determined by nucleotide sequence analysis and bpdS gene disruption (15). In this study, we found the two-component regulatory system encoded by RHA1 bphST not only by nucleotide sequence analysis but also by deletion and frameshift mutation analysis with a reporter assay in IAM1399. The involvement of bphS in growth on biphenyl was confirmed by a complementation experiment, which excluded the polar effect of gene disruption. Thus, this study illustrated the functional roles and induction characteristics of bphST gene products and the exact involvement of bphS in growth on biphenyl.
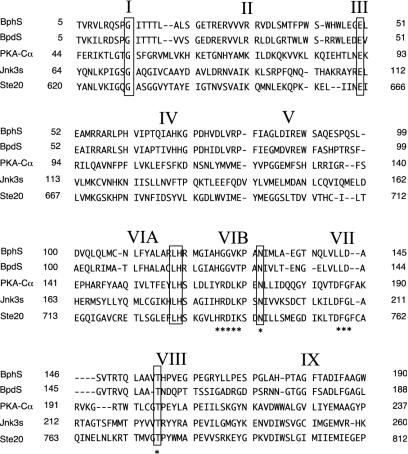
The amino terminus of BphS was found to contain a domain that is similar to the serine/threonine kinases (Fig. 9), as well as BpdS (15). We compared the BphS and BpdS amino-terminal domains with PKA-Cα, which is the best-characterized serine/threonine kinase to date (8). BphS has most of the key residues that are highly conserved in serine/threonine kinases. T156 seems to be the counterpart of PKA-Cα T196, which was found to be the phosphorylation site. BphS lacks counterparts of DFG in subdomain VII and RD in the consensus sequence H/YRDLKXXN in subdomain VIB. Because the D residue in the consensus sequence H/YRDLKXXN was estimated to be a catalytic base, which is essential for the catalytic reaction (8), the amino-terminal domain of BphS may not function as a serine/threonine kinase. The carboxyl-terminal region of BphS shares residues with the HPK7 subfamily (7) of sensor kinase proteins such as DegS, UhpB, and VsrA, as pointed out for BpdS (15). It has the conserved residues in the H-box, N-box, D-box, and G-box of histidine kinases, as proposed by Stock et al. (34, 35). H1411 in the H-box and the G-box from residue G1563 to residue V1594 are thought to be an autophosphorylation site and a nucleotide-binding site, respectively.
FIG. 9.
Comparative features of the amino terminus of BphS. The boxes indicate invariant residues in sequences. The amino terminus of BphS is aligned with those of protein kinases, including human cAMP-dependent protein kinase catalytic subunit PKA-Cα (National Center for Biotechnology Information protein database accession number P17621), Saccharomyces cerevisiae Ste20 (AAA35039), human mitogen-activated protein kinase JNK3 (Jnk3s) (U76020), and BpdS (AAB52543). The roman numerals indicate the positions of conserved subdomains in eukaryotic serine/threonine kinases (8). The asterisks indicate the residues mentioned in the text.
BphT shares residues with the response regulator proteins of two-component regulatory systems, such as DegU, UhpA, and VsrD, as well as BpdT (15). Among the residues that are conserved, D8, D9, and D54 appear to correspond to the residues which have been proposed to constitute an acid pocket for phosphorylation (26). D54 may serve as the main phosphorylation site, and K104 is a seemingly invariant residue that has been proposed to play a key role in the response activity (26).
In the presence of the BphST regulatory system in heterologous host strain IAM1399, PbphA1 was induced by a variety of aromatic compounds, including biphenyl, ethylbenzene, benzene, toluene, xylenes, cumene, p-cymene, and chlorinated benzenes, suggesting that the BphST regulatory system has a significantly broad spectrum of inducers. The inducer spectrum of the todX promoter in P. putida DOT-T1 was described without characterization of the responsible regulatory system (22). Using a heterologous host strain, we examined the exact BphST-dependent induction, which is expected to reflect the exact features of the BphST regulatory system. However, it seems to be impossible to compare the inducing activities of substrate compounds because the solubility in the medium and the permeability of inducers through the cell membrane are estimated to be different. PbphA1 was not induced by succinate, benzoate, and p-chlorobiphenyl. The inability of p-chlorobiphenyl to induce PbphA1 agrees with the requirement of biphenyl for cometabolic PCB degradation as an inducer of PbphA1.
RT-PCR analysis of the bphA1A2A3A4C1B and bphST genes indicated that the induced transcription by biphenyl continues from gene to gene. These results did not exclude transcription termination within a gene. Our results imply the operonic structure of bphA1A2A3A4C1BST but do not exclude the possibility that some unidentified promoter(s) other than PbphA1, which is located in the region from bphA1 to bphB, is also responsible for the induction of bphST transcription by biphenyl. Like expression in RHA1, it has been suggested that expression of the bpdC1C2BADE and bpdST genes in Rhodococcus sp. strain M5 is induced by biphenyl (15). In the presence of biphenyl transcription seems to terminate between bpdE and bpdS in M5, suggesting that there is an inducible promoter for bpdS in the adjacent upstream region. We found that in RHA1 there is a constitutive promoter in the adjacent upstream region of bphS (PbphS), which seems to be responsible for the transcription of bphS in the absence of biphenyl. In the presence of biphenyl, RT-PCR analysis of the region between bphB and bphS suggested that the induced transcription of bphB extends to bphS to a considerable extent. Thus, an inducible promoter for bphS is thought to reside in the distant upstream region of bphS. Based on the results of the RT-PCR analysis of the intergenic regions of bphA1 to bphS, PbphA1 seems to be a good candidate for this inducible promoter for bphS. These results led us to propose the following probable scheme of induction by bphST. In the absence of biphenyl, bphST genes are constitutively transcribed from the adjacent PbphS promoter at the basal level. In the presence of biphenyl, biphenyl activates the bphS product (BphS), which then activates the bphT product (BphT) by phosphorylation. The activated BphT promotes transcription initiation from PbphA1 and induces expression of the bphA1A2A3A4C1B and bphST genes.
Acknowledgments
We thank René De Mot for the kind gift of plasmid pFAJ2574.
This study was supported in part by the Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) in Japan.
REFERENCES
- 1.Achong, G. R., A. M. Rodriguez, and A. M. Spormann. 2001. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J. Bacteriol. 183:6763-6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, D., R. Masse, and M. Sylvestre. 1990. Cloning and expression of genes involved in 4-chlorobiphenyl transformation by Pseudomonas testosteroni: homology to polychlorobiphenyl-degrading genes in other bacteria. Gene 86:53-61. [DOI] [PubMed] [Google Scholar]
- 3.Beltrametti, F., D. Reniero, S. Backhaus, and B. Hofer. 2001. Analysis of transcription of the bph locus of Burkholderia sp. strain LB400 and evidence that the ORF0 gene product acts as a regulator of the bphA1 promoter. Microbiology 147:2169-2182. [DOI] [PubMed] [Google Scholar]
- 4.Coschigano, P. W., and L. Y. Young. 1997. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl. Environ. Microbiol. 63:652-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Mot, R., I. Nagy, A. De Schrijver, P. Pattanapipitpaisal, G. Schoofs, and J. Vanderleyden. 1997. Structural analysis of the 6 kb cryptic plasmid pFAJ2600 from Rhodococcus erythropolis NI86/21 and construction of Escherichia coli-Rhodococcus shuttle vectors. Microbiology 143:3137-3147. [DOI] [PubMed] [Google Scholar]
- 6.Furukawa, K., and A. M. Chakrabarty. 1982. Involvement of plasmids in total degradation of chlorinated biphenyls. Appl. Environ. Microbiol. 44:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grebe, T. W., and J. B. Stock. 1999. The histidine protein kinase superfamily. Adv. Microb. Physiol. 41:139-227. [DOI] [PubMed] [Google Scholar]
- 8.Hanks, S. K., and T. Hunter. 1995. Protein kinases. 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576-596. [PubMed] [Google Scholar]
- 9.Hashimoto, Y., M. Nishiyama, F. Yu, I. Watanabe, S. Horinouchi, and T. Beppu. 1992. Development of a host-vector system in a Rhodococcus strain and its use for expression of the cloned nitrile hydratase gene cluster. J. Gen. Microbiol. 138:1003-1010. [DOI] [PubMed] [Google Scholar]
- 10.Hauschild, J. E., E. Masai, K. Sugiyama, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1996. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl. Environ. Microbiol. 62:2940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofer, B., S. Backhaus, and K. N. Timmis. 1994. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene 144:9-16. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi, Y., Y. Yasukochi, Y. Nagata, M. Fukuda, and M. Takagi. 1994. Nucleotide sequence and functional analysis of the meta-cleavage pathway involved in biphenyl and polychlorinated biphenyl degradation in Pseudomonas sp. strain KKS102. J. Bacteriol. 176:4269-4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J. Bacteriol. 171:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa, W., A. Suzuki, T. Hoaki, E. Masai, and M. Fukuda. 2001. Multiplicity of aromatic ring hydroxylation dioxygenase genes in a strong PCB degrader, Rhodococcus sp. strain RHA1, demonstrated by denaturing gradient gel electrophoresis. Biosci. Biotechnol. Biochem. 65:1907-1911. [DOI] [PubMed] [Google Scholar]
- 15.Labbé, D., J. Garnon, and P. C. Lau. 1997. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J. Bacteriol. 179:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau, P. C., Y. Wang, A. Patel, D. Labbé, H. Bergeron, R. Brousseau, Y. Konishi, and M. Rawlings. 1997. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc. Natl. Acad. Sci. 94:1453-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leoni, L., P. Ascenzi, A. Bocedi, G. Rampioni, L. Castellini, and E. Zennaro. 2003. Styrene-catabolism regulation in Pseudomonas fluorescens ST: phosphorylation of StyR induces dimerization and cooperative DNA-binding. Biochem. Biophys. Res. Commun. 303:926-931. [DOI] [PubMed] [Google Scholar]
- 18.Leuthner, B., and J. Heider. 1998. A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol. Lett. 166:35-41. [DOI] [PubMed] [Google Scholar]
- 19.Masai, E., K. Sugiyama, N. Iwashita, S. Shimizu, J. E. Hauschild, T. Hatta, K. Kimbara, K. Yano, and M. Fukuda. 1997. The bphDEF meta-cleavage pathway genes involved in biphenyl/polychlorinated biphenyl degradation are located on a linear plasmid and separated from the initial bphACB genes in Rhodococcus sp. strain RHA1. Gene 187:141-149. [DOI] [PubMed] [Google Scholar]
- 20.Masai, E., A. Yamada, J. M. Healy, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mondello, F. J. 1989. Cloning and expression in Escherichia coli of Pseudomonas strain LB400 genes encoding polychlorinated biphenyl degradation. J. Bacteriol. 171:1725-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosqueda, G., M. I. Ramos-Gonzalez, and J. L. Ramos. 1999. Toluene metabolism by the solvent-tolerant Pseudomonas putida DOT-T1 strain, and its role in solvent impermeabilization. Gene 232:69-76. [DOI] [PubMed] [Google Scholar]
- 23.Mouz, S., C. Merlin, D. Springael, and A. Toussaint. 1999. A GntR-like negative regulator of the biphenyl degradation genes of the transposon Tn4371. Mol. Gen. Genet. 262:790-799. [DOI] [PubMed] [Google Scholar]
- 24.Ohtsubo, Y., M. Delawary, K. Kimbara, M. Takagi, A. Ohta, and Y. Nagata. 2001. BphS, a key transcriptional regulator of bph genes involved in polychlorinated biphenyl/biphenyl degradation in Pseudomonas sp. KKS102. J. Biol. Chem. 276:36146-36154. [DOI] [PubMed] [Google Scholar]
- 25.Olsson, O., C. Koncz, and A. A. Szalay. 1988. The use of the luxA gene of the bacterial luciferase operon as a reporter gene. Mol. Gen. Genet. 215:1-9. [DOI] [PubMed] [Google Scholar]
- 26.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 27.Ramos-Gonzalez, M. I., M. Olson, A. A. Gatenby, G. Mosqueda, M. Manzanera, M. J. Campos, S. Vichez, and J. L. Ramos. 2002. Cross-regulation between a novel two-component signal transduction system for catabolism of toluene in Pseudomonas mendocina and the TodST system from Pseudomonas putida. J. Bacteriol. 184:7062-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakai, M., E. Masai, H. Asami, K. Sugiyama, K. Kimbara, and M. Fukuda. 2002. Diversity of 2,3-dihydroxybiphenyl dioxygenase genes in a strong PCB degrader, Rhodococcus sp. RHA1. J. Biosci. Bioeng. 93:421-427. [DOI] [PubMed] [Google Scholar]
- 29.Sakai, M., K. Miyauchi, N. Kato, E. Masai, and M. Fukuda. 2003. 2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 69:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seto, M., K. Kimbara, M. Shimura, T. Hatta, M. Fukuda, and K. Yano. 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:3353-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seto, M., E. Masai, M. Ida, T. Hatta, K. Kimbara, M. Fukuda, and K. Yano. 1995. Multiple polychlorinated biphenyl transformation systems in the gram-positive bacterium Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 61:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimizu, S., H. Kobayashi, E. Masai, and M. Fukuda. 2001. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 67:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shingler, V., M. Bartilson, and T. Moore. 1993. Cloning and nucleotide sequence of the gene encoding the positive regulator (DmpR) of the phenol catabolic pathway encoded by pVI150 and identification of DmpR as a member of the NtrC family of transcriptional activators. J. Bacteriol. 175:1596-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 35.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Velasco, A., S. Alonso, J. L. Garcia, J. Perera, and E. Diaz. 1998. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J. Bacteriol. 180:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe, T., R. Inoue, N. Kimura, and K. Furukawa. 2000. Versatile transcription of biphenyl catabolic bph operon in Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 275:31016-31023. [DOI] [PubMed] [Google Scholar]
- 38.Yamada, A., H. Kishi, K. Sugiyama, T. Hatta, K. Nakamura, E. Masai, and M. Fukuda. 1998. Two nearly identical aromatic compound hydrolase genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 64:2006-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]