Abstract
The mammalian target of rapamycin complex 1 (mTORC1) has the ability to sense a variety of essential nutrients and respond by altering cellular metabolic processes. Hence, this protein kinase complex is poised to influence adaptive changes to nutrient fluctuations toward the maintenance of whole-body metabolic homeostasis. Defects in mTORC1 regulation, arising from either physiological or genetic conditions, are believed to contribute to the metabolic dysfunction underlying a variety of human diseases, including type 2 diabetes. We are just now beginning to gain insights into the complex tissue-specific functions of mTORC1. Here, we detail our current knowledge of the physiological functions of mTORC1 in controlling systemic metabolism, with a focus on advances obtained through genetic mouse models.
mTOR complex 1: sensing local and systemic nutrient status
The ability of organisms to adapt to fluctuations in nutrient availability is fundamental to the fitness of a species. Organisms respond to nutrient fluctuations by altering the balance between energy-producing catabolic processes and energy-consuming anabolic processes. In eukaryotes, the metabolic response to nutrients is tightly coordinated by nutrient- and energy-sensing signaling pathways. Much progress has been made in our understanding of the cell-intrinsic wiring of these key signaling pathways. Conversely, how these pathways maintain metabolic homeostasis in organisms as complex as mammals is poorly defined and requires consideration of tissue-specific functions. The mammalian target of rapamycin (mTOR) lies at the heart of a nutrient-sensing signaling network that controls cellular metabolism. Genetic studies in mice are beginning to reveal the diverse functions of the mTOR signaling network in specific metabolic tissues to ultimately control whole-body metabolic homeostasis.
The target of rapamycin (TOR) proteins are evolutionarily conserved protein kinases found in nearly all eukaryotic cells [1]. TOR proteins form two distinct protein complexes, referred to as TOR complex 1 and 2 (TORC1 and TORC2), which differ in their regulation, function, and sensitivity to the compound rapamycin (Box1). TORC1 is the primary nutrient-sensing complex and is, therefore, the focus of this review. While the sensing mechanisms are not yet fully defined, the activation status of TORC1 is closely tied to the availability of certain nutrients (e.g., amino acids, glucose, and oxygen) in organisms from yeast to human. States of nutrient depletion lead to an attenuation of TOR protein kinase activity within TORC1, whereas nutrient rich conditions activate TORC1 to promote anabolic cell growth (Reviewed in [2]).
Box 1: The complexity of mTOR complexes and the effects of rapamycin.
It is believed that all mammalian cells contain two distinct mTOR-containing complexes, referred to as mTORC1 and mTORC2 (Reviewed in [1]). In addition to the mTOR kinase, the core components are Raptor and mLST8 for mTORC1 and Rictor, mSIN1, and mLST8 for mTORC2, with a number of other non-essential components found in both complexes. Relative to mTORC1, very little is known regarding the upstream regulation and downstream functions of mTORC2. Insulin and growth factors acutely stimulate the kinase activity of mTORC2 (Reviewed in [98]), but the mechanism is currently unknown. The best-characterized downstream substrate of mTORC2 is Akt, which it directly phosphorylates on S473 within a highly conserved hydrophobic motif. Like the mTORC1 substrate S6K, Akt is a member of the protein kinase A, G, and C (AGC) family. mTORC2 also appears to be the major kinase responsible for phosphorylation of the hydrophobic motif on other AGC kinases, including PKCα [43, 99, 100] and the serum and glucocorticoid-inducible kinase (SGK1 [101]). Hydrophobic motif phosphorylation on these AGC kinases contributes to their stability (PKC) and/or activation (Akt, SGK, and S6K). In addition, the phosphorylation of a second conserved motif on Akt (T450) and PKCα (T638), referred to as the turn motif, is also dependent on mTORC2 [99, 100]. This motif is phosphorylated constitutively and is thought to be involved in the proper folding of these kinases.
Much of our knowledge of mTOR function has been gained from experiments using rapamycin, an exquisitely specific inhibitor of mTOR (reviewed in [102]). The specificity of rapamycin for mTOR stems from its mode of action: this large macrolide antibiotic is bound by a cellular prolyl isomerase, FKBP12, and it is the rapamycin-FKBP12 complex that binds to mTOR. However, unlike most kinase inhibitors, rapamycin is an allosteric inhibitor, binding to a region N-terminal to the kinase domain of mTOR. It is believed that rapamycin can only gain access to this region on mTOR within mTORC1, and therefore, mTORC2 is largely resistant to acute inhibition with rapamycin. Nonetheless, it is important to note that prolonged exposure of cells to rapamycin hinders mTORC2 assembly, and in many cell types, including endothelial cells and adipocytes, inhibits mTORC2 activity [103]. In addition to this caveat, the development and characterization of mTOR kinase domain inhibitors, which inhibit both mTORC1 and mTORC2, have revealed rapamycin-resistant functions of mTORC1 [102]. For instance, while rapamycin strongly inhibits S6K1 phosphorylation by mTORC1, it only partially and transiently inhibits its phosphorylation of 4E-BP1. However, mTOR kinase domain inhibitors completely block mTORC1 signaling to both of these established downstream targets and have more pronounced effects on protein synthesis. Therefore, rapamycin is a powerful tool for analyzing the function of mTORC1 in different settings, but should not be used in isolation to make conclusions regarding whether or not a process is mediated by mTORC1.
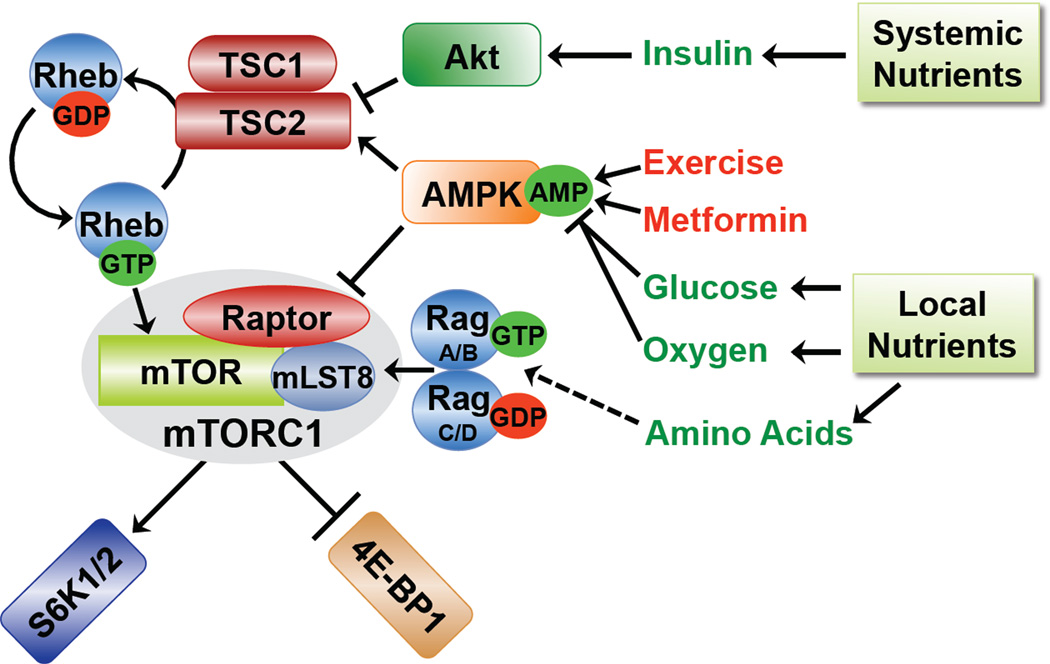
A major breakthrough in understanding the nutrient-dependent regulation of mammalian TORC1 (mTORC1) came from the discovery of a small G protein switch lying directly upstream. When in its GTP-bound form, the Ras-related small G protein Rheb is a potent and essential activator of mTORC1. Rheb is tightly controlled by the GTPase-activating protein (GAP) TSC2 (also referred to as tuberin). In a complex with its binding partner TSC1 (also referred to as hamartin), TSC2 stimulates the intrinsic GTPase activity of Rheb, thereby promoting the conversion from its GTP-bound “on” state to its GDP-bound “off” state. Through this molecular mechanism, the TSC1-TSC2 complex blocks mTORC1 activation in response to a diverse array of signals (Figure 1; reviewed in [3]). For instance, insulin, IGF-1, and other growth factors and cytokines all stimulate mTORC1, at least in part, by inhibiting TSC2. The PI3K-Akt signaling pathway is primarily responsible for this regulation, and the protein kinase Akt directly phosphorylates and inhibits TSC2, within the TSC1-TSC2 complex [4, 5]. This allows Rheb-GTP to accumulate and activate mTORC1. Cellular energy levels also impinge on this upstream TSC-Rheb circuit to regulate mTORC1. An imbalance in the production and consumption of ATP, due to glucose deprivation, acute hypoxia, an increase in anabolic metabolism (e.g., protein or lipid biosynthesis), or tissue-specific processes (e.g., muscle contraction during exercise) leads to a rise in intracellular AMP levels and activation of the AMP-dependent protein kinase (AMPK; reviewed in [6]). In response to energy depletion, AMPK phosphorylates TSC2 on sites distinct from Akt, which activates TSC2, leading to inhibition of mTORC1 [7, 8]. AMPK also inhibits mTORC1 activation through direct phosphorylation of the mTORC1 subunit Raptor [9]. The mechanism by which mTORC1 senses the presence of amino acids is poorly understood, but like all known stimuli that activate mTORC1, Rheb is required. Amino acid re-feeding of starved cells results in docking of mTORC1 to small G proteins of the Rag family, which function as a heterodimer of RagA or B with RagC or D [10, 11]. In response to amino acids, mTORC1 appears to bind to Rag heterodimers at the lysosome, where it is believed that Rheb ultimately stimulates its activity [12]. Therefore, multiple pathways have evolved to tightly control the activation status of mTORC1 in response to nutrients (Figure 1). These signals stem from both systemic nutrients, via hormonal responses (e.g., insulin and IGF-1), and local or intracellular nutrients.
Figure 1. Nutrient sensing and upstream regulation of mTORC1.
The presence of extracellular (or systemic) and intracellular nutrients are sensed by mTORC1 and stimulate its activation. In response to insulin, Akt inhibits TSC2, thereby allowing Rheb-GTP to accumulate and activate mTORC1. Nutrients and other factors influencing the energy status of the cell affect intracellular levels of AMP, which activates AMPK. AMPK exerts inhibitory effects on mTORC1 through activation of TSC2 and phosphorylation of Raptor. In addition, the presence of intracellular amino acids is sensed by mTORC1 through an undefined pathway affecting the Rag GTPases. Finally, the two best-characterized direct downstream targets of mTORC1, S6K1/2 and 4E-BP1, are shown.
It is worth noting that the mechanistic studies defining the upstream regulation of mTORC1 have been primarily in cell culture models, where the levels of nutrients and hormones that the cell perceives can be easily manipulated. However, these experimental conditions (e.g., complete amino acid or glucose starvation followed by acute refeeding) could only be replicated in mammals under extreme physiological conditions. The relevance of these in vitro paradigms to mTORC1 regulation in different mammalian tissues under homeostatic conditions is not yet known.
Downstream of mTORC1: Protein synthesis and beyond
In mammals, two main classes of direct mTORC1 substrates have emerged (reviewed in [13]): the ribosomal S6 kinases (S6K1 and S6K2) and the eukaryotic initiation factor 4E (eIF4E)-binding proteins (4EBP1 and 4EBP2). mTORC1 phosphorylates the hydrophobic motif on the S6Ks (T389 on the 70-kD isoform of S6K1), which is essential for subsequent activating phosphorylation events. The S6Ks phosphorylate a number of downstream targets, the best characterized of which is the ribosomal protein S6. While our understanding of the molecular consequence of S6 phosphorylation is incomplete, it is the most frequently used readout of mTORC1 activation in cells and tissues. S6K has an expanding number of downstream substrates, some of which are involved in promoting mRNA translation [13]. However, mTORC1 exerts a more direct control on translation through its regulation of the 4E-BPs, which bind to eIF4E at the 5’-cap of mRNAs and block translation initiation. mTORC1 phosphorylates the 4E-BPs on multiple residues, triggering their release from eIF4E and allowing subsequent binding of translation initiation factors. In addition to these acute effects on mRNA translation, mTORC1 signaling is believed to enhance the general protein synthesis capacity of the cell by promoting ribosome biogenesis (reviewed in [14]). In parallel, mTORC1 plays a conserved role in suppressing autophagy, a nutrient-recycling process in which cellular macromolecules and organelles are lysosomally degraded into their constituent components (reviewed in [15]). By analogy to other major signaling hubs in mammalian cells, it seems highly likely that many more direct downstream targets of mTORC1 will be identified in the near future.
Interestingly, a number of cell-based studies examining global changes in gene expression suggest that mTORC1 controls specific aspects of cellular metabolism through the regulation of transcription (e.g., [16–18]). The implications are that mTORC1 impinges on metabolic pathways through transcription factors that regulate the expression of metabolic enzymes. For instance, mTORC1 signaling enhances the translation of hypoxia-inducible factor (HIF1α; [19–21]), which can transcriptionally activate genes encoding glucose transporters and glycolytic enzymes [22, 23]. Consistent with this regulation, mTORC1 activation can promote glucose uptake and flux through glycolysis [18]. In addition, the sterol-regulatory element-binding proteins (SREBP1 and SREBP2), which induce the expression of a large number of genes involved in de novo lipid and sterol biosynthesis, have emerged as major effectors of mTORC1 signaling, and mTORC1 activation stimulates lipogenesis [18, 24]. The influence of these cell intrinsic changes in metabolism downstream of mTORC1 on organismal metabolic phenotypes is poorly understood. However, it is clear from mouse genetic studies that mTORC1 signaling affects metabolic processes beyond protein synthesis.
mTORC1-dependent feedback mechanisms and cell intrinsic insulin resistance
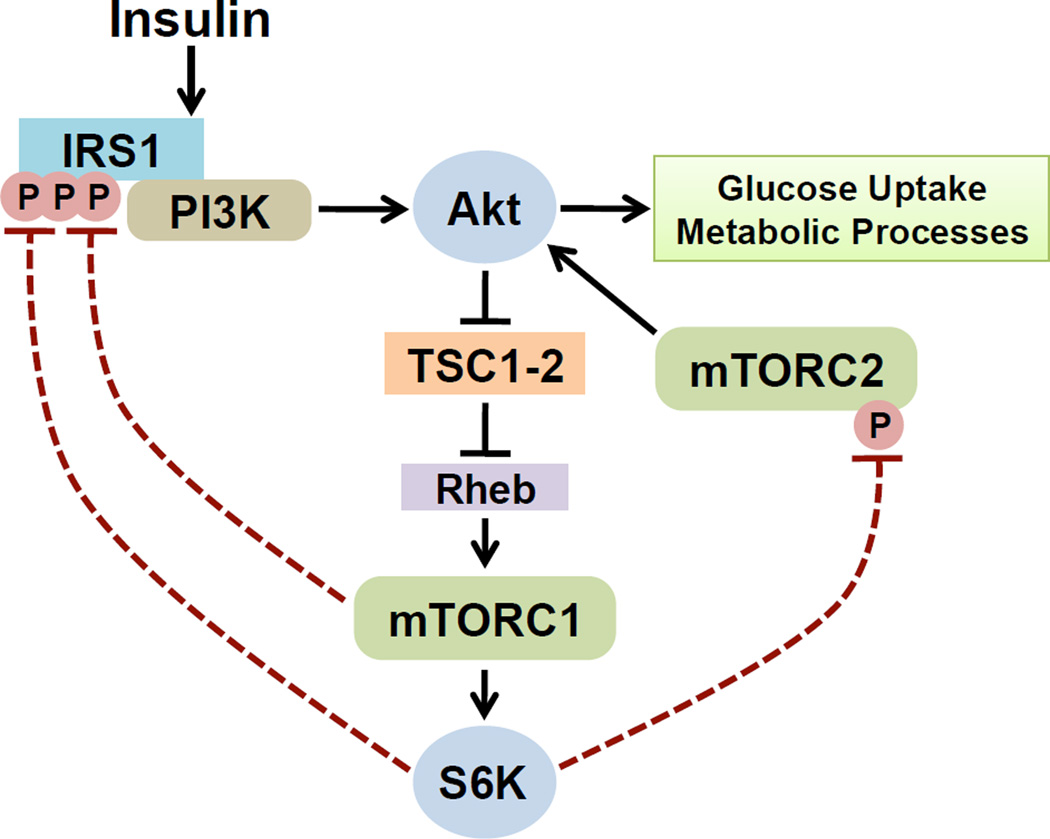
A critical element of mTORC1 signaling is its feedback effects on upstream pathways. Both mTORC1 and its downstream target S6K1 can exert negative regulatory inputs into upstream signaling molecules (Figure 2). The best characterized of these are the insulin receptor substrate (IRS) proteins, which are required to activate the PI3K-Akt pathway downstream of the insulin receptor. There are a number of rapamycin-sensitive phosphorylation sites on IRS1, and several of these serine residues appear to be phosphorylated directly by mTORC1 or S6K1 [25–29]. The net effect of these phosphorylation events on IRS1 is the suppression of PI3K activation by this scaffolding adaptor. Another feedback mechanism involves S6K1-mediated phosphorylation of the core mTORC2 component Rictor on T1135, which leads to an attenuation of Akt phosphorylation and activation [30, 31]. It is worth noting that there are likely to be additional points of feedback control from mTORC1 to the upstream components of the insulin signaling pathway. This is illustrated by the fact that there is a clear negative regulatory effect of TORC1 and S6K signaling on insulin-stimulated activation of Akt in Drosophila [32, 33], but the inhibitory phosphorylation sites on neither IRS1 nor Rictor are conserved in the corresponding fly orthologs.
Figure 2. mTORC1 signaling attenuates AKT activation through negative feedback mechanisms.
Inhibitory phosphorylation events on IRS1 and the mTORC2 component Rictor by mTORC1 and/or S6K1 suppress the ability of insulin to stimulate Akt, a major metabolic effector of insulin signaling. Through such mechanisms, mTORC1 signaling can decrease insulin sensitivity. Importantly, this feedback effect on AKT signaling has been observed in all metabolic tissues examined (summarized in Table 1).
While these mTORC1-dependent feedback mechanisms have likely evolved to assure that the insulin response is transient in nature, elevated levels of mTORC1 signaling can strongly attenuate the cellular response to insulin. This is best illustrated in cells lacking a functional TSC1-TSC2 complex, where Rheb-GTP levels are greatly elevated and mTORC1 signaling is constitutively activated. IRS1 and IRS2 are phosphorylated on many inhibitory serine residues in TSC-deficient cells, and these cells are unresponsive to insulin unless subjected to prolonged treatment with rapamycin [28, 34, 35]. The TSC1-TSC2 complex also exerts a positive regulatory effect on mTORC2 that is independent of its inhibition of Rheb and mTORC1 [36]. This further attenuates Akt activation upon loss of the TSC genes. While a clear link between mTORC1 activation and insulin resistance is seen in cell culture models, much less is known about the physiological role of these feedback mechanisms in vivo. However, mTORC1 activity is elevated in the liver and muscle of rodent models of hyperinsulinemia and obesity, and this correlates with the development of insulin resistance [29, 37–40]. As discussed in the following sections, recent mouse models affecting mTORC1 and its downstream targets are beginning to shed light on the importance of these cell intrinsic feedback mechanisms for the development of systemic insulin resistance.
Pharmacological versus genetic manipulation of mTORC1 signaling in rodent models
Given the defined roles of mTORC1 in nutrient sensing and control of cellular physiology, there is a real push to understand the functions of mTORC1 in regulating the physiological state of mammals. To this end, gain and loss of function models have been developed to delineate the role of mTORC1 signaling in rodents. One limitation of these models comes from the finding that null alleles of the mTORC1 components (mTOR or RAPTOR [41–43]) or its key upstream regulators (TSC1 and TSC2 [44, 45]) result in very early embryonic lethality. It is clear that rapamycin is a powerful tool in the study of the physiological functions of mTORC1. However, studies examining the effects of systemic administration of rapamycin on different species of adult rodents have yielded variable results, ranging from hyperglycemia to hypoglycemia, with inconsistent effects on insulin levels [46–48]. There is agreement amongst studies that rapamycin blocks weight gain and decreases adiposity [46–50]. However, the effects of rapamycin on circulating levels of insulin, triglycerides, and glucose may be more sensitive experimental variables, including rodent species, genotype, diet, age, and dose and duration of rapamycin treatment, rendering it difficult to make unifying conclusions from these disparate studies. The most common side effects reported for clinical use of rapamycin and its analogs in humans are hyperlipidemia and glucose intolerance [51]. The major caveat to the in vivo use of rapamycin is that one cannot readily identify the tissue(s) responsible for the beneficial or detrimental effects of mTORC1 inhibition on complex systemic metabolic parameters. Therefore, genetic approaches including knockouts of non-essential downstream targets, conditional knockouts affecting mTORC1 or its negative regulator the TSC1-TSC2 complex, transgenics, and viral delivery of cDNAs are all needed to grasp this complexity. The findings from these genetic gain and loss of function models are providing important insights into the tissue-specific functions of mTORC1, and these are discussed in the next section and summarized in Table 1.
Table 1.
Summary of metabolic phenotypes resulting from perturbation of mTORC1 in metabolic tissues.
| Tissue | Model1 | mTORC1 activity2 |
AKT signaling3 |
Phenotype Reported for Given Tissue | References |
|---|---|---|---|---|---|
| Pancreas | Tsc1-cKO (RIP) | Gain | ↓ | Hyperinsulinemia, increased β-cell mass | [53] |
| Tsc2-cKO (RIP) | Gain | ↓ | Hyperinsulinaemia, increased β-cell mass, hypoglycemia, improved glucose tolerance | [54, 55] | |
| Rheb-Tg (RIP) | Gain | ↓ | Increased β-cell mass, improved glucose tolerance | [56] | |
| S6k1−/− | Loss | NR | Hypoinsulinaemia, decreased β-cell mass, decreased glucose tolerance | [58] | |
| rpS6P−/− | Loss | NR | Hypoinsulinemia, decreased β-cell size, increased β-cell number, decreased glucose tolerance | [59] | |
| Muscle | Raptor-cKO (HSA) | Loss | ↑ | Lean, decreased muscle mass, muscle dystrophy, decreased oxidative capacity, increased glycogen | [60] |
| mTor-cKO (HSA) | Loss | ↑ | Decreased muscle mass, muscle dystrophy, decreased oxidative capacity, increased glycogen | [61] | |
| S6k1−/− | Loss | ↑(HFD) | Decreased muscle mass, increased mitochondrial content, increased glycogen, decreased lipid | [37, 62, 63] | |
| Adipose | Raptor-cKO (aP2) | Loss | ↑(HFD) | Lean, increased mitochondrial uncoupling, increased insulin sensitivity and glucose tolerance | [74] |
| S6k1−/− | Loss | ↑(HFD) | Lean, increased mitochondrial mass and lipolysis, increased insulin sensitivity and metabolic rate, decreased adipogenesis | [37, 73] | |
| Eif4ebp1−/−, Eif4ebp2−/− | Gain | ↓ | Increased adiposity and body weight, decreased lipolysis, reduced metabolic rate; insulin resistance, glucose inolerance, and, increased fatty acid reesterification on HFD |
[72] | |
| Liver | Ad-RaptorΔCT [KK-Ay obese mice] | Loss | ↑ | Enhanced glucose tolerance, increased insulin sensitivity | [79] |
| Eif4ebp1−/−, Eif4ebp2−/− | Gain | ↓ | Increased liver weight and steatosis on HFD | [72] | |
| Hypothalamus | Tsc1-cKO (RIP) | Gain | NR | Hyperphagic obesity | [57] |
| Tsc1-cKO (POMC) | Gain | NR | Hyperphagic obesity, decreased axonal projections to the paraventricular nucleus | [57] | |
| S6k1−/− | Loss | NR | Resistant to anorectic effects of leptin and CNTF | [89] | |
| Ad-CA S6k [Rat] | Gain | ↓ | Decreased body weight and food intake, decreased metabolic rate, decreased weight/fat gain on HFD, hypothalamic insulin resistance | [90, 97] | |
| Ad-DN S6k [Rat] | Loss | ↑ | Increased body weight and food intake, enhanced insulin sensitivity in hypothalamus | [90, 97] |
Mouse models unless otherwise indicated in brackets; cKO, conditional knockout; Tg, transgene; Ad, adenovirus. Promoters driving Cre (for cKOs) or Tg expression are indicated in parentheses.
Denotes loss or gain of mTORC1 activity or downstream signaling events.
Effects on AKT signaling in the tissue of interest, often used as a measure of cell intrinsic insulin sensitivity. NR, not reported;
HFD, high fat diet; MBH, mediobasal hypothalamus; CA, constitutively active; DN, dominant negative; CNTF, Ciliary neurotrophic factor
Toward understanding the metabolic tissue-specific functions of mTORC1
Pancreas
The endocrine pancreas functions as a sensor and regulator of circulating glucose levels, and alterations in the physiological function of the pancreas often reflect changes in islet mass [52]. Given its function as both a nutrient sensor and a regulator of cell growth, it is not surprising that mTORC1 has been shown to play a major role in the pancreatic control of insulin secretion and glucose homeostasis, largely through the regulation of β-cell size. Mouse models with constitutive activation of mTORC1 signaling in β-cells, through conditional deletion of Tsc1 (RIP-Cre) or Tsc2 (RIP-Cre) or transgenic overexpression of Rheb (RIP-Rheb), display increased β-cell mass, hyperinsulinemia, and improved glucose tolerance [53–56]. One point of contention in these models is the long-term effects of chronic mTORC1 activation in β-cells. Contrary to their findings in younger mice, Shigeyama et al. report that older mice lacking the TSC1-TSC2 complex in β-cells (>40 weeks) have decreased β-cell number and progressive hypoinsulinemia and hyperglycemia [55], similar to what is observed in the progression of type 2 diabetes [52]. However, using a very similar model, Rachdi et al. saw sustained improvement in glucose homeostasis in aged mice [54]. While some of these models are also complicated by hypothalamic expression of RIP-Cre [53, 57], the collective data from these independent studies strongly support a role for mTORC1 in pancreatic insulin secretion. This conclusion is fully substantiated by systemic loss of function models: mice treated with rapamycin or knockouts of the mTORC1 target S6K1 (S6k1−/−) exhibit decreased β-cell mass and reduced glucose-stimulated insulin secretion, resulting in hypoinsulinemia and glucose intolerance [46, 58]. Studies in S6k1−/− mice indicate that S6K1 is the primary target of mTORC1 in the control of β-cell mass and insulin production. Interestingly, ribosomal S6 knock-in mice (rpS6P−/−) lacking the sites phosphorylated by S6K1 and S6K2 display β-cell and insulin phenotypes similar to the S6k1−/− mice [59], suggesting that this downstream substrate is involved in the S6K-dependent regulation of these processes. This is notable, as a molecular function for these long-identified sites on S6 has not been defined. Another important feature of the S6k1−/− mice is that, despite their hypoinsulinemia, these mice maintain normal blood glucose levels, likely due to enhanced peripheral insulin sensitivity.
Muscle
The role of mTORC1 in muscle has recently been advanced by the characterization of several loss-of-function animal models. Skeletal muscle-specific deletion of Raptor (HSA-Cre) [60] or mTor (HSA-Cre) [61] results in reduced muscle mass with increased glycogen content and decreased oxidative capacity compared to controls. Interestingly, muscle from S6k1−/− or S6k1−/− S6k2−/− mice show similar phenotypes [62, 63]. Conditional deletion of Raptor or mTOR, but not the mTORC2 component Rictor, in skeletal muscle results in mice with reduced body weight and progressively dystrophic muscles. The reduced muscle mass in these mice is accompanied by weakness and decreased contractile properties, decreased glycogenolysis and glycolysis, and mitochondrial defects with reduced oxidative capacity despite an apparent switch to slow-twitch fibers. Concomitantly, loss of mTORC1 in skeletal muscle enhances Akt activation and basal glucose uptake, consistent with alleviation of mTORC1-dependent feedback mechanisms affecting upstream insulin signaling in these muscles (Figure 2). Muscles from S6k1−/− and S6k1−/− S6k2−/− mice exhibit a milder phenotype, without signs of severe dystrophy or fiber-type switching [64]. Interestingly, in myotube cultures from the single or double S6K knockout mice, basal AMP:ATP ratios are increased leading to activation of AMPK and increased mitochondrial fatty acid oxidation [63]. Surprising, the data suggest that elevated AMPK activity is responsible for the decreased myotube size upon loss of S6K. A role for AMPK in the phenotypes exhibited by the skeletal muscle knockouts of mTOR and Raptor has not been examined. It is interesting to note that the S6k1−/− mice have been described to have increased mitochondria in their skeletal muscle with enhanced oxidative capacity [37], while the skeletal muscle knockouts of Raptor and mTOR have defects in mitochondrial metabolism [60, 61]. Consistent with mTORC1 signaling promoting mitochondrial biosynthesis and/or function in skeletal muscle, mTORC1 has been suggested to positively regulate the transcription factor PGC1a, a master regulator of mitochondrial biogenesis [17]. Collectively, these genetic models indicate that mTORC1 plays a major role in the maintenance of skeletal muscle integrity and metabolism. While the effects on muscle fiber size are controlled, at least in part, through S6K, mTORC1 has additional S6K-independent functions promoting oxidative metabolism in muscle.
Adipose
In addition to its function as an energy storage depot, adipose tissue is an active endocrine organ and plays a key role in the control of metabolic homeostasis [65]. Cell culture studies have shown that mTORC1 activation is both necessary and sufficient to drive adipocyte differentiation through the upregulation of PPARγ, the master regulator of this process [66–71]. In vivo data also support a role for mTORC1 signaling in adipogenesis, as well as in the metabolism of mature adipocytes. S6k1−/− mice display increased energy expenditure and are resistant to age and diet-induced obesity, possessing smaller adipocytes with increased mitochondria and enhanced basal rates of lipolysis [37]. As predicted from loss of S6K1-mediated feedback mechanisms, adipose tissue from these mice display increased activation of Akt in response to insulin. Reciprocally, mice lacking 4EBP1 and 2 (Eif4ebp1/2−/−), which are normally inhibited by mTORC1, are more prone to obesity and display increased adiposity, reduced lipolysis, a reduction in overall energy expenditure, and insulin resistance [72]. However, as the 4EBP knockout mice display increased S6K1 activation in metabolic tissues, it was hypothesized that these phenotypes result from a gain of S6K1 function. Supporting this model, the triple knockout mice lacking S6K1, 4EBP1, and 4EBP2 appear to be resistant to diet-induced obesity, similar to the S6k1−/− mice [73]. It was recently shown that S6K1 is dispensable for terminal adipocyte differentiation but is required at an early stage of commitment to an adipocyte progenitor lineage, suggesting an additional mechanism for the resistance of the S6k1−/− mice to diet-induced obesity [73]. However, complete loss of mTORC1 signaling in terminally differentiated adipocytes, through conditional knockout of Raptor (aP2-Cre), yields a strikingly similar phenotype to that of the S6k1 knockout [74]. The adipose-specific Raptor knockout mice are lean with improved glucose tolerance and insulin sensitivity, and are resistant to diet-induced obesity [74]. The reduced adiposity in these mice results from both fewer and smaller adipocytes with increased energy consumption through upregulation of the mitochondrial uncoupling protein UCP1 [74], which is also seen in the adipose tissue of S6k1−/− mice [37]. There is increasing evidence that mTORC1 signaling can promote lipogenesis and inhibit lipolysis [18, 24, 75, 76], suggesting the potential for additional mechanisms contributing to the profound decrease in adiposity in these mouse models. The close phenotypic links between mice lacking mTORC1 in mature adipocytes and those lacking the downstream targets of mTORC1 in all tissues, suggest that mTORC1 activity in adipose tissue plays a critical role in the regulation of systemic metabolic homeostasis.
Liver
A primary function of the liver is the maintenance of glucose homeostasis during periods of fasting [77]. Thus, the liver must be acutely sensitive to fluctuations in nutrients and hormones, such as glucagon and insulin. Despite this key role in systemic glucose and lipid metabolism, to date, there have been relatively few mouse models developed to examine the metabolic functions of mTORC1 in this organ. Nevertheless, current evidence implicates mTORC1 in the attenuation of insulin signaling and promotion of lipid biosynthesis in the liver. Indeed, elevated levels of hepatic mTORC1 signaling have been correlated with decreased insulin sensitivity in the liver [38, 78]. Consistent with mTORC1 and S6K1-mediated feedback mechanisms contributing to hepatic insulin resistance, adenoviral delivery of a proposed dominant-negative Raptor mutant (which lacks approximately 400 amino acids at its C-terminus and blocks the phosphorylation of S6K1 but not 4EBP1 when overexpressed in the liver) enhances insulin-stimulated PI3K-Akt signaling and improves glucose tolerance [79].
An additional consequence of insulin signaling in the liver is the regulation of lipid metabolism, and several recent studies support a role for mTORC1 in this process. In cell culture models, mTORC1 has been shown to be necessary and sufficient for the pathways downstream of insulin to induce the SREBP transcription factors [18, 24], which play a critical role in hepatic lipid and sterol biosynthesis. Consistent with these mechanistic studies, rapamycin abolishes the induction of SREBP1c expression by insulin in primary rat hepatocytes [80, 81]. Furthermore, Eif4ebp1/2−/− knockout mice display increased hepatic steatosis on a high-fat diet relative to control mice, but this phenotype is confounded by their increased susceptibility to obesity [72]. While these collective findings emphasize the importance of mTORC1 in the metabolic functions of the liver, genetic loss and gain of function models manipulating mTORC1 signaling specifically in the liver will be necessary to clarify these roles and their impact on whole body energy homeostasis.
Hypothalamus
Neuronal populations within the mammalian hypothalamus respond to the nutritional status of the organism to control food intake and energy expenditure [82, 83]. The anorectic effects of insulin and leptin in the hypothalamus are well accepted, but only recently has mTORC1 been recognized as a critical regulator of this response [84–88]. Although it is expressed throughout the brain, organismal energy status appears to affect mTORC1 signaling specifically in the hypothalamus [86]. Hypothalamic mTORC1 signaling is reduced in the fasting state, while anorectic signals such as leptin and ciliary neurotrophic factor (CNTF) require mTORC1 activity to exert their inhibitory effects on feeding behavior [86, 89]. Interestingly, stimulation of mTORC1 signaling through intracerebroventricular administration of L-leucine also decreases food intake in mice, effects that are reversed by rapamycin [86]. Consistent with mTORC1 signaling eliciting a hypophagic response, adenoviral delivery of constitutively active S6K1 to the mediobasal hypothalamus of rats suppresses food intake, whereas dominant negative S6K1 induces food intake [90]. Furthermore, S6k1−/− mice were shown to be resistant to leptin and CNTF and, when normalized to body weight, appear to be hyperphagic [37, 89]. These studies suggest a clear role for hypothalamic mTORC1 in sensing and relaying signals to decrease food intake. However, constitutive activation of mTORC1, via conditional deletion of Tsc1, in either a broad set of neurons (RIP-Cre) or specifically in the anorexigenic POMC neurons (POMC-Cre) results in increased food intake and corresponding weight gain [57, 91]. As mTORC1 is known to control axonal guidance and synaptic plasticity [92, 93], it is possible that these counterintuitive phenotypes reflect defects in neuronal morphology upon disruption of the TSC1-TSC2 complex. Indeed, conditional activation of mTORC1 in these neurons results in enlarged POMC soma and decreased axonal projections leading to the paraventricular nucleus [57]. Finally, differential regulation of mTORC1 has been observed in distinct neuronal populations within the hypothalamus, indicating that the physiological regulation and effects of mTORC1 activation could vary greatly depending on which specific hypothalamic neurons that are targeted [86, 88]. Therefore, future genetic models manipulating mTORC1 activity in these distinct neuronal populations will greatly enhance our knowledge of this potential link between nutrient sensing and feeding behavior.
Conclusions and outstanding questions
The function of mTORC1 as a nutrient sensor and regulator of anabolic processes has been well established in cell culture models. However, we are just beginning to understand how its cell intrinsic regulation and function translate into the control of systemic metabolism. From the rodent models discussed here, it appears that in response to food intake, mTORC1 activation enhances nutrient mobilization into peripheral tissues through increased insulin secretion from the pancreas, while also enhancing lipid storage in adipose tissue. At the same time, mTORC1 signaling appears to play a role in the hypothalamus to suppress further food intake, thereby creating a systemic negative feedback loop to maintain nutrient homeostasis. However, more sophisticated genetic models are needed to fully understand the consequences of mTORC1 activation in specific metabolic tissues and to reveal critical points of cross talk with other signaling pathways regulating metabolism.
Currently, we know very little regarding the hierarchy of mechanisms regulating mTORC1 activity in vivo and how fluctuations in circulating levels of specific nutrients and hormones, as well as signals from the sympathetic nervous system, influence mTORC1 signaling properties in distinct metabolic tissues. The importance of understanding this regulation is underscored by the fact that aberrant mTORC1 activation in peripheral insulin-responsive tissues appears to contribute to the development of insulin resistance, and defects in the proper control of mTORC1 could be a major factor in the pathophysiology of type 2 diabetes and other common metabolic disorders. However, further investigation is required to more clearly elucidate the contributions of mTORC1 signaling to the development of systemic insulin resistance. Furthermore, the role of mTORC1-regulated cellular processes established primarily through cell culture studies, such as protein and lipid synthesis and autophagy, on organismal metabolic phenotypes initiated by aberrant mTORC1 signaling is poorly understood.
On the surface, mTORC1 would appear to be a good drug target for improving peripheral insulin sensitivity. In fact, the most commonly prescribed anti-diabetes drug, metformin, strongly inhibits mTORC1 signaling [94, 95]. However, whether mTORC1 inhibition contributes to the beneficial effects of metformin on systemic glucose metabolism is unknown. While rapamycin treatment results in decreased adiposity in mice [49, 50], its potential effectiveness in treating metabolic diseases is not clear. An important consideration in the systemic use of rapamycin is the role of mTORC1 in insulin production, where rapamycin could impede the ability of the pancreas to secrete insulin in response to glucose. Rapamycin significantly extends life span when administered to older mice [96], but the health benefits leading to longevity are currently unknown. At the molecular level, it is also possible that some of the effects of rapamycin on life span could be through inhibition of mTORC2 (Box 1). Further characterization of tissue-specific targets and processes downstream of mTORC1 could reveal opportunities to develop therapeutics that more selectively target mTORC1 signaling. Reciprocally, supplements aimed at activating mTORC1 are being marketed to body builders for muscle enhancement. It is possible that strategies to activate mTORC1 in skeletal muscle could prove beneficial for muscular dystrophies. However the effects of mTORC1-activating supplements or compounds on other tissues, such as fat and liver, where they might increase lipid accumulation or promote insulin resistance, are unknown. Finally, given the recent popularity of high protein diets predicted to enhance mTORC1 activation, it will be important to determine the influence on mTORC1 functions in different tissues, with an eye toward understanding the long-term metabolic effects of such diets.
The studies reviewed in this work have greatly advanced our knowledge of mTORC1 functions in the context of whole body metabolism. However, we are clearly still in the early stages of connecting our ever-expanding knowledge of the cell intrinsic regulation and function of mTORC1 to its role in controlling systemic metabolic homeostasis.
Acknowledgements
Research on the regulation and metabolic function of mTORC1 in the Manning laboratory is supported by grants to B.D.M. from the National Institutes of Health (CA122617) and the American Diabetes Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wullschleger S, et al. TOR Signaling in Growth and Metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Sengupta S, et al. Regulation of the mTOR Complex 1 Pathway by Nutrients, Growth Factors, and Stress. Molecular Cell. 40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning BD, et al. Identification of the Tuberous Sclerosis Complex-2 Tumor Suppressor Gene Product Tuberin as a Target of the Phosphoinositide 3-Kinase/Akt Pathway. Molecular Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 5.Inoki K, et al. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 6.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 7.Inoki K, et al. TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 8.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Gwinn DM, et al. AMPK Phosphorylation of Raptor Mediates a Metabolic Checkpoint. Molecular Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sancak Y, et al. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim E, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancak Y, et al. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 14.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 15.Kroemer G, et al. Autophagy and the Integrated Stress Response. Molecular Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng T, et al. The Immunosuppressant Rapamycin Mimics a Starvation-Like Signal Distinct from Amino Acid and Glucose Deprivation. Mol. Cell. Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 18.Düvel K, et al. Activation of a Metabolic Gene Regulatory Network Downstream of mTOR Complex 1. Molecular Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong H, et al. Modulation of Hypoxia-inducible Factor 1α Expression by the Epidermal Growth Factor/Phosphatidylinositol 3-Kinase/PTEN/AKT/FRAP Pathway in Human Prostate Cancer Cells: Implications for Tumor Angiogenesis and Therapeutics. Cancer Research. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 20.Laughner E, et al. HER2 (neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1{alpha} (HIF-1 {alpha}) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol. Cell. Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson CC, et al. Regulation of Hypoxia-Inducible Factor 1{alpha} Expression and Function by the Mammalian Target of Rapamycin. Mol. Cell. Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semenza GL, et al. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 23.Hu C-J, et al. Differential Roles of Hypoxia-Inducible Factor 1 {alpha} (HIF-1 {alpha}) and HIF-2{alpha} in Hypoxic Gene Regulation. Mol. Cell. Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porstmann T, et al. SREBP Activity Is Regulated by mTORC1 and Contributes to Akt-Dependent Cell Growth. Cell Metabolism. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning BD. Balancing Akt with S6K. The Journal of Cell Biology. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington LS, et al. Restraining PI3K: mTOR signalling goes back to the membrane. Trends in Biochemical Sciences. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Tzatsos A, Kandror KV. Nutrients Suppress Phosphatidylinositol 3-Kinase/Akt Signaling via Raptor-Dependent mTOR-Mediated Insulin Receptor Substrate 1 Phosphorylation. Mol. Cell. Biol. 2006;26:63–76. doi: 10.1128/MCB.26.1.63-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shah OJ, Hunter T. Turnover of the Active Fraction of IRS1 Involves Raptor-mTOR- and S6K1-Dependent Serine Phosphorylation in Cell Culture Models of Tuberous Sclerosis. Mol. Cell. Biol. 2006;26:6425–6434. doi: 10.1128/MCB.01254-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tremblay Fd.r, et al. Identification of IRS-1 Ser-1101 as a target of S6K1 in nutrient- and obesity-induced insulin resistance. Proceedings of the National Academy of Sciences. 2007;104:14056–14061. doi: 10.1073/pnas.0706517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dibble CC, et al. Characterization of Rictor Phosphorylation Sites Reveals Direct Regulation of mTOR Complex 2 by S6K1. Mol. Cell. Biol. 2009;29:5657–5670. doi: 10.1128/MCB.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Julien L-A, et al. mTORC1-Activated S6K1 Phosphorylates Rictor on Threonine 1135 and Regulates mTORC2 Signaling. Mol. Cell. Biol. 2010;30:908–921. doi: 10.1128/MCB.00601-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radimerski T, et al. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes & Development. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarbassov DD, et al. Phosphorylation and Regulation of Akt/PKB by the Rictor-mTOR Complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 34.Harrington LS, et al. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. The Journal of Cell Biology. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah OJ, et al. Inappropriate Activation of the TSC/Rheb/mTOR/S6K Cassette Induces IRS1/2 Depletion, Insulin Resistance, and Cell Survival Deficiencies. Current Biology. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 36.Huang J, et al. The TSC1-TSC2 Complex Is Required for Proper Activation of mTOR Complex 2. Mol. Cell. Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 38.Khamzina L, et al. Increased Activation of the Mammalian Target of Rapamycin Pathway in Liver and Skeletal Muscle of Obese Rats: Possible Involvement in Obesity-Linked Insulin Resistance. Endocrinology. 2005;146:1473–1481. doi: 10.1210/en.2004-0921. [DOI] [PubMed] [Google Scholar]
- 39.Korsheninnikova E, et al. Sustained activation of the mammalian target of rapamycin nutrient sensing pathway is associated with hepatic insulin resistance, but not with steatosis, in mice. Diabetologia. 2006;49:3049–3057. doi: 10.1007/s00125-006-0439-5. [DOI] [PubMed] [Google Scholar]
- 40.Ueno M, et al. Regulation of insulin signalling by hyperinsulinaemia: role of IRS-1/2 serine phosphorylation and the mTOR/p70 S6K pathway. Diabetologia. 2005;48:506–518. doi: 10.1007/s00125-004-1662-6. [DOI] [PubMed] [Google Scholar]
- 41.Gangloff Y-G, et al. Disruption of the Mouse mTOR Gene Leads to Early Postimplantation Lethality and Prohibits Embryonic Stem Cell Development. Mol. Cell. Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami M, et al. mTOR Is Essential for Growth and Proliferation in Early Mouse Embryos and Embryonic Stem Cells. Mol. Cell. Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guertin DA, et al. Ablation in Mice of the mTORC Components raptor, rictor, or mLST8 Reveals that mTORC2 Is Required for Signaling to Akt-FOXO and PKC[alpha], but Not S6K1. Developmental Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Rennebeck G, et al. Loss of function of the tuberous sclerosis 2 tumor suppressor gene results in embryonic lethality characterized by disrupted neuroepithelial growth and development. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:15629–15634. doi: 10.1073/pnas.95.26.15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobayashi T, et al. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fraenkel M, et al. mTOR Inhibition by Rapamycin Prevents β-Cell Adaptation to Hyperglycemia and Exacerbates the Metabolic State in Type 2 Diabetes. Diabetes. 2008;57:945–957. doi: 10.2337/db07-0922. [DOI] [PubMed] [Google Scholar]
- 47.Rovira J, et al. Effect of mTOR inhibitor on body weight: from an experimental rat model to human transplant patients. Transplant International. 2008;21:992–998. doi: 10.1111/j.1432-2277.2008.00710.x. [DOI] [PubMed] [Google Scholar]
- 48.Houde VP, et al. Chronic Rapamycin Treatment Causes Glucose Intolerance and Hyperlipidemia by Upregulating Hepatic Gluconeogenesis and Impairing Lipid Deposition in Adipose Tissue. Diabetes. 2010;59:1338–1348. doi: 10.2337/db09-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang G-R, et al. Rapamycin Protects Against High Fat Diet–Induced Obesity in C57BL/6J Mice. Journal of Pharmacological Sciences. 2009;109:496–503. doi: 10.1254/jphs.08215fp. [DOI] [PubMed] [Google Scholar]
- 50.Chang GR, et al. Long-term Administration of Rapamycin Reduces Adiposity, but Impairs Glucose Tolerance in High-Fat Diet-fed KK/HlJ Mice. Blackwell Publishing Ltd.; 2009. pp. 188–198. [DOI] [PubMed] [Google Scholar]
- 51.Stallone G, et al. Management of side effects of sirolimus therapy. Transplantation. 2009;87:S23–S26. doi: 10.1097/TP.0b013e3181a05b7a. [DOI] [PubMed] [Google Scholar]
- 52.Butler AE, et al. β-Cell Deficit and Increased β-Cell Apoptosis in Humans With Type 2 Diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 53.Mori H, et al. Critical roles for the TSC-mTOR pathway in β-Cell function. Am J Physiol Endocrinol Metab. 2009;297:E1013–E1022. doi: 10.1152/ajpendo.00262.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rachdi L, et al. Disruption of Tsc2 in pancreatic β cells induces β cell mass expansion and improved glucose tolerance in a TORC1-dependent manner. Proceedings of the National Academy of Sciences. 2008;105:9250–9255. doi: 10.1073/pnas.0803047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shigeyama Y, et al. Biphasic Response of Pancreatic β-Cell Mass to Ablation of Tuberous Sclerosis Complex 2 in Mice. Mol. Cell. Biol. 2008;28:2971–2979. doi: 10.1128/MCB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamada S, et al. Upregulation of the Mammalian Target of Rapamycin Complex 1 Pathway by Ras Homolog Enriched in Brain in Pancreatic β-Cells Leads to Increased β-Cell Mass and Prevention of Hyperglycemia. Diabetes. 2009;58:1321–1332. doi: 10.2337/db08-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mori H, et al. Critical Role for Hypothalamic mTOR Activity in Energy Balance. Cell Metabolism. 2009;9:362–374. doi: 10.1016/j.cmet.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pende M, et al. Hypoinsulinaemia, glucose intolerance and diminished β-cell size in S6K1-deficient mice. Nature. 2000;408:994–997. doi: 10.1038/35050135. [DOI] [PubMed] [Google Scholar]
- 59.Ruvinsky I, et al. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes & Development. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bentzinger CF, et al. Skeletal Muscle-Specific Ablation of raptor, but Not of rictor, Causes Metabolic Changes and Results in Muscle Dystrophy. Cell Metabolism. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Risson V, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. The Journal of Cell Biology. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohanna M, et al. Atrophy of S6K1−/− skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol. 2005;7:286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- 63.Aguilar V, et al. S6 Kinase Deletion Suppresses Muscle Growth Adaptations to Nutrient Availability by Activating AMP Kinase. Cell Metabolism. 2007;5:476–487. doi: 10.1016/j.cmet.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Shima H, et al. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohamed-Ali V, et al. Adipose tissue as an endocrine and paracrine organ. Int J Obes Relat Metab Disord. 1998;22:1145–1158. doi: 10.1038/sj.ijo.0800770. [DOI] [PubMed] [Google Scholar]
- 66.Yeh WC, et al. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11086–11090. doi: 10.1073/pnas.92.24.11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bell A, et al. Rapamycin Inhibits Human Adipocyte Differentiation in Primary Culture. Obesity. 2000;8:249–254. doi: 10.1038/oby.2000.29. [DOI] [PubMed] [Google Scholar]
- 68.Cho HJ, et al. Regulation of adipocyte differentiation and insulin action with rapamycin. Biochemical and Biophysical Research Communications. 2004;321:942–948. doi: 10.1016/j.bbrc.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 69.Gagnon A, et al. Rapamycin-sensitive phase of 3T3-L1 preadipocyte differentiation after clonal expansion. John Wiley & Sons, Inc.; 2001. pp. 14–22. [DOI] [PubMed] [Google Scholar]
- 70.El-Chaar D, et al. Inhibition of insulin signaling and adipogenesis by rapamycin: effect on phosphorylation of p70 S6 kinase vs eIF4E-BP1. Int J Obes Relat Metab Disord. 2004;28:191–198. doi: 10.1038/sj.ijo.0802554. [DOI] [PubMed] [Google Scholar]
- 71.Kim JE, Chen J. Regulation of Peroxisome Proliferator-Activated Receptor-γ Activity by Mammalian Target of Rapamycin and Amino Acids in Adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 72.Le Bacquer O, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. The Journal of Clinical Investigation. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carnevalli LS, et al. S6K1 Plays a Critical Role in Early Adipocyte Differentiation. Developmental cell. 2010;18:763–774. doi: 10.1016/j.devcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Polak P, et al. Adipose-Specific Knockout of raptor Results in Lean Mice with Enhanced Mitochondrial Respiration. Cell Metabolism. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 75.Laplante M, Sabatini DM. An Emerging Role of mTOR in Lipid Biosynthesis. Current Biology. 2009;19:R1046–R1052. doi: 10.1016/j.cub.2009.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chakrabarti P, et al. Mammalian Target of Rapamycin Complex 1 Suppresses Lipolysis, Stimulates Lipogenesis, and Promotes Fat Storage. Diabetes. 2010;59:775–781. doi: 10.2337/db09-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cherrington AD. Banting Lecture 1997. Control of glucose uptake and release by the liver in vivo. Diabetes. 1999;48:1198–1214. doi: 10.2337/diabetes.48.5.1198. [DOI] [PubMed] [Google Scholar]
- 78.Mordier S, Iynedjian PB. Activation of mammalian target of rapamycin complex 1 and insulin resistance induced by palmitate in hepatocytes. Biochemical and Biophysical Research Communications. 2007;362:206–211. doi: 10.1016/j.bbrc.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 79.Koketsu Y, et al. Hepatic overexpression of a dominant negative form of raptor enhances Akt phosphorylation and restores insulin sensitivity in K/KAy mice. Am J Physiol Endocrinol Metab. 2008;294:E719–E725. doi: 10.1152/ajpendo.00253.2007. [DOI] [PubMed] [Google Scholar]
- 80.Brown NF, et al. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 2007;56:1500–1507. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 81.Li S, et al. Bifurcation of insulin signaling pathway in rat liver: mTORC1 required for stimulation of lipogenesis, but not inhibition of gluconeogenesis. Proceedings of the National Academy of Sciences. 2010;107:3441–3446. doi: 10.1073/pnas.0914798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hisayuki Funahashi FT, Guan Jian-Lian, Kageyama Haruaki, Yada Toshihiko, Shioda Seiji. Hypothalamic neuronal networks and feeding-related peptides involved in the regulation of feeding. Anatomical Science International. 2003;78:123–138. doi: 10.1046/j.0022-7722.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 83.Schwartz MW, et al. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- 84.Weigle DS, et al. Recombinant ob protein reduces feeding and body weight in the ob/ob mouse. The Journal of Clinical Investigation. 1995;96:2065–2070. doi: 10.1172/JCI118254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Woods SC, et al. Chronic intracerebroventricular infusion of insulin reduces food intake and body weight of baboons. Nature. 1979;282:503–505. doi: 10.1038/282503a0. [DOI] [PubMed] [Google Scholar]
- 86.Cota D, et al. Hypothalamic mTOR Signaling Regulates Food Intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 87.Campfield LA, et al. Recombinant Mouse OB Protein: Evidence for a Peripheral Signal Linking Adiposity and Central Neural Networks. Science. 1995;269:546–549. doi: 10.1126/science.7624778. [DOI] [PubMed] [Google Scholar]
- 88.Catania C, et al. mTORC1 signaling in energy balance and metabolic disease. Int J Obes. 2010 doi: 10.1038/ijo.2010.208. [DOI] [PubMed] [Google Scholar]
- 89.Cota D, et al. The Role of Hypothalamic Mammalian Target of Rapamycin Complex 1 Signaling in Diet-Induced Obesity. J. Neurosci. 2008;28:7202–7208. doi: 10.1523/JNEUROSCI.1389-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blouet C, et al. Mediobasal Hypothalamic p70 S6 Kinase 1 Modulates the Control of Energy Homeostasis. Cell Metabolism. 2008;8:459–467. doi: 10.1016/j.cmet.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin X, et al. Dysregulation of insulin receptor substrate 2 in β cells and brain causes obesity and diabetes. The Journal of Clinical Investigation. 2004;114:908–916. doi: 10.1172/JCI22217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes & Development. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 93.Jaworski J, Sheng M. The growing role of mTOR in neuronal development and plasticity. Molecular Neurobiology. 2006;34:205–219. doi: 10.1385/MN:34:3:205. [DOI] [PubMed] [Google Scholar]
- 94.Dowling RJO, et al. Metformin Inhibits Mammalian Target of Rapamycin Dependent Translation Initiation in Breast Cancer Cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- 95.Shaw RJ, et al. The Kinase LKB1 Mediates Glucose Homeostasis in Liver and Therapeutic Effects of Metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ono H, et al. Activation of hypothalamic S6 kinase mediates diet-induced hepatic insulin resistance in rats. The Journal of Clinical Investigation. 2008;118:2959–2968. doi: 10.1172/JCI34277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sparks CA, Guertin DA. Targeting mTOR: prospects for mTOR complex 2 inhibitors in cancer therapy. Oncogene. 2010;29:3733–3744. doi: 10.1038/onc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ikenoue T, et al. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Facchinetti V, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.García-martínez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 102.Guertin DA, Sabatini DM. The Pharmacology of mTOR Inhibition. Sci. Signal. 2009;2 doi: 10.1126/scisignal.267pe24. pe24-. [DOI] [PubMed] [Google Scholar]
- 103.Sarbassov DD, et al. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Molecular Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]