Abstract
A locus containing the clpP and clpL genes in the lactic acid bacterium Oenococcus oeni was studied. Real-time reverse transcription-PCR analysis revealed different induction factors involved in expression of these genes during stress. According to the conditions, clpP and clpL genes could be transcripted as two distinct transcripts or cotranscripted. The clpP promoter depended on the CtsR regulator, but surprisingly the clpL promoter did not. The amount of the clpL transcript depended on mRNA stability. This clp ATPase gene is at least controlled at the posttranscriptional level.
Stress response plays a key role in the cell adaptation of all organisms to environmental conditions. To survive environmental changes, cells synthesize proteins, including both chaperones and proteases, above all to prevent accumulation of abnormal proteins (20). These stress proteins are also involved in various cellular regulations during growth (28). Recent studies have focused on a large family of proteins, named Clp (for caseinolytic protein), which is well conserved in both eukaryotic and prokaryotic organisms. Many Clp proteins are ATPases and may play a part in determining the half-life of regulatory proteins (29, 32, 33, 41).
The Clp ATPase proteins are divided into two classes. Members of the first class, (ClpA, ClpB, ClpC, ClpD, ClpE, and ClpL), also known as HSP100 proteins, contain two ATP nucleotide-binding domains (NBDs). The second class includes smaller proteins containing only one NBD, such as ClpX (31). It is widely accepted that Clp ATPases can function either as molecular chaperones or as regulator components of the proteolytic complex (38). This complex consists of two types of subunit: the proteolytic ClpP subunit and the Clp ATPase subunit. When ClpP is associated with a Clp ATPase, it can degrade larger specific substrates (7, 11, 23, 39). In Bacillus subtilis or Lactococcus lactis, ClpP is required for growth at high temperature (8, 10). The clpP gene of B. subtilis and Listeria monocytogenes is induced after several stresses (10, 21, 35). Besides stress, ClpP and associated Clp ATPases have been linked to many cellular development processes in bacteria (9, 15, 19, 25, 27).
The majority of clp genes are heat inducible, and their regulation is often described as CtsR dependent in gram-positive bacteria. In B. subtilis, clpP and clpC genes are class III heat shock genes, whose transcription is repressed by CtsR. The CtsR repressor recognizes a directly repeated heptanucleotide operator (4). In many cases, a gene similar to ctsR and a CtsR operator located upstream from the clp genes have been found in several gram-positive bacteria (19, 34).
Oenococcus oeni, a gram-positive bacterium most often responsible for malolactic fermentation in wine, is tolerant to various stresses. O. oeni, which is introduced into starter cultures, is able to grow after alcoholic fermentation under acidic conditions and in the presence of high concentrations of ethanol. Among the O. oeni stress genes, the expression of hsp18, trxA, and clpX has been explored (16-18). These studies made it possible to characterize the pattern of expression of these genes during growth or under stress conditions. The clpX gene, which is preferentially expressed at the beginning of the exponential phase, is heat inducible, but its regulation remains unknown. However, it has been suggested that the long 5′-untranslated region could be involved in stability of mRNA (18). The present work focuses on an O. oeni locus containing the clpP and clpL genes. The expression of this clp locus was characterized during growth and following different stress conditions. Moreover, elements of transcriptional regulation of this locus were investigated.
The clpP gene is located on the same locus as the clpL gene.
Based on knowledge about the O. oeni I.O.B 8413 genome, thanks to a sequencing project involving our laboratory, the Genome Express S.A. (France) and the Œnological Institut of Bordeaux (France), the annotation step revealed the presence of a gene product that had 70 and 65% identities with ClpP of B. subtilis and L. lactis, respectively. This clpP gene (609 bp in length) encodes a protein of 202 amino acid residues with a calculated molecular mass of 21.5 kDa and a predicted pI of 6.88. A typical putative ribosome-binding site (RBS) is located upstream from the putative ATG codon at an appropriate distance. The residues Ser, His, and Asp (at positions 96, 121, and 172, respectively), which constitute the catalytic triad of the serine protease ClpP in Escherichia coli, are conserved in the ClpP sequence of O. oeni (24, 37).
No typical sequence of the Rho-independent transcriptional terminator sequence was found downstream from the clpP gene. Nevertheless, a putative open reading frame (ORF) gene (2,163 bp in length) encoding a 720-amino-acid protein was predicted 303 nucleotides downstream from the clpP gene. This ORF is preceded by a putative RBS, 8 bases upstream of the putative start codon. The encoded protein has a predicted molecular mass of 79 kDa and a predicted pI of 6.14. Blastprot analysis against the SwissProt databank revealed identities of 57% with the ClpL of Lactococcus lactis and 68% with the ClpL of Streptococcus pneumoniae. A likely Rho-independent transcription terminator stem-loop sequence (AAAAAATCCCTAAAAATTATTTTTGGGGATTTTTT; ΔG = −12.5 kcal·mol−1) is located downstream from the stop codon (TAA) of clpL. To our knowledge, O. oeni is the first microorganism discovered to have such a genetic organization between these two genes. In general, the organization tig-clpP-clpX is found in gram-negative bacteria (26). In the case of several gram-positive bacteria, the clpP gene is located in a chromosomal region with no clp ATPase gene nearby (6, 8, 10, 30). Until now, very little information about the physiological role of ClpL has been available.
Differential expression of the clpP and clpL genes was observed during the growth phase.
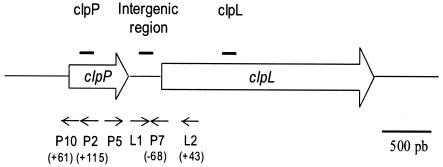
Clp proteins are known for being implicated in regulatory mechanisms during growth. The ClpP protease especially affects the stability of key regulators during growth phases (28). Thus, Northern blot analyses with specific intragenic probes for clpP and clpL (Table 1) were performed with total RNAs extracted from cells in different growth phases. Probes were radiolabeled by using a random-primer DNA labeling kit (Invitrogen), and their position was given in Fig. 1. The strain O. oeni IOB84.13 (2) was grown at 30°C in modified FT80 medium (pH 5.3) (1). Then RNAs were purified with Tri-reagent (Sigma) after disruption of the cells in a Fastprep cell disintegrator FP120 Instrument Savant (BIO 101) for 6 × 20 s at 6,000 × g.
TABLE 1.
Primers used in this study
| Name | Sequence (5′→3′) | Functiona |
|---|---|---|
| P5 | CGGTACCAAAGGCAAGCGTTTTAT | Probe P, QRT-PCR, RT-PCR |
| P6 | CTCTTCCGAGTCTTCAAAAGTTGAT | Probe P, QRT-PCR |
| L4 | GCGCGACAAGGCAAACTAGAT | Probe L, QRT-PCR |
| L5 | GCCGGAACATTTCCATTGAT | Probe L, QRT-PCR |
| L1 | AATTATTAAGAAGCCGTTTGACT | RT-PCR |
| P8 | GGCAAACTTAATTACTTCACA | Probe I |
| P9 | GGCCTTTATGTCAATTTCtAT | Probe I |
| L2 | CCCGAATTCGAATGGATCATTATTACTGTTATT | Primer extension with clpL, RT-PCR |
| P7 | TTTGAGTGGCTTGTGCGAGAGC | Primer extension with clpL, RT-PCR |
| Pi2 | CCATTTGATCTTCGACTTCTC | Primer extension with clpP |
| P10 | CGACTTGGAAGATCATAACT | Primer extension with clpP |
| Ldh1 | GCCGCAGTAAAGAACTTGATG | QRT-PCR |
| Ldh2 | TGCCGACAACACCAACTGTTT | QRT-PCR |
| 16S1L | TCGTAAAGCACTGTTGTAAGG | QRT-PCR |
| 16S2R | CCTATTGCGAGCCCTGTATGC | QRT-PCR |
| Olcg7 | GGGGAATTCCCGGAGGCCAGTTTAAG | pDLclpP construction |
| Olcg8 | GGGGGATCCCCTTGAACCAAGATAATTCGG | pDLclpP construction |
| ClpPL1 | CTACGGGAATTCCTTATTAAGAAGCCGTTTGACTTC | pDLclpL construction |
| ClpPL2 | CTACGGGGATCCGATTGAATCCTCCTATCAATCT | pDLclpL construction |
QRT-PCR, quantitative real-time RT-PCR.
FIG. 1.
Schematic diagram of the clpP-clpL locus and positions of probes (black lines) and primers (arrows) used in this study. Positions relative to ATG are indicated in parentheses. pb, base pairs.
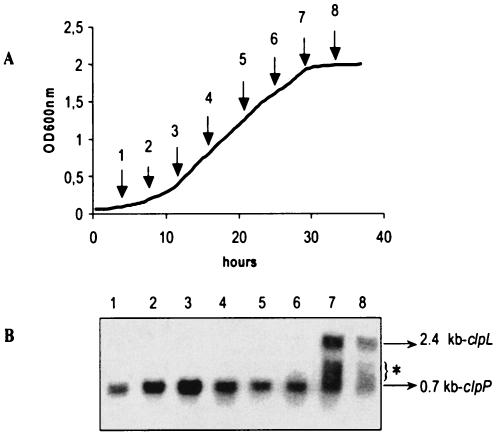
A 0.7-kb transcript was detected with the clpP probe, whereas the clpL probe allowed us to detect a larger transcript of 2.4 kb (data not shown). The clpP and clpL mRNAs had different sizes, and in this way, they could be detected by the same Northern blot analysis. Differences in the relative amounts of the clpP and the clpL transcript during growth are shown in Fig. 2. Contrary to clpP, the clpL transcript was very weakly detected during growth, and its level increased dramatically at entry into the stationary phase. Curiously, as found by Ingmer et al. (14) and Lemos and Burne (19) with clp ATPase probes, Northern blot analyses showed that with the clpL-specific probe, more than one transcript size was detected, contrary to the results obtained with the clpP-specific probe (Fig. 2, 3B, and 4C).
FIG. 2.
Expression of clpP and clpL as a function of growth phase. (A) Growth curve of O. oeni. Arrows indicate cells samples for RNA extraction. (B) Northern blot analysis of RNA samples 1 to 8 using the clpP- and clpL-specific probes P and L (at each of the eight sampling times indicated in panel A). In all cases, 20 μg of RNA was applied per lane. This experiment was carried out two times. Note that with longer time exposure, a 2.4-kb signal was weakly detected during the mid-exponential phase. The sizes of the transcripts detected are indicated, with 0.7 kb corresponding to clpP and 2.4 kb corresponding to clpL. An asterisk indicates transcript detected with the clpL probe, the size of which did not correlate with the entire clpL gene.
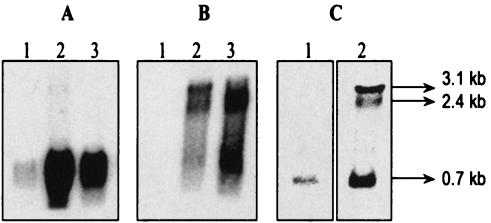
FIG. 3.
Expression of clpP and clpL genes under stress conditions. Total RNAs were isolated from O. oeni cells grown at 30°C (lane 1) and submitted to heat shock (30 min at 42°C) (lane 2) or ethanolic shock (30 min at 10% [vol/vol]) (lane 3). Northern blot analyses of RNAs were performed with clpP probe (A); clpL probe (B); or a mixture of clpP, clpL, and intergenic region-specific probes (C). The sizes of the transcripts detected are indicated. In all cases, 15 μg of RNA was applied per lane.
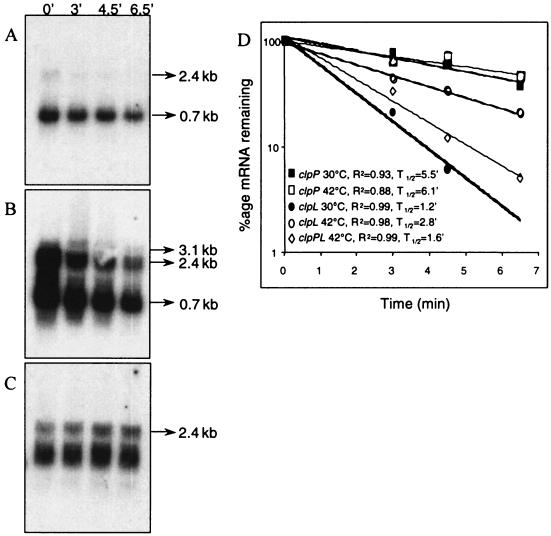
FIG. 4.
Northern blotting performed to determine the relative rates of degradation of transcripts produced from the clp locus. The decay was measured in the exponential phase (A) at an OD600 of 0.7, with a heat stress at 42°C during 30 min (B) at an OD600 of 0.7, and in the stationary phase (C) at an OD600 of 1.8. The RNAs were prepared after 0, 3, 4.5, or 6.5 min after rifampin addition (250 μg ml−1). Northern blot analyses of RNAs were performed with a mixture of the three clpP, clpL, and intergenic region-specific probes. In all cases, 15 μg of RNA was applied per lane. This experiment was carried out two times. (D) Semilogarithmic plot of (i) clpP mRNA decay at OD600 of 0.7 (solid squares) or after heat-shock (open squares), (ii) clpL mRNA decay at an OD600 of 0.7 (solid circles) or after heat shock (open circles), and (iii) cotranscript mRNA decay at an OD600 of 0.7 after heat shock (open diamonds). The correlation coefficients (R2) and half-life (T1/2) were determined for each regression analysis.
The clpP gene was expressed during all stages of growth at a high basal level, but its level reached its maximum in the exponential phase. The clpL transcript was highly detected in the stationary phase, as previously described for S. pneumoniae (30). Jobin et al. found a high level of clpX mRNA in the early log phase (18). The differential expression of clpX and clpL genes may reflect the possibility of different complexes between ClpP and Clp ATPase subfamilies.
A larger transcript was detected under stress conditions.
The transcriptional analyses of clpP and clpL were performed under two stress conditions for 30 min: heat at 42°C or the presence of 10% (vol/vol) ethanol. Northern blotting was performed with clpP and clpL probes. As shown in Fig. 3, the signals of the 0.7- and 2.4-kb transcripts, corresponding to clpP and clpL transcripts, respectively, were more intense after stress treatments. Thus, both genes are induced by stress. A stronger increase in clpP transcript level was obtained after heat stress compared to that obtained after ethanolic stress, whereas signal corresponding to clpL transcript was more intense after ethanolic stress. Moreover, under heat stress conditions, a larger transcript of 3.1 kb was detected with clpL and clpP probes (Fig. 3A and B). This transcript was better detected when a mix of specific probes of the intergenic region clpP-clpL, clpL, and clpP genes was used (Fig. 3C). The size of this transcript corroborates the hypothesis of the cotranscription of both genes clpP and clpL. A Northern blot analysis with probes of the regions located upstream from clpP and downstream from clpL and reverse transcription-PCR (RT-PCR) analysis with appropriate primers (Table 1) confirmed the presence of a cotranscript (data not shown). This finding differs from previous works on clpP gene expression in other gram-positive bacteria, which have shown that clpP transcription was monocistronic (8, 9, 30).
clpL and clpP transcripts were differentially induced under stress conditions.
In order to measure the stress induction factor of the clp genes, a quantitative RT-PCR experiment was set up. O. oeni cDNAs were synthesized by using the Superscript II RT-PCR system (Invitrogen) using random hexamers as recommended. Specific cDNAs were amplified by real-time PCR with appropriate primers (Table 1) using the PCR Master mix SYBR Green I (Invitrogen). The presence of intact cellular mRNAs and the uniform efficiency of each RT reaction have been checked. Amplifications were performed on a Bio-Rad I cycler. The specificity of real-time PCR for each primer pair was determined with a melting curve. The efficiency of real-time amplification is calculated by the formula E = [10(1/−s) − 1] × 100, where s is the slope of standard curve. The results were calculated by the comparative critical threshold (ΔΔCT) method, in which the amount of target RNA is adjusted to a reference relative to an internal calibrated target RNA. The constitutive ldhD gene was chosen as an internal control for these experiments. The intragenic region of ldhD, clpP, and clpL was amplified using Ldh1/Ldh2, P5/P6, and L4/L5 oligonucleotide pairs, respectively (Table 1). The comparison of the ΔCT values between ldhD in unstressed and stressed cells confirmed that the transcription of this gene is not influenced by heat shock or ethanolic shock. As expected, higher levels of clpP and clpL messengers were detected in stressed cells compared to unstressed cells. After several calculations, we concluded that the amounts of clpP and clpL transcripts increased by 16- and 38-fold, respectively, after 30 min at 42°C. After 30 min in the presence of ethanol (10% [vol/vol]), the induction factors were 4 for clpP and 53 for clpL. As observed by Northern blot analysis, the folds of induction depended on the kind of stress. The difference between clpP and clpL induction rates after stress implied differential stability of transcripts and/or the presence of an initiation transcriptional site upstream from the clpL gene.
Stability of transcripts modulate the expression level of the clpL gene.
To examine the possibility that environmental conditions affected the stability of clp transcripts and particularly clpL mRNA, the rate of these transcripts was investigated under three conditions: the mid-exponential phase (optical density at 600 nm [OD600] of 0.7), part of the mid-exponential-phase culture subjected to heat shock (30 min, 42°C), and the stationary phase (OD600 of 1.6). Total RNAs were extracted from cells treated with rifampin (final concentration of 250 μg· ml−1) to prevent the initiation of transcription. Northern blot analysis was performed with RNAs extracted at different times after rifampin addition (Fig. 4). To compare the amounts of the three transcripts directly, Northern blotting was performed with a mix of the three specific probes of clpP, clpL, and the intergenic region. To calculate half-lives, the amount of clp transcripts is adjusted to the amount of 16S rRNA transcript. A linear regression analysis of the data is given in Fig. 4D. As shown in Fig. 4C and D, under stress conditions, clpP transcript presented a half-life of approximately 6 min, whereas clpL transcript and clpPL cotranscript had half-lives of 3 and 1.5 min, respectively. On the other hand, the half-lives of clpP mRNA were approximately the same in the exponential phase and after heat shock. The increase in the amount of clpP mRNA under heat shock conditions was weakly influenced by the rate of mRNA decay and strongly influenced by RNA synthesis. In contrast, the increase in the amount of clpL under heat shock conditions and especially in the stationary phase appeared to be affected more by the rate of mRNA decay (Fig. 4C and D). Indeed, clpL transcript seemed very stable during the stationary phase (Fig. 4C) compared to the exponential phase (Fig. 4A). However, the Northern blot analysis was not sensitive enough to determine precisely the half-life of this transcript during the exponential phase. Thus, the half-life of clpL transcript was calculated thanks to real-time PCR. For this experiment, the 16S rRNA transcript was chosen as an internal control. The results confirmed that the half-lives of clpL transcript were about 1 min in the exponential phase, 3 min under heat shock, and at least 10 min in the stationary phase. Thus, the stability of the clpL mRNA acts as a form of posttranscriptional regulation.
The clpP promoter depended on CtsR but the clpL promoter did not.
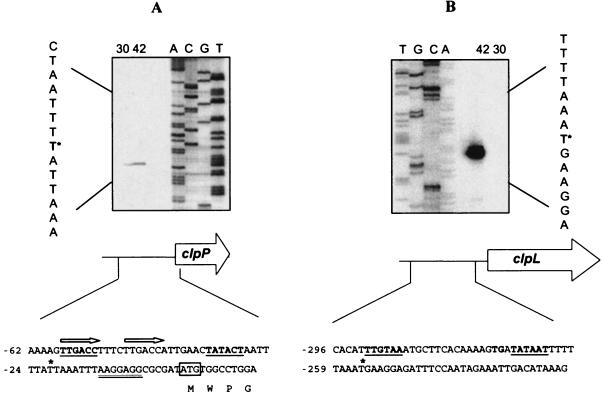
In order to characterize the promoter region of clpP and to highlight potential transcription initiation in the intergenic region, primer extension analyses were carried out with specific primers for clpP or clpL (Table 1). Primer extensions were performed from RNA extracted from an exponential culture either before or after heat shock treatment. Total RNAs (5 μg) were mixed with 2 pmol of each primer (Table 1). RT was performed with 1 U of Superscript II RNase H− reverse transcriptase (Invitrogen) as recommended by the manufacturer in the presence of [α-32P]dATP (Perkin Elmer). The 5′-end mRNA of the clpL gene was located 256 bp upstream from the putative start codon (Fig. 5B). This signal was detected only from RNA extracted from heat-shocked cells. Two hexanucleotides (TTGTAA and TATAAT) separated by 17 nucleotides presented high similarities to the −35 and −10 sequences of several promoters of O. oeni (13). The −10 sequence could be extended, since it is preceded by TGN (36). The transcription of clpL could be initiated from this site, supporting the hypothesis of an independent transcription of clpL gene. The long untranslated sequence at the 5′ end of clpL mRNA is not uncommon in other bacteria, but its role remains obscure. Nevertheless, this sequence could be involved in stability as described previously (5, 18, 22).
FIG. 5.
Determination of the 5′ end of clp transcripts. The RT reaction was performed with 5 μg of total RNA isolated from O. oeni grown at 30°C or heat shocked for 1 h at 42°C. Lanes A, C, G, and T show the dideoxy sequencing ladder obtained with the same primer as that used for the reverse transcriptase reaction. The asterisk indicates the base representing the 5′ end of clpP mRNA or clpL mRNA in the sequence shown to the left. The RBS is double underlined. The putative −10 and −35 sequences are underlined and in boldface. An asterisk indicates the transcription start site. The ATG start codon was boxed. Arrows indicate the probable operator sites for CtsR. Positions relative to the translation initiation site are indicated to the left. (A) Determination of the 5′ end of the clpP transcript with oligonucleotide Pi2. (B) Determination of the 5′ end of the clpL transcript with oligonucleotide L2.
A 5′-end mRNA was located 22 bp upstream from the putative start codon of clpP (Fig. 5A). Promoter sequence with significant similarity to the housekeeping promoter recognition sequence was found upstream from this transcription signal. Analysis of this promoter region revealed a directly tandem heptanucleotide sequence (5′-TTTGACCTTTCTTGACC-3′) overlapping the potential −35 sequence and showing high similarities to the operator sequence recognized by the repressor ctsR (4). Analysis of the genome sequence of O. oeni has shown the presence of a gene product having 52% identity to the CtsR of B. subtilis. These findings led us to think about the possibility of CtsR-dependent regulation.
However, no CtsR consensus sequence was found in the intergenic region clpP-clpL. Differences observed in profile expression between clpP and clpL could be explained by several mechanisms of regulation. To investigate the CtsR-dependent regulation of clpP suggested by the CtsR consensus sequence in the promoter region, transcriptional fusions were built in pDL vector (40). Promoter regions of clpP and clpL amplified by PCR with appropriate primers (Table 1) were cloned upstream of the bgaB gene encoding the Bacillus stearothermophilus β-galactosidase in pDL vector. Recombinant vectors were transferred in B. subtilis strain 168 and B. subtilis strain QB 4991, which had the ctsR gene deleted (3), and β-galactosidase activity was measured as described by Grandvalet et al. (12). A high level of activity (8,000 Miller units · mg−1) was measured for clpL′-bgaB, which confirmed the functionality of this promoter. The clpP′-bgaB fusion was expressed at a low level in the wild-type strain (74 Miller units · mg−1). In the ΔctsR genetic background, the level of clpL′-bgaB expression remained the same (7,300 Miller units · mg−1), whereas clpP′-bgaB fusion increased 200-fold (15,800 Miller units · mg−1). These results clearly indicated that CtsR of B. subtilis negatively regulated the clpP gene, whereas transcription from clpL promoter was not dependent on CtsR. It is interesting to note that if CtsR regulates the clpP gene in O. oeni, its repression was only partial, since a high basal expression of clpP was detected by Northern blot analysis. The CtsR-dependent repression in O. oeni could be modulated, and in this case, it is presumably due to the preponderant role of ClpP during the growth phase.
Taken together, these results give evidence for the presence of multiple levels of control of the clpP-clpL locus in O. oeni. We would like to point out the fact that findings strongly suggest the existence of a clpL promoter. (i) Analysis by primer extension allowed us to identify a potential transcription start site upstream from clpL. (ii) A transcription event involving the clpL gene was demonstrated in B. subtilis. (iii) The clpL level highly increased in the stationary phase, while clpP transcript decreased in this phase. However, this promoter did not appear to be dependent on CtsR. Moreover, neither an alternative sigma factor nor the hrcA gene has been identified in the O. oeni genome so far. Thus, future work will be focused on identification of the mechanism involved in clpL gene regulation.
Nucleotide sequence accession number.
The nucleotide sequence has been submitted to the EMBL nucleotide sequence database under accession no. AJ606044.
Acknowledgments
We thank D. Garmyn for constant interest throughout the work and for helpful discussions. We thank T. Msadek for providing B. subtilis strains and the pDL vector.
This study was supported by the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Université de Bourgogne, the Institut National de la Recherche Agronomique, and the Conseil Régional de Bourgogne.
REFERENCES
- 1.Cavin, J. F., H. Prevost, J. Lin, P. Schmitt, and C. Divies. 1989. Medium for screening Leuconostoc oenos strains defective in malolactic fermentation. Appl. Environ. Microbiol. 55:751-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavin, J. F., P. Schmitt, A. Arias, J. Lin, and C. Divies. 1988. Plasmid profiles in Leuconostoc species. Microbiol. Aliment. Nutr. 5:55-62. [Google Scholar]
- 3.Derre, I., G. Rapoport, K. Devine, M. Rose, and T. Msadek. 1999. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol. Microbiol. 32:581-593. [DOI] [PubMed] [Google Scholar]
- 4.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 5.Emory, S. A., and J. G. Belasco. 1990. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 172:4472-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fedhila, S., T. Msadek, P. Nel, and D. Lereclus. 2002. Distinct clpP genes control specific adaptive responses in Bacillus thuringiensis. J. Bacteriol. 184:5554-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, E. G., D. G. Ennis, M. Gonzalez, A. S. Levine, and R. Woodgate. 1996. Regulation of SOS mutagenesis by proteolysis. Proc. Natl. Acad. Sci. USA 93:10291-10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frees, D., and H. Ingmer. 1999. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol. Microbiol. 31:79-87. [DOI] [PubMed] [Google Scholar]
- 9.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 10.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandvalet, C., M. Gominet, and D. Lereclus. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805-1813. [DOI] [PubMed] [Google Scholar]
- 13.Guzzo, J., M. P. Jobin, F. Delmas, L. C. Fortier, D. Garmyn, R. Tourdot-Marechal, L. Byong, and C. Divies. 2000. Regulation of stress response in Oenococcus oeni as a function of environmental changes and growth phase. Int. J. Food Microbiol. 55:27-31. [DOI] [PubMed] [Google Scholar]
- 14.Ingmer, H., F. K. Vogensen, K. Hammer, and M. Kilstrup. 1999. Disruption and analysis of the clpB, clpC, and clpE genes in Lactococcus lactis: ClpE, a new Clp family in gram-positive bacteria. J. Bacteriol. 181:2075-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jobin, M.-P., F. Delmas, D. Garmyn, C. Diviès, and J. Guzzo. 1997. Molecular characterization of the gene encoding an 18-kilodalton small heat shock protein associated with the membrane of Leuconostoc oenos. Appl. Environ. Microbiol. 63:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jobin, M. P., D. Garmyn, C. Divies, and J. Guzzo. 1999. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145:1245-1251. [DOI] [PubMed] [Google Scholar]
- 18.Jobin, M.-P., D. Garmyn, C. Diviès, and J. Guzzo. 1999. The Oenococcus oeni clpX homologue is a heat shock gene preferentially expressed in exponential growth phase. J. Bacteriol. 181:6634-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemos, J. A. C., and R. A. Burne. 2002. Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184:6357-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindquist, S., and E. A. Craig. 1988. The heat-shock proteins. Annu. Rev. Genet. 22:631-677. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lundberg, U., A. von Gabain, and O. Melefors. 1990. Cleavages in the 5′ region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J. 9:2731-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makovets, S., A. J. Titheradge, and N. E. Murray. 1998. ClpX and ClpP are essential for the efficient acquisition of genes specifying type IA and IB restriction systems. Mol. Microbiol. 28:25-35. [DOI] [PubMed] [Google Scholar]
- 24.Maurizi, M. R., W. P. Clark, S. H. Kim, and S. Gottesman. 1990. ClpP represents a unique family of serine proteases. J. Biol. Chem. 265:12546-12552. [PubMed] [Google Scholar]
- 25.Msadek, T., V. Dartois, F. Kunst, M. L. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 26.Østerås, M., A. Stotz, S. Schmid Nuoffer, and U. Jenal. 1999. Identification and transcriptional control of the genes encoding the Caulobacter crescentus ClpXP protease. J. Bacteriol. 181:3039-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 28.Porankiewicz, J., J. Wang, and A. K. Clarke. 1999. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol. Microbiol. 32:449-458. [DOI] [PubMed] [Google Scholar]
- 29.Pummi, T., S. Leskelä, E. Wahlström, U. Gerth, H. Tjalsma, M. Hecker, M. Sarvas, and V. P. Kontinen. 2002. ClpXP protease regulates the signal peptide cleavage of secretory preproteins in Bacillus subtilis with a mechanism distinct from that of the Ecs ABC transporter. J. Bacteriol. 184:1010-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson, G. T., W.-L. Ng, J. Foley, R. Gilmour, and M. E. Winkler. 2002. Global transcriptional analysis of clpP mutations of type 2 Streptococcus pneumoniae and their effects on physiology and virulence. J. Bacteriol. 184:3508-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schirmer, E. C., J. R. Glover, M. A. Singer, and S. Lindquist. 1996. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 21:289-296. [PubMed] [Google Scholar]
- 32.Schweder, T., K.-H. Lee, O. Lomovskaya, and A. Matin. 1996. Regulation of Escherichia coli starvation sigma factor (σs) by ClpXP protease. J. Bacteriol. 178:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turgay, K., J. Hahn, J. Burghoorn, and D. Dubnau. 1998. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17:6730-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varmanen, P., H. Ingmer, and F. K. Vogensen. 2000. ctsR of Lactococcus lactis encodes a negative regulator of clp gene expression. Microbiology 146:1447-1455. [DOI] [PubMed] [Google Scholar]
- 35.Volker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Volker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 36.Voskuil, M. I., and G. H. Chambliss. 1998. The −16 region of Bacillus subtilis and other Gram-positive bacterial promoters. Nucleic Acids Res. 26:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, J., J. A. Hartling, and J. M. Flanagan. 1997. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91:447-456. [DOI] [PubMed] [Google Scholar]
- 38.Wawrzynow, A., B. Banecki, and M. Zylicz. 1996. The Clp ATPases define a novel class of molecular chaperones. Mol. Microbiol. 21:895-899. [DOI] [PubMed] [Google Scholar]
- 39.Wiegert, T., and W. Schumann. 2001. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 183:3885-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, G., and S.-L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the σA-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, Y., and S. Gottesman. 1998. Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180:1154-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]