Abstract
Synthetic regulatory proteins such as tetracycline (tet)-controlled transcription factors are potentially useful for repression as well as ectopic activation of endogenous genes and also for probing their regulatory mechanisms, which would offer a versatile genetic tool advantageous over conventional gene targeting methods. Here we provide evidence supporting this concept using Cd4 as a model. CD4 is expressed in Double Positive (DP) and CD4 cells but irreversibly silenced in CD8 cells. The silencing is mediated by heterochromatin established during CD8 lineage development via transient action of the Cd4 silencer; once established, the heterochromatin becomes self-perpetuating independently of the Cd4 silencer. Using a tet-sensitive Cd4 allele harboring a removable Cd4 silencer, we found that a tet-controlled repressor recapitulated the phenotype of Cd4–deficient mice, inhibited Cd4 expression in a reversible and dose-dependent manner, and could surprisingly replace the Cd4 silencer to induce irreversible Cd4 silencing in CD8 cells, thus suggesting the Cd4 silencer is not the (only) determinant of heterochromatin formation. On the other hand, a tet-controlled activator reversibly disrupted Cd4 silencing in CD8 cells. The Cd4 silencer impeded this disruption but was not essential for its reversal, which revealed a continuous role of the silencer in mature CD8 cells while exposing a remarkable intrinsic self-regenerative ability of heterochromatin after forced disruption. These data demonstrate an effective approach for gene manipulation and provide insights into the epigenetic Cd4 regulatory mechanisms that are otherwise difficult to obtain.
Introduction
Conventional gene knockout (KO) technologies based on deletion of DNA sequences suffer from several limitations. First, the KO is irreversible, making it impossible to determine if, for example, the malignancies and neurological disorders observed in p53 and MeCP2 KO mice, respectively, can be cured by restoring gene functions, a question of obvious biological and clinical relevance. Since the KO is irreversible, additional mouse models had to be created to address this question(1-3). Second, gene KO is in general unfit for inducing ectopic gene expression and hence for determining, for example, whether a master transcription factor (such as GATA3, T-bet or Th-PoK/cKrox) is sufficient for reprogramming cell identity; one often resorts to transfection or transgenic mice to address such questions (4-6). Third, while gene KO can reveal biological roles of a DNA sequence such as a cis-acting regulatory element, it provides little insight into its mechanisms of action.
Several methods have been devised to achieve reversible gene regulation. In one method, endogenous genes are modified so that their expression is now driven by regulatable synthetic transcription activators expressed from the endogenous regulatory elements, thus allowing for reversible gene regulation, but it is difficult to recapitulate the expression levels of the endogenous genes with the synthetic activators (7-10). Regulated expression of shRNA is also useful for reversible gene repression, but the repression is usually incomplete (2). Finally, removable transcription stop cassettes can be inserted into target genes, leading to constitutive KO that can be conditionally rescued (1, 3). This method, however, is not fit for inducing gene repression. Furthermore, none of these methods are fit for achieving ectopic expression or probing the molecular mechanisms of gene regulation.
A potential method for overcoming all three limitations in the conventional gene KO strategies would be to manipulate the endogenous genes using conditional regulatory proteins, the most popular of which are tetracycline (tet)-sensitive transcription factors such as tet-regulated Transcription Silencer (tTS) and tet-regulated Transcription Activator tTA (11, 12). tTS and tTA consist of the same bacterial tet-sensitive DNA binding domain fused to the mammalian KRAB repressor domain or the viral VP16 activation domain, respectively, and both proteins bind the cognate Tet-Response Element (TRE) only in the absence of Tet or its derivative Doxycycline (Dox). In addition, a modified version of tTA (rtTA) binds the TRE only in the presence of Tet or Dox (13). Surprisingly, although the Tet system is routinely used for reversible regulation of simple target/reporter genes bearing the TRE and a minimal promoter (11, 12), little is known about whether the system can directly control endogenous genes via perturbation of their natural regulatory mechanisms, and whether the perturbation, if achieved, can be exploited to probe the regulatory mechanisms. Indeed, two decades after the introduction of the Tet system, it has been applied to only three endogenous genes (Hoxa2, Htr1a and Mlc1) and in a rather preliminary manner (14-18). Specifically, tTS is shown to repress all three genes, with the reversibility of repression demonstrated only for Hoxa2 and Htr1a. Furthermore, ectopic expression was shown only for Mlc1, and mechanistic studies performed for none. A detailed characterization of the Tet system at these genes is hampered by a technical difficulty: the expression of these genes must be measured in embryos (for Hoxa2) or adult brains (for Htr1a and Mlc1). Additional studies using genes whose expression is readily quantifiable are therefore warranted to better characterize the system.
Two major subsets of T lymphocytes in the adaptive immune system are the CD4 helper and CD8 cytotoxic T cells that express the antigen coreceptor CD4 or CD8, respectively. CD4 acts to strengthen TCRαβ-MHC-II interaction and to help trigger downstream signaling cascade following TCRαβ ligation, thus playing essential roles in T cell activation and function. CD4 is also essential for CD4 lineage development. Intrathymic T cell development starts with DN (Double Negative) cells lacking CD4 or CD8 expression (Fig. 1A) (19, 20). DN clls develop through four successive stages, DN1-4. DN4 cells then up-regulate CD8 to become Immature Single Positive (ISP) cells before they express CD4 to become DP (Double Positive) cells. DP cells express an entire repertoir of TCRαβ, but only those cells expressing TCRαβ with moderate affinity/avidity for self-MHC are signaled to survive. These cells then develop into CD4 and CD8 Single Positive (SP) thymocytes based on the specificity of TCRαβ, with the MHC-II and MHC-I restricted TCRαβ coupled to the CD4 and CD8 lineages, respectively (21, 22). The survival and further maturation of DP cells are both controlled by TCR signaling and hence dependent on the antigen coreceptors, with CD4 selectively required for signaling by MHC-II-restricted TCRαβ and hence for CD4 lineage development.
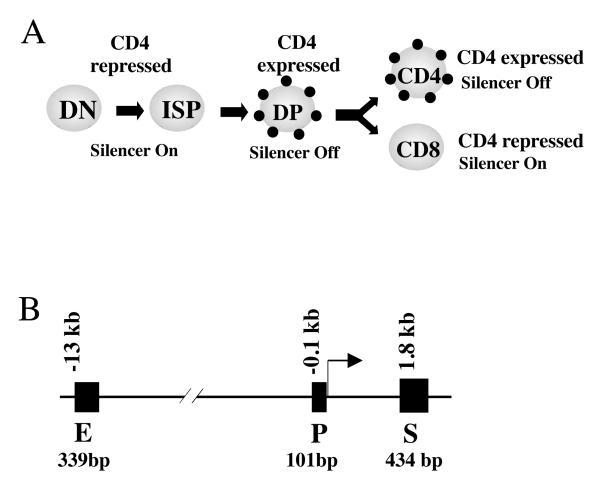
Fig. 1. Regulation of Cd4 expression during T cell development.
(A) The expression of CD4 (small dots), in conjunction with that of CD8 (not shown), marks the four major stages in T cell development, namely, Double Negative (DN), Immature CD8 SP (ISP), Double Positive (DP) and mature CD4/CD8 cells. CD4 is expressed in DP and CD4 cells, but repressed in the rest of the T cell types by the Cd4 silencer.
(B) The regulatory elements in the Cd4 locus. The Cd4 promoter (P) and enhancer (E) are constitutively active, but its activity in CD8 cells are suppressed by the Cd4 silencer (S) located 1.8 kb downstream of the transcription start site (+1). The sizes of the regulatory elements are indicated below the DNA.
Given the biological importance of Cd4, much effort has been devoted to studying its mechanisms of regulation (23, 24). Cd4 transcription is driven by the Cd4 promoter and enhancer, but repressed by the Cd4 silencer (Fig. 1B). The silencer is inactive in DP and CD4 cells, allowing CD4 expression in these cells, but is active in DN and CD8 cells where it inhibits the Cd4 promoter/enhancer; in the absence of the silencer, both DN and CD8 cells express CD4. Interestingly, although the silencer is required to establish and maintain Cd4 repression DN cells, it seems only required to establish repression in CD8 cells during CD8 lineage development, because the silencer can be conditionally deleted in mature CD8 cells without causing CD4 derepression under physiological conditions (24-27). Thus, the Cd4 silencer induces two types of repression in T cells: reversible repression in DN cells and irreversible repression or epigenetic silencing in CD8 cells. Epigenetic Cd4 silencing is thought to be mediated by certain self-perpetuating repressive heterochromatin structure at the Cd4 locus, although little is known about the molecular feature of this structure (24-28). The Cd4 silencer is perhaps the only silencer in mammalian cells known to be capable of epigenetic silencing. It is unclear how the Cd4 silencer establishes epigenetic silencing in the CD8 lineage, whether the Cd4 silencer has any potential role in maintaining Cd4 repression in CD8 cells under special, more challenging conditions, or how stable the heterochromatin is. Such questions are difficult to address using conventional genetic or molecular approaches.
Here, we characterized the behavior of the Tet system at the Cd4 gene in detail, and used the system to address the mechanistic questions mentioned above. Our work is made possible in part by the ability to quantify CD4 expression at the single cell level in a non-lethal manner. Our data illustrate the power of the Tet system for controlling the expression of endogenous genes and probing their physiological regulatory mechanisms.
Materials and Methods
Mice
The construct for producing the Cd4* allele was generated using standard molecular biology techniques on the pEZ FRT plasmid backbone (a gift of K. Rajewsky), where the left and right arms flanking the Cd4 silencer were PCR-amplified from C57B6 mice. The construct including the PCR-generated DNA was verified by sequencing. ES cells (from 129Sv strain) were selected on G418 and screened by PCR. Founder mice carrying the targeted allele were mated with a line ubiquitously expressing Flpe (a gift from Dr. Flavell) to delete the neomycine resistance cassette, thus generating the Cd4* allele. To repress Cd4*, the mice were mated with a transgenic line widely expressing tTS from the b-actin promoter (a gift from M. Mallo)(15). To activate Cd4*, the Cd4* mice were mated with a transgenic line ubiquitously expressing rtTA from the Rosa-26 locus (a gift from R. Jaenisch (29)). As the Rosa26-rtTA and the Cd4 loci are both situated on chromosome 6 (but separated by ~1 million base pairs or 1 cM), to obtain Cd4*/*; rtTA+/+ mice homozygous for the Cd4* allele and the rtTA transgene, we used meiotic recombination to generate a rearranged chromosome 6 harboring both alleles. After screening ~100 pups from Cd4*/+; rtTA+/− interbreeding, we obtained two Cd4*/*; rtTA+/− founders, which were used to produce the Cd4*/*; rtTA+/+ offspring. No difference in CD4 regulation was detected between the two lineages, and so one of lineages was used for all subsequent experiments. Mice expressing ER-Cre (from the Rosa 26 promoter) were provided by Dr. Thomas Ludwig (Columbia University) (30-32). All mice analyzed were on the C57/B6x129Sv background.
FACS analysis of lymphocytes
Adult (4-8 weeks-old) mice were used. In general, cells were stained with anti-CD4-APC, anti-CD8-PE-Cy7, anti-B220-PE and anti-TCR-FITC or anti-CD3-pacific blue. However, to determine CD4 expression throughout T cell development (Figs. 3A and 4A), thymocytes were stained with anti-CD4-APC, anti-CD8-PE-Cy7, anti-CD25-PE, anti-CD44-FITC and anti-CD3-pacific blue and the data, collected on the LSRII flow cytometers, were analyzed as described (33-35). The FACS plots shown in the paper were representatives of at least total 3 mice per condition; for the most part, these mice were analyzed in several independent experiments.
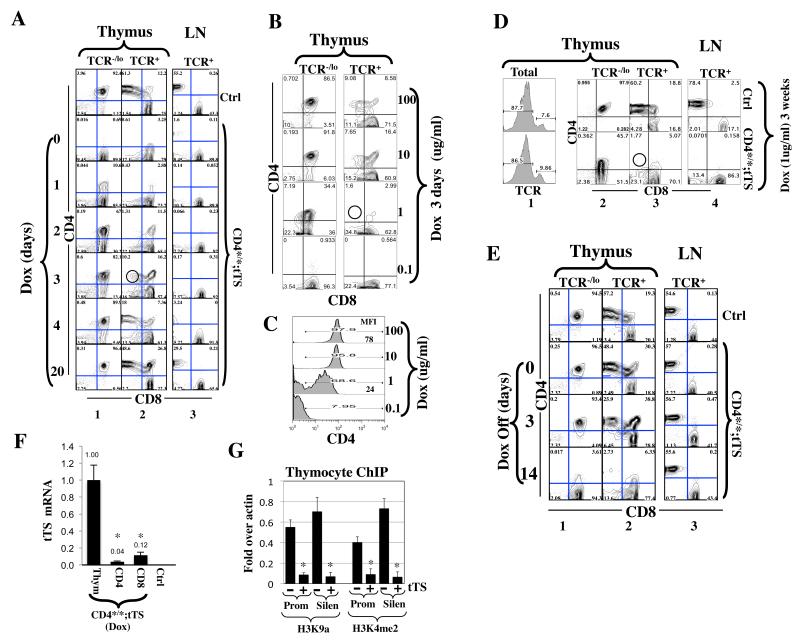
Fig. 3. Reversible and graded Cd4> repression in Cd4*/*; tTS+/− mice.
(A)tTS reversibly repressed Cd4. Mice were grown to adulthood in the absence of Dox. Dox treatment (100ug/ml in drinking water) was then sequentially initiated on five mice before they were simultaneously sacrificed and analyzed together with a control (Cd4*/*) mouse; by then, individual mice have been kept on Dox for various days as indicated. Thymocytes and lymph node (LN) cells were stained for TCR, CD4 and CD8 before TCR−/lo and TCR+ subsets of thymocytes (columns 1-2) and TCR+ LN cells (column 3) were analyzed for CD4/CD8 expression.
(B-C)Dose-dependent Cd4 repression in DP cells. Mice were exposed for 3 days to drinking water containing various concentrations of Dox and analyzed as in (A). Fig. 3C quantifies CD4 expression in “DP” (TCR−/lo CD8+) cells, where the numbers in blue are the mean fluorescence intensity (MFI). The red circle in (B) denotes the absence of TCR+CD4+CD8lo intermediate cells.
(D)Same as (B), except mice were exposed to Dox (1 ug/ml) for 3 weeks. The red circle denotes the absence of TCR+CD4+CD8lo intermediate cells.
(E)tTS acting in adult mice terminated ongoing CD4 transcription in DP but not CD4 cells. Mice were raised in the presence of Dox (5 ug/ml). Dox was discontinued upon weaning and mice analyzed at various times thereafter. Cd4*/* mice were used as control (Ctrl).
(F)qRT-PCR comparing tTS expression in thymocytes, peripheral CD4 and CD8 cells isolated from Cd4*/*; tTS+/− mice raised as in Dox was discontinued upon weaning and cells harvested 14 days thereafter. Two mice were analyzed, with the averaged values plotted relative to the expression level in thymocytes. Thymocytes from Cd4*/* mice were used as a negative control (Ctrl), where the tTS message was undetectable as expected. Asterisks indicate significant differences (p<0.02) in tTS expression between thymocytes and CD4 or CD8 cells.
(G)tTS induced epigenetic changes at the Cd4 locus in thymocytes. Cells were isolated from Cd4*/* mice (−) or Cd4*/*; tTS+/− mice (+). H3K9a and H3K4me2 levels at the CD4 promoter (Prom) and silencer (Silen) were plotted relative to that at the b-actin. Values were averaged from two mice, with the error bars indicating standard deviations between the two samples and asterisks indicate statistically significant effects of tTS (p<0.05).
Fig. 4. rtTA induced ectopic CD4 expression.
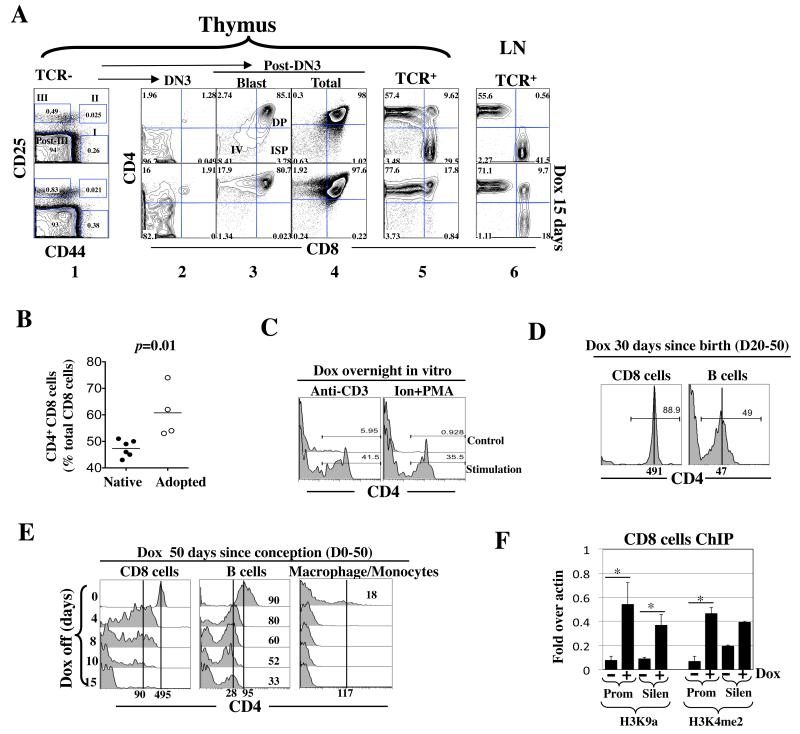
(A) Adult Cd4*/+; rtTA+/− mice were exposed to Dox (1 mg/ml) for 15 days. Thymocytes were stained for TCR, CD4, CD8, CD25 and CD44 and analyzed as described (33-35). Briefly, the TCR−/lo cells were first resolved into 4 subsets, DN1 (I), DN2 (II), DN3 (III) and post-DN3 (Post-III) based on CD25/44 expression (column 1) before each subset was analyzed for CD4/CD8 expression (columns 2-5); the post-DN3 cells consist of DN4 (IV), immature single positive (ISP) and DP cells, where the DN4 and ISP cells are rare proliferating blasts that are clearly detectable only by gating on the large cells within the post-DN3 compartment (column 3). Shown is a representative of three mice from a single experiment.
(B) CD4 induction in CD8 cells in the peripheral blood in adult Cd4*/*; rtTA+/+ mice (left) or in Rag2 KO mice adoptively transferred with CD8 cells isolated from Cd4*/*; rtTA+/+ mice (right). Mice were exposed to Dox (1 mg/ml) for 17 days. Blood was drawn from the tail vein at various times and the cells stained for CD4, CD8 and TCR before analysis. Data are pooled from two independent experiments. Each symbol represents a mouse.
(C) TCR activation facilitated rtTA-mediated Cd4 induction in CD8 cells. Peripheral blood cells from Cd4*/*; rtTA+/+ mice were stimulated overnight in vitro with plate-bound anti-CD3/28 (left) or 1 uM ionomycin plus 10ng/ml PMA (right) in the presence of Dox (100ng/ml). CD4 expression in CD8 cells was analyzed.
(D) Cd4*/*; rtTA+/+ mice were exposed to Dox from birth to adulthood before analysis of CD4 expression in the peripheral blood.
(E) Cd4*/*; rtTA+/+ mice were exposed to Dox from conception to adulthood. Dox was then discontinued and CD4 expression in peripheral blood cells monitored, where macrophages/monocyte were identified by their large size.
(F) rtTA induced epigenetic changes at the Cd4 promoter and silencer in peripheral CD8 cells. Cd4*/*; rtTA+/+ mice were exposed to Dox for 1 week (+), and the peripheral CD8 cells expressing CD4 were isolated and analyzed as described in Fig. 3G. CD8 cells from the mice lacking Dox exposure (−) were used as a control. Asterisks indicate statistically significant effects of Dox (p<0.05).
Adoptive transfer
CD8 cells were magnetically purified from Cd4*/*; rtTA+/+ mice using Dynabeads Flowcomp mouse CD8 purification kit (Invitrogen). The purity was ~95%. 1 million cells (in 100 ul PBS) per mouse were injected retro-orbitally into Rag2 KO mice, and Dox exposure (1mg/ml) initiated one month thereafter.
RT-PCR
LN CD4 and CD8 cells were purified to near homogeneity using FlowComp Dynabeads (Invitrogen). RNA was isolated with RNAeasy plus (Qiagen). tTS and CD4 mRNA were quantified using QuantiTect SYBR Green RT-PCR Kits (Qiagen). PCR were done in triplicate, and the values normalized to β-actin. PCR primers are:
tTS: CTCGCCCAGAAGCTAGGTGTAGAGC and CGTAATAATGGCGGCATACTATCAG
Cd4: AAGTCTCGAGCCCTCATATACACACA and CTCGGCACATGGTGGTCTCCTTGAGC
β-actin: GTCCACCCGCGAGCACAGCTTCTT and CTTTGCACATGCCGGAGCCGTTGTC
ChIP
ChIP was performed essentially as described (28), but with important modifications that significantly reduced nonspecifically precipitated signals. Briefly, lymph node cells were fixed with 1% formaldehyde at room temperature for 5 min, and cells resuspended in RIPA (0.8) before chromatin was sheared to ~300 bp with a Bioruptor sonicator. Chromatin from 1 million cells was diluted with equal volume of RIPA (0) before the addition of histone antibodies (0.5 ug, SABiosciences), salmon sperm DNA (final 1 ug/ml) and BSA (final 1 ug/ml). The samples were rocked at 4°C overnight, and precipitated removed by centrifugation. 10 ul Dynal protein G magnetic beads preblocked with salmon DNA and BSA were added to the supernatant and the samples rotated for 3 hrs. The beads were washed sequentially in RIPA (0.3), RIPA (0) and TE. Then 10 ul DirectPCR DNA Extraction Buffer (Viagen Biotech) containing 1 ug/ul proteinase K was added, and the samples incubated 60°C overnight. The samples were diluted with 40 ul H2O and used directly for real time multiplex PCR which quantified the Cd4 promoter and silencer together with the internal control b-actin. Input DNA was quantified in parallel, which yielded % Input for both target sequences and b-actin. The % input of the ChIP DNA was then plotted relative to the % input of b-actin as “ fold over actin”. Of note, using the magnetic beads, DNA nonspecifically precipitated by rabbit IgG-beads was negligible and hard to quantify, and thus ignored. Anti-H3K9ac and H3K4me2 were from SABiosciences (#GAM-1209 and GAM-3203, respectively). The primer pairs and probes (Biosearch Technologies) are:
CD4 silencer primers: GCAGCCACTGAACCACAAG, TCTGTTGCCTAGCTTCGTATAAAC,
CD4 silencer probe: CAL Fluor Red 610 TCGCTTAGAGGGTCACATCTCTAGGA
CD4 promoter primers: TGAGGGCTGGCTTACGTC, TTTGCCTTCACAGATGTTCGTA
CD4 promoter probe: Quasar 670 CGTGCAGAGGAGCCTCACGAC
b-actin primers: CAGGCCTAGTAACCGAGACATTG and GCTCCGTAGGCCCAGATGT
b-actin probe: FAM TGTCCACAAGGGCGGAGGCTATT
Conditional Cd4 silencer deletion
Tamoxifen solution (20mg/ml) was prepared by dissolving 200 mg tamoxifen (free base, sigma T5648) to 0.5 ml ethanol before adding 9.5 ml autoclaved peanut oil. The solution was sonicated and stored at −20°C. To delete the Cd4 silencer, 100 ul of the solution was injected i.p into adult Cd4*/*; ER-Cre+/− mice once a day for 3 consecutive days. To quantify the deletion efficiency, 50 ul of peripheral blood was drawn. RBC was lysed and CD8 cells purified to near homogeneity using FlowComp Dynabeads (Invitrogen). The cells were resuspended in 10 ul DirectPCR Lysis buffer (Viagen). QuantiTect SYBR Green PCR Kits (Qiagen) was used to quantify the Cd4 silencer and enhancer, the latter serving as an internal control. The primers have been described before (28).
DNA methylation of the Cd4 promoter
Genomic DNA (1ug) was bisulfite-converted using Epitech Bisulfite kit (Qiagen). The resulting DNA was amplified using PyroMark PCR Kit (Qiagen) with a primer pair targeting the Cd4 promoter; the primer sequences are GTGGGAGGGAGGGATTTT and CCAATCCTTCCTTCACTCTACTTCT. An internal primer (GGAGGGATTTTTGAGG) was then used to sequence the amplicon in the PyroMark Q24 system (Qiagen).
Results
The tet-sensitive Cd4 allele bearing a removable Cd4 silencer
To use the Tet system to manipulate Cd4 expression and explore its regulatory mechanisms, we created a bifunctional Cd4 allele (Cd4*) bearing the TRE and a floxed Cd4 silencer, which allows for Dox-mediated transcription regulation as well as Cre-mediated silencer deletion (Fig. 2A). The TRE was inserted immediately upstream of the Cd4 silencer, effectively creating an extended version of the Cd4 silencer, which could presumably avoid non-specific disruption of Cd4 regulation by the TRE insertion while allowing TRE-bound factors to impact the Cd4 promoter/enhancer via long-range interactions between the regulatory elements (36).
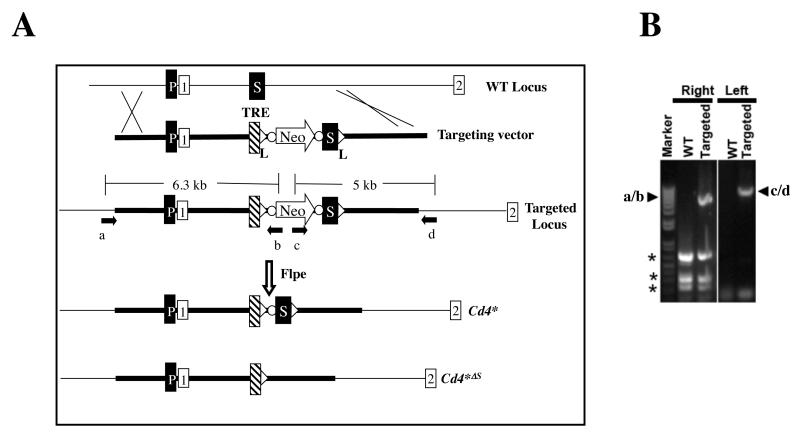
Fig. 2. Generation of Cd4* mice.
(A) The strategy. The Cd4 locus contains the Cd4 promoter (P) and silencer (S), the latter located between the first and second exons (while boxes). In the targeting construct, the TRE, together with a neomycin expression cassette (Neo), is inserted immediately upstream of the silencer. The Neo cassette is flanked by FRT sequences (dots) to allow for its Flpe-mediated excision and the subsequent generation of the Cd4* allele. The silencer in the Cd4* allele is floxed (triangles), allowing for Cre-mediated excision and the generation of the Cd4*ΔSallele. The arrows denote primers used for screening ES cells, where primers a/b and c/d amplify the junction sequences of the left and right integration sites, respectively.
(B) PCR analysis of a correctly targeted ES clone. Arrowheads indicate the PCR amplicons produced by primers a/b and c/d depicted in Fig. 2A. The amplicons were verified by sequencing. DNA from parental ES cells (WT) was used as a control. Asterisks, nonspecific amplicons. This ES cell clone was used to produce mice carrying the Cd4* allele.
tTS reproduces the phenotype in Cd4 KO mice and causes reversible and graded Cd4 repression in DP cells
We bred mice homozygous for the Cd4* allele and also carrying a transgene that widely expresses tTS from the human β-actin promoter (Cd4*/*; tTS+/−) (15). We found these mice recapitulated the phenotype of the Cd4 KO mice, namely CD4 lineage elimination due to loss of CD4 expression in DP cells (37), as indicated by the lack of TCR+ CD4 cells (columns 2-3, second row) and conversion of the normal DP (CD4+CD8+TCR−/lo) into CD4− CD8+TCR−/lo cells (Fig. 3A, column 1, second row). Furthermore, as in the case of Cd4 KO mice, a TCR+ CD4− CD8−/lo population with unknown function was present in the thymus and periphery (columns 2-3). Cd4 repression was reversible upon tTS inactivation by Dox: CD4 expression was restored on 10% of DP cells within 24 hr following Dox administration (100 ug/ml) and in most DP cells by day 4, with concomitant emergence of CD4 SP thymocytes (Fig. 3A, columns 1-2). By day 20, the thymus was indistinguishable from control mice, and substantial numbers of peripheral CD4 cells had accumulated (Fig. 3A, column 3). Of note, on Day 3, the “CD4 SP” thymocytes weakly expressed CD8 (Fig. 3A, red circle, row 5, column 2), suggesting they were the “intermediate cells”, i.e., positively selected DP cells that have recently terminated CD8 transcription (22, 38).
Having shown that tTS can fully repress Cd4 in DP cells, we examined the feasibility of graded Cd4 repression, by treating adult Cd4*/*; tTS+/− mice with decreasing concentrations of Dox (from 100 to 0.1 ug/ml) for 3 days. Dox as low as 10 ug/ml was able to fully inactivate tTS and restore Cd4 expression in >95% DP cells, while Dox at 1 ug/ml successfully produced a partial effect, in that CD4 was expressed only in 69% of DP cells and only to a low level (MFI was 24 units as compared with 78 units induced by Dox at 10 ug/ml) (Fig. 3B-C). Curiously, while high doses of Dox produced the TCR+CD4+CD8lo “intermediate cells” as previously described, such cells were absent in mice exposed to Dox at 1 ug/ml (red circle, Fig. 3B, row 3, column 2) even after prolonged (3 weeks) exposure (Fig. 3D). As CD4 transcription in the intermediate cells is weaker than DP cells (38) and thus perhaps more vulnerable to tTS, the “intermediate cells” might have completely lost CD4 expression and merged into the CD4−CD8−/lo TCR+ compartment that normally exist in Cd4*/*; tTS+/− mice. Indeed, as shown in Fig. 3B, cells in this compartment did seem to accumulate concomitantly with the loss of intermediate cells, reaching 35% in mice with 1 ug/ml Dox exposure, as opposed to 11- and 15% in mice with 100- and 10 ug/ml Dox exposures, respectively.
The data above indicate that tTS can prevent CD4 expression if acting constitutively throughout development. To determine whether tTS can also terminate ongoing CD4 transcription in preexisting CD4+ cells, Cd4*/*; tTS+/− newborns were raised to adulthood in the presence of Dox (5 ug/ml in drinking water to the mothers), which inactivated tTS and allowed for normal T cell development (Fig. 3E, column 1, Day 0). Dox was then removed and mice analyzed at various times thereafter. CD4 expression on DP cells began to decrease within 3 days following Dox withdrawal (column 1, Day 3), and was completely lost by 14 days (column 1), concomitant with the depletion of TCR+CD4 SP cells (column 2). Surprisingly, even at Day 14, there was no decrease in CD4 expression in peripheral CD4 cells at the protein (Fig. 3E, column 3) or RNA level (Fig. S1A). One possibility is that CD4 cells in vivo are mostly resting cells with compact chromatin, which may render the TRE inaccessible to tTS. This does not seem to be the case, because the CD4 cells, driven into proliferation with anti-CD3/CD28, retained CD4 expression after 5 days of culture in vitro (Fig. S1B). Surprisingly, tTS expression, driven by the b-actin promoter, was ~10-20-fold lower in peripheral T cells than thymocytes (Fig. 3F), which might explain the apparent tTS resistance in CD4 cells. Finally, we examined the effects of tTS at the chromatin level. The Cd4 promoter and silencer were acetylated in thymocytes (mostly DP cells), as we previously reported (28), and both elements were also enriched in H3K4me2, as expected (Fig 3G). Importantly, tTS depleted H3K9a and H3K4me2 at both the Cd4 promoter and the silencer in thymocytes, consistent with its being a transcription repressor (Fig. 3G).
We conclude that tTS can reproduce the phenotype in Cd4 KO mice, but is more versatile in that it can repress Cd4 (in DP albeit not CD4 cells) reversibly and dose-dependently.
Reversible ectopic Cd4 induction by rtTA
To determine whether rtTA can induce ectopic CD4 expression, we used a transgene (rtTA) that ubiquitously expresses rtTA from the Rosa-26 promoter (29). In adult Cd4*/+; rtTA+/− mice, following 15 days of Dox exposure (1mg/ml, which is around saturating dose (39)), CD4 was highly induced in all CD4− thymocyte subsets except the earliest ones, i.e., in DN4, ISP and CD8 SP but not DN1-3 cells (Fig. 4A, columns 1-5 and data not shown). In contrast to the thymocytes, peripheral CD8 cells were rather resistant to Dox, and Cd4 induction occurred in a stochastic, all-or-none rather than uniform manner, with ~30% of the cells fully expressing CD4 while the rest fully repressing CD4 (column 6), consistent with the pattern of gene de-repression resulting from probabilistic heterochromatin disruption. Finally, B cells and macrophages were completely resistant to Dox (not shown).
We next sought to increase the efficiency of ectopic Cd4 induction. Since these mice described above were heterozygous for the Cd4* allele and the rtTA transgene, we wondered whether homozygocity could potentiate the effect of rtTA (by doubling its cellular concentrations). We therefore generated mice homozygous for both the Cd4* allele and the rtTA transgene (Cd4*/*; rtTA+/+). As the Rosa26-rtTA and the Cd4 loci are both located on chromosome 6, we used meiotic recombination to create the Cd4*/*; rtTA+/+ mice (see Materials and Method). We then administered a saturating amount of Dox (1mg/ml) to adults and monitored Cd4 induction in peripheral blood. Dox induced Cd4 in CD8 cells in slowly and in an all-or-none manner, with CD4+ CD8 cells becoming detectable within two days and gradually reaching 47%+/−1.2 on Day 17 (Fig. 4B, filled circles) with MFI being approximately 320 (data not shown), which is indeed more efficient than the heterozygous Cd4*/+; rtTA+/− mice. However, since Dox also induced Cd4 in CD8 SP thymocytes which were continuously exported into the periphery, the peripheral CD8 cell pool was contaminated with the recent thymic emigrants, thus confounding data interpretation. To address this issue, CD8 cells were isolated from Cd4*/*; rtTA+/+ mice and adoptively transferred into Rag2 KO mice. The mice were challenged with Dox one month thereafter, when CD8 cells had populated the peripheral blood (Fig. S2). Interestingly, Dox induced Cd4 on the transferred cells more efficiently than in Cd4*/*; rtTA+/+ mice, with the Cd4-expresssing cells reaching 63+/−6% on Day 17 (Fig. 4B, open circles). The higher induction frequency perhaps reflects the fact that adoptively transferred CD8 cells had undergone and perhaps were still undergoing homeostatic proliferation in the recipients, which might facilitate gene expression by, eg., decompacting chromatin. To explore this further, peripheral blood cells were stimulated overnight with an anti-CD3/28 antibody in vitro in the presence of Dox. Anti-CD3/28, known to activate TCR signaling and globally decondense chromatin in T cells, indeed greatly facilitated Cd4 induction, leading to CD4 expression in >40% of CD8 cells as compared with ~6% in the absence of stimulation (Fig. 4C, left). As expected, ionomycine and PMA, which can also activate TCR signaling, similarly enhanced Cd4 inducibility (Fig. 4C, right).
We conclude that Dox can rather effectively induce ectopic Cd4 expression in peripheral CD8 cells when adult Cd4*/*; rtTA+/+ mice were exposed to Dox for 17 days, and the induction is further enhanced by TCR signaling. However, neither the 17-days Dox-treatment of Cd4*/*; rtTA+/+ mice in vivo nor ionomycine plus PMA stimulation in vitro could effectively induce Cd4 in B cells or macrophages (not shown). In contrast, when mice were exposed to Dox for one month starting at birth, namely from Day 20-50 post-conception, Cd4 was successfully induced in 49% of B cells, although the expression level was low (MFI=47); under this condition, 89% of CD8 cells expressed CD4 to very high levels (Fig. 4D). Cd4 induction was most robust if Dox exposure was initiated prenatally: in mice exposed from conception till one month after birth (Day 0-50), CD4 was expressed in all CD8 cells and 90% of B cells to higher levels (MFI=495 and 95 units for CD8 and B cells, respectively; Fig. 4E, left and middle, top histograms), and furthermore, CD4+ macrophages/monocyte also emerged (Fig. 4E, top right). Interestingly, upon Dox withdrawal, while Cd4 induction was rapidly (within four days) reversed in macrophages/monocytes (Fig. 4E, right), in B and CD8 cells, the reversal proceeded in two distinct phases: within the first four days after Dox withdrawal, CD4 expression was rapidly and rather uniformly reduced by 3-6 fold (MFI from 495 to 90 in CD8 and from 95 to 28 in B cells), which was followed by the second phase (Day 4-15) where the CD4 expression level no longer declined but the frequencies of CD4-expressing cells dwindled, the kinetics of which was slow especially in B cells where CD4 expression still lingered on in 33% of B cells as late as 15 days after Dox withdrawal (Fig. 4E, left and middle); the mechanisms of the reversal in the two phases may differ (see Discussion).
Finally, we examined histone modifications in CD8+CD4+ cells produced after 1 week of Dox exposure. In unexposed CD8 cells, H3K9a and H3K4me2 were depleted at the Cd4 promoter and silencer, whereas both markers were increased following Dox stimulation (Fig. 4F). Thus, the effect of rtTA (in CD8 cells) is opposite to the effects of tTS (in thymocytes; Fig. 3G), consistent with their respective roles in Cd4 regulation.
We conclude that rtTA can reversibly induce ectopic CD4 expression in multiple cell types, but the efficiencies are cell-type specific and also dependent on functional states of the cells.
tTS triggers epigenetic Cd4 silencing in CD8 and early T cells
We next used the Tet system to address biological questions regarding Cd4 silencing. One of the most interesting features of Cd4 regulation during T cell development is that the Cd4 silencer can induce irreversible Cd4 repression (i.e., epigenetic silencing) in CD8 cells. It has been proposed that the silencer achieves this feat by recruiting special repressors to establish heritable heterochromatin at the Cd4 locus; the heterochromatin, once formed, can then maintain Cd4 repression independently of the Cd4 silencer (24-26). However, it remains possible that the Cd4 locus in CD8 cells is uniquely prone to the formation of heterochromatin. In this scenario, even a repressor such as tTS typically only capable of reversible repression might cause epigenetic Cd4 silencing in CD8 cells.
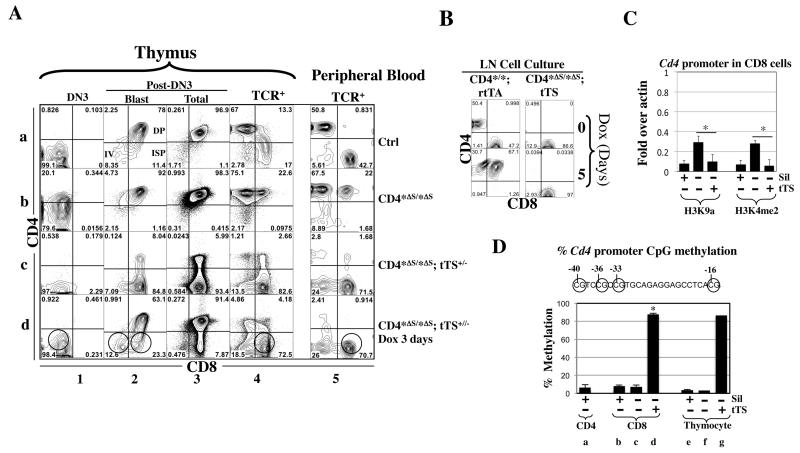
To test this idea, we created the Cd4* allele lacking the Cd4 silencer (Cd4*ΔS, Fig. 2A) by deleting the Cd4 silencer in the germline using a transgenic line that constitutively expresses Cre (gift from Dr. R. Flavell). Silencer deletion de-repressed Cd4 in all CD4− T cells, including the early (DN3, DN4, ISP) and CD8 T cells (Fig. 5A, row b). tTS restored Cd4 repression in these cells while also inhibiting Cd4 in DP cells (column 3, row c; note that some DP cells escaped the repression perhaps because the mice were hemizygous for the tTS transgene as opposed to the homozygous mice in Fig. 3A). To determine whether the tTS-mediated repression was reversible, we treated the mice with Dox (100 ng/ml) for 3 days. Dox restored CD4 expression in DP but importantly, not in the other T cells including CD8 cells, demonstrating that tTS had epigenetically silenced Cd4 in these cells (circles, row d). Physiological Cd4 silencing induced by the Cd4 silencer is known to be mitotically stable, surviving multiple rounds of cell division(27). To determine the mitotic stability of tTS-induced silencing, LN cells from Cd4*Δs/*Δs; tTS mice were stimulated in vitro with plate-bound anti-CD3/CD28, and cells cultured for 5 days in the presence of Dox (100ng/ml) and IL-2. Cells from Cd4*/*; rtTA mice were used as a control for the effect of Dox. In these mice, Dox induced Cd4 in all CD8 cells, as expected (Fig. 5B, left). In sharp contrast, in CD8 cells from Cd4*Δs/*Δs; tTS mice, Cd4 remained completely silenced under this condition, demonstrating tTS-triggered silencing was mitotically stable as in the case of the physiological silencing.
Fig. 5. tTS epigenetically silenced Cd4 in CD8 and early T cells at the Cd4* allele lacking the Cd4 silencer.
(A) Effects of tTS on CD4 expression in mice homozygous for the CD4*Δs allele. Cd4*/* mice were used as control (Ctrl). Cells were analyzed as in Fig. 4A. The red and green circles mark the cells with irreversible and reversible Cd4 repression, respectively. Note that in the presence of tTS, the fluorescence from the anti-CD4 antibody in DN3 cells was even weaker than the control (column 1, rows c vs. a), which is unrelated to Cd4 repression because the DN3 cells in the control mice do not express CD4.
(B)Mitotic stability of tTS-induced silencing. LN cells from Cd4*/*; rtTA+/+ or Cd4*Δs/*Δs; tTS+/− mice were stimulated with anti-CD3/CD28 and cultured in the presence of Dox (100ng/ml) and IL-2. Of note, after 5 days of culture, CD8 cells had preferentially expanded relative to CD4 cells (compared left top with left bottom; see also Fig. S1B). Cells were analyzed as in Fig. S1B.
(C)ChIP assays comparing the effects of Cd4 silencer vs. tTS on histone modifications at the Cd4 promoter in CD8 cells. CD8 cells were isolated from Cd4*Δs/*Δs (Sil−tTS−) or Cd4*Δs/*Δs; tTS+/− (Sil−tTS+) mice, with the former CD8 cells all de-repressing CD4. Values were averaged from 2-3 mice. The values for the control CD8 cells (Sil+ tTS−) were copied from Fig. 4F. Asterisks indicate significant effects of tTS (p<0.05).
(D) Pyrosequencing measuring DNA methylation at the Cd4 promoter (−101 to +1) harboring four CpGs (circles). The methylation levels at these CpG were similar and the values averaged. For each condition, two mice are analyzed and the mean values, except that Conditions f and g involve one mouse. Mouse genotypes are labeled as in Fig. 5C. Asterisks indicate significant effects of tTS.
To address the mechanism of action of tTS, we first compared the effects of tTS vs. the Cd4 silencer on H3K9a and H3K4me2 at the Cd4 promoter in CD8 cells. The markers were absent in normal CD8 cells (as described in Fig. 4F) and enriched by silencer deletion, but this enrichment was abolished by tTS (Fig. 5C). Thus, tTS can replace the Cd4 silencer to remove the two markers. To gain further mechanistic insights, we examined CpG methylation at the Cd4 promoter. It was reported that the Cd4 promoter is highly methylated in CD8 cells (27), and so we wondered whether tTS could similarly induce Cd4 promoter methylation. Unexpectedly, we found little methylation in CD8 cells in the control mice with an intact Cd4 silencer (Fig. 5G, group b), the level (~10%) being comparable to that in CD4 cells (group a) and total thymocytes (group e). Also, deleting the Cd4 silencer did not further reduce the methylation (group c), indicating the silencer was not responsible for this baseline-level methylation. In contrast, in the presence of tTS, we observed near maximal (~87%) Cd4 promoter methylation in CD8 cells (group d) and thymocytes (group g). This result suggests that tTS induced Cd4 promoter de novo methylation. Alternatively, under the physiological condition, the Cd4 promoter may be “naturally” methylated in the DN cells but becomes de-methylated in DP cells and remains so in CD8 cells. In this scenario, tTS acts not by inducing de novo methylation, but by preventing de-methylation, and future studies are needed to distinguish between the two possibilities. Despite this uncertainty, the data strongly suggest that the mode of action of tTS is different from that of the Cd4 silencer, even though tTS and the silencer can both trigger irreversible epigenetic silencing, which has mechanistic implications (see Discussion). Of note, CpG methylation, if necessary, was not sufficient for tTS-mediated epigenetic silencing, given that tTS also methylated the Cd4 promoter in DP cells and yet the repression in DP cells was reversible.
We conclude that tTS can replace the Cd4 silencer to trigger epigenetic Cd4 silencing in CD8 cells even though tTS is typically a reversible repressor and seems to repress Cd4 via a mechanism distinct from that used by the Cd4 silencer.
Roles of the Cd4 silencer vs. heterochromatin in maintaining epigenetic Cd4 silencing in CD8 cells
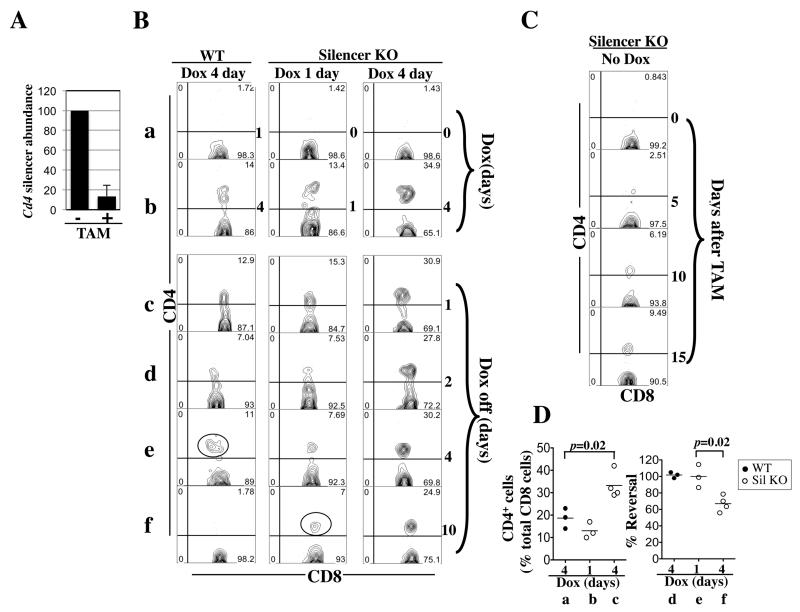
We sought to expose the role, if any, of the Cd4 silencer in maintaining Cd4 repression in CD8 cells. To this end, we used rtTA to challenge the Cd4 locus before and after conditional deletion of the Cd4 silencer in CD8 cells. Conditional deletion was achieved with a transgene ubiquitously expressing ER-Cre, a fusion protein between Cre and modified estrogen receptor that is normally trapped in the cytoplasm until stimulation by the estrogen analog Tamoxifen (TAM) (30-32). In mice carrying the Cd4* allele and expressing both rtTA and ER-Cre (Cd4*/+; rtTA+/−; ER-Cre+/-), three consecutive TAM injections induced Cd4 silencer deletion in virtually all of peripheral CD8 cells (Fig. 6A). Twenty-four hours after the last injection, we challenged the mice with Dox (1mg/ml) for various times and monitored CD4 expression in peripheral blood CD8 cells. In the control mice without TAM treatment, the frequencies of CD4+CD8 cells remained at the baseline level one day after Dox administration and reached ~ 19+/−3% on Day 4 (Fig. 6B, left column, rows a-b; Fig. 6D, group a). In contrast, in the mice with prior TAM treatment and hence silencer deletion, the frequency of CD4+CD8 cells already reached ~13+/−2% on Day 1 and was further increased to ~33+/−3% on Day 4 (row b, middle and right columns; Fig. 6D, group b-c). Thus, rtTA-mediated Cd4 induction was much faster in the absence of the silencer, revealing an important role of the silencer in protecting against rtTA-mediated disruption of Cd4 repression in CD8 cells. In contrast to CD8 cells, silencer deletion did not facilitate Dox-induced CD4 derepression in B cells, consistent with the fact that the silencer is inactive in B cells (not shown).
Fig. 6. Induction and reversal of CD4 expression in CD8 cells in Cd4*/+; rtTA+/−; ER-Cre+/− mice.
(A) qPCR quantifying Cd4 silencer in peripheral blood CD8 cells before (−) and after (+) TAM injection. TAM (2 mg) was injected daily for three days. The Cd4 silencer abundance 24 hours after the last injection was displayed as the percentage of that before injection. Data were averaged from three mice.
(B) Five-weeks-old mice were exposed to Dox (1mg/ml) for four days (column 1) or pretreated with TAM to remove the Cd4 silencer before Dox challenges for one or four days (columns 2-3). The FACS plots show the CD4/CD8 expression pattern of CD8 cells (CD8+TCR+) cells in the peripheral blood at various time points (as indicated at the right side of the plots) during the induction (rows a-b) and reversal (rows c-f) of CD4 expression. The cells marked by red circle in column 1 and 2 are probably recent thymic emigrants.
(C) Same as (A) except the mice were treated with TAM only.
(D) Data from two independent experiments were pooled to show the effects of silencer deletion on the induction (left) and reversal (right) of Cd4 expression in CD8 cells. The induction efficiency is the percentage of CD4+ CD8 cells among total CD8 cells at the end of Dox challenge. The reversal efficiency is the percentage of the CD4+ CD8 cells that had managed to restore Cd4 repression on Day 10 after Dox withdrawal, calculated after correcting for the averaged percentage of CD4+ CD8 cells observed in silencer-deleted mice lacking Dox challenge. A total of 3-4 mice in each group were analyzed.
We next determined whether the silencer is also important for restoring Cd4 repression after forced disruption by rtTA. In the control mice, CD4 expression was fully reversed by Day 10 following Dox withdrawal (Fig. 6B, left column, row f; Fig. 6D, group d). In fact, the reversal seemed complete within four days: although on Day 4 following Dox withdrawal, CD4 expression persisted in ~10% of CD8 cells (Fig. 6B, left column, row e, red circle), the expression level was higher than Day 1 or 2, suggesting these CD4+CD8 cells were not preexisting CD8 cells with lingering Cd4 expression but rather were recently generated CD8 cells that were unable to establish Cd4 silencing because of rtTA interference during development. Importantly, in TAM-treated mice with a brief (1-day) Dox exposure, Cd4 expression was reversed in a manner hardly distinguishable from the control mice (Fig. 6B, middle column), except for the presence of a CD4+CD8+ population (comprising 7% of CD8 cells) on Day 10 after Dox withdrawal (red circle, middle column, row e). These cells, however, were not derived from preexisting CD8 cells, but perhaps were recently exported CD8 cells that aberrantly expressed CD4 due to TAM-mediated premature Cd4 silencer deletion occurring before the establishment of Cd4 silencing, as indicated by the following data: in mice treated with TAM alone, the CD4+CD8+ cells slowly accumulated in the periphery, reaching ~6% of the CD8 population on Day 10 (Fig. 6C). Correcting for these recent thymic emigrants, we found that in mature CD8 cells where the silencer had been conditionally deleted, Cd4 expression induced by a brief (1-day) Dox exposure was readily reversible in the absence of the silencer (Fig. 6D, group e). Thus, the silencer was dispensable for effective restoration of Cd4 repression if the repression was disrupted by a brief Dox exposure. However, the silencer became important if Dox exposures were prolonged: in mice challenged with Dox for four days, CD4 expression persisted in 25% of CD8 cells on Day 10 after Dox withdrawal, which amounted to 15% of mature CD8 cells after correcting for the recent thymic emigrants (column 4, row f). Considering that immediately following the 4-day Dox exposure, CD4 was expressed on 35% of CD8 cells (column 4, row b), we estimate that ~40% of the silencer-deleted Cd4* alleles whose Cd4 repression were disrupted by the 4-day Dox challenge failed to restore Cd4 repression within 10 days after Dox withdrawal. Thus, the Cd4 silencer is important for efficient restoration of Cd4 repression after the prolonged, 4-day Dox challenge. However, under this condition, ~60% of the silencer-deleted Cd4* allele managed to restore the repression (Fig. 6D, group f; averaged value being 67+/−5%).
These data expose a role of the Cd4 silencer in stabilizing Cd4 repression in CD8 cells, but also reveal a remarkable potential of the heterochromatin to regenerate itself and restore Cd4 repression after forced disruption.
Discussion
This study represents the first in-depth characterization of the Tet system in the context of an endogenous gene and the first use of this system to probe its regulatory mechanism. Generally speaking, our study demonstrates the feasibility of controlling endogenous genes and probing their regulatory mechanisms via recruitment of synthetic regulatory proteins. This recruitment-based approach should have diverse applications. For example, one can recruit a histone-modifying enzyme to explore the role of a relevant histone mark, a nuclease to study DNA repair, or GFP for real-time imaging. Of note, since the Tet system can deliver any effector protein, it is adaptable for diverse purposes. Below we will discuss the characteristics of the Tet system when applied to Cd4 and the mechanistic insights this system generated regarding Cd4 regulation; such insights are difficult to obtain with any other tool.
Tet system as a tool for regulating endogenous genes: advantages, limitations and potential solutions
We show that the Tet system can reproduce the phenotype in Cd4 KO mice and is additionally capable of reversible Cd4 repression and reversible ectopic Cd4 activation, thus confirming its advantages over conventional gene KO methods. The limitation of the Tet system, as revealed in our study, is that the efficacy of gene regulation is variable among different cell-types. However, this variability may not be intrinsic to the Tet system (see further). Of note, efficiency of Cre-mediated deletion can also be cell-type specific resulting perhaps from variable accessibility of the LoxP sites (see e.g., (34, 40)), and just as in the case of the Cre-Lox system, the limitation in the Tet system is overweighed by the advantages; indeed, the Tet system is indispensable for certain applications and an attractive alternative to the conventional gene targeting methods. Below we will discuss in detail the cell-type specific variation in the efficiency of the Tet system.
First, tTS could repress Cd4 in DP but unexpectedly not in CD4 cells. While this may suggest that tTS function is cell-type specific, which would be an intrinsic weakness of the Tet system, we found that tTS expression in CD4 cells was 10-20 fold lower than that in thymocytes, pointing to a different but readily manageable culprit. tTS transcription in the transgene used in our study is directed by the ubiquitous human β-actin promoter, with the transgene flanked by the chicken β-globin insulator. Despite this, in situ hybridization assays reveal that while tTS is widely expressed in the mouse embryos, the expression levels vary significantly in different regions of the embryos (Fig. 1E in reference (15), consistent with variable tTS expression in thymocytes vs. peripheral T cells. Different mouse lines are therefore needed to direct uniform, high-level tTS expression.
Second, rtTA could readily induce ectopic Cd4 expression in some cell types (DN4, ISP and CD8) but not others (DN1-3 thymocytes, B cells and macrophages). This might reflect variation in rtTA expression, but the following data suggest a more significant possibility. Specifically, the ability of rtTA to induce Cd4 in various T cell subsets paralleled the effects of Cd4 silencer KO: in mice with germline silencer deletion, Cd4 was highly expressed in DN4, ISP and CD8 cells but not DN1-3 cells, presumably because the latter do not possess strong endogenous activators of Cd4 transcription (Fig. 5A). Since endogenous activators can facilitate rtTA function, their variation among different cell types can contribute to the variable rtTA sensitivity. Furthermore, endogenous repressors of Cd4 transcription, which can antagonize rtTA, may also be variable among different cell types, which might further diversify rtTA sensitivity. We should stress that despite this variability in the efficiency, rtTA successfully induced Cd4 in the diverse cell types examined.
Finally, rtTA-induced Cd4 expression was reversed very slowly in B cells as compared with CD8 or macrophages. Interestingly, in both B and CD8 cells, the reversal proceeded in two phases: a uniform drop in CD4 expression in all cells to a stable level, followed by shut-off of the remaining expression in a stochastic, all-or-none manner. The reversal in the first phase was rapid in both B and CD8 cells, and the rapid kinetics resembled that at simple tet-responsive transgenes (41-43), suggesting the reversal resulted from the departure of rtTA. But how was CD4 expression sustained thereafter, how was it stochastically terminated, and why was this termination so slow in B cells? We imagine that both B and CD8 cells contain endogenous activators of Cd4 transcription. These activators are presumably excluded from the Cd4 locus by the heterochromatin, but rtTA can disrupt heterochromatin (44-46) and open up the Cd4 locus, which would allow the activators to bind Cd4 and sustain its expression after rtTA’s departure. However, the damaged heterochromatin tends to regenerate itself to restore Cd4 repression (see further), which is expected to occur in the observed stochastic manner as in the case of other heterochromatin-mediated phenomena such as position effect variegation. This heterochromatin-mediated reversal is facilitated by the Cd4 silencer in CD8 cells but not in B cells where the silencer is inactive, thus explaining the slow reversal kinetics in B cells.
In summary, the Tet system is not equally effective in regulating Cd4 in different cell types, which might reflect tissue-specific tTS/rtTA expression and/or the extremely complex cell-type specific Cd4 regulatory mechanisms. The former problem is extrinsic to the Tet system, while the latter may be insignificant for genes with less complex regulatory mechanisms. Indeed, so far, the Tet system has been applied to 3 other endogenous genes in mice beside Cd4 (Hoxa2, Htr1a and Mlc1), and whenever tested, tTS and rtTA can effectively alter target gene expression (see Introduction). These data, together with ours, strongly suggest general applicability of the Tet system, which will be greatly facilitated by transgenic lines with various rtTA and tTA expression patterns, many of the lines already available (11). Furthermore, the efficiency of Tet system may be enhanced by, e.g., moving the TRE closer to the promoter, increasing the copy number of the cognate DNA binding sites for tet-controlled factors (there are 5 copies in our TRE), or altering the physiological state of the target cells (Fig. 4C). In addition, we found that a few days of exposures of Cd4*/*; rtTA+/+ mice to Dox during fetal development suffices to greatly facilitate subsequent rtTA-mediated Cd4 induction in adult mice (M/S under review), suggesting another method for improving the efficiency of ectopic induction.
The Cd4 promoter/enhancer in CD8 cells is predisposed to epigenetic silencing: the silencer is not the key?
tTS induced epigenetic Cd4 silencing in CD8 and early (DN and ISP) T cells. Paradoxically, tTS-mediated repression of simple transgenes in adult mice and tumor lines is reversible, as its repression of Hoxa2 in early mouse embryos (15, 47-49) and Cd4 in DP cells. Although tTS can irreversibly repress the ubiquitin and β-actin promoters in transgenic mice if it acts during first few days of embryogenesis (50), the epigenetic Cd4 silencing did not require fetal tTS action (not shown). These data raise the possibility that the Cd4 locus in CD8 and early T cells is predisposed to epigenetic silencing so that even a reversible repressor such as tTS can induce epigenetic silencing. The fact that tTS but not the Cd4 silencer affects CpG methylation in CD8 cells reinforces the notion that epigenetic Cd4 silencing can be induced via different mechanisms. We thus propose that the Cd4 silencer, just like tTS, is not the (only) determinant of epigenetic Cd4 silencing, which is testable by asking whether the silencer can trigger epigenetic silencing at heterologous genes in CD8 cells. It is of great interest to understand the putative silencer-independent mechanisms that predispose the Cd4 locus to epigenetic silencing.
Cd4 silencer, heterochromatin resilience and maintenance of Cd4 repression in CD8 cells
In CD8 cells, Cd4 silencer deletion facilitated rtTA-mediated disruption of Cd4 repression and could hamper its restoration, indicating that the Cd4 silencer continuously exerts repressive effects on CD4 transcription. Although the heterochromatin at the Cd4 locus is sufficient for Cd4 repression under normal conditions, the Cd4 silencer may become additionally required under pathological conditions such as defective heterochromatin formation or aberrant expression of Cd4 transcriptional activators.
While the Cd4 silencer could facilitate restoration of Cd4 repression after Dox challenges, the heterochromatin at the Cd4 locus was at least partially sufficient for the restoration, particularly when the Dox exposures were brief. Since Cd4 induction necessitates the disruption of the heterochromatin structure, the data reveal remarkable resilience of this structure that allows it to regenerate or “heal” itself after rtTA-inflicted damages. To our knowledge, this is the first demonstration of such resilience of heterochromatin. rtTA and the VP16 activation domain it carries has been used to disrupt heterochromatin at randomly integrated reporter templates in tumor cell lines (44-46) but the reversibility of the disruption has not been explored, let alone the contribution of heterochromatin resilience to this reversibility. Indeed, these systems are not fit for addressing heterochromatin resilience because the mechanisms that establish heterochromatin at the reporter genes persist after the establishment, analogous to the situation at the WT Cd4 locus that carries the Cd4 silencer, which would obscure the intrinsic property of heterochromatin. Of note, for the same reason, the ability of B cells to restore Cd4 repression after rtTA-mediated disruption does not prove the resilience of heterochromatin (or other forms of repressive chromatin), because the repressive chromatin structure is not established by the Cd4 silencer but rather by an unknown, presumably persistent mechanism. The molecular basis of heterochromatin resilience is a mystery, and the Cd4 locus in CD8 cells provides an excellent model for addressing this fundamental question, which has implications for the mechanisms of epigenetic memory and plasticity.
Supplementary Material
Acknowledgement
We thank T. Ludwig (Columbia University, New York, NY) for ER-Cre mice and K. Rajewsky for plasmids.
Footnotes
Author contributions
TC conceived the project and wrote the manuscript. MW and SL created the Cd4* mice. All authors participated in the analysis of the mice.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 2.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–60. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–7. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 5.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 6.Jenkinson SR, Intlekofer AM, Sun G, Feigenbaum L, Reiner SL, Bosselut R. Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J Exp Med. 2007;204:267–72. doi: 10.1084/jem.20061982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng H, Horie K, Madisen L, Pavlova MN, Gragerova G, Rohde AD, Schimpf BA, Liang Y, Ojala E, Kramer F, Roth P, Slobodskaya O, Dolka I, Southon EA, Tessarollo L, Bornfeldt KE, Gragerov A, Pavlakis GN, Gaitanaris GA. An inducible and reversible mouse genetic rescue system. PLoS genetics. 2008;4:e1000069. doi: 10.1371/journal.pgen.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin MK, Levorse JM, Ingram RS, Tilghman SM. The temporal requirement for endothelin receptor-B signalling during neural crest development. Nature. 1999;402:496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 9.Bond CT, Sprengel R, Bissonnette JM, Kaufmann WA, Pribnow D, Neelands T, Storck T, Baetscher M, Jerecic J, Maylie J, Knaus HG, Seeburg PH, Adelman JP. Respiration and parturition affected by conditional overexpression of the Ca2+-activated K+ channel subunit, SK3. Science. 2000;289:1942–6. doi: 10.1126/science.289.5486.1942. [DOI] [PubMed] [Google Scholar]
- 10.Gross C, Zhuang X, Stark K, Ramboz S, Oosting R, Kirby L, Santarelli L, Beck S, Hen R. Serotonin1A receptor acts during development to establish normal anxiety-like behaviour in the adult. Nature. 2002;416:396–400. doi: 10.1038/416396a. [DOI] [PubMed] [Google Scholar]
- 11.Schonig K, Bujard H, Gossen M. The power of reversibility regulating gene activities via tetracycline-controlled transcription. Methods in enzymology. 2010;477:429–53. doi: 10.1016/S0076-6879(10)77022-1. [DOI] [PubMed] [Google Scholar]
- 12.Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu Rev Genet. 2002;36:153–73. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 13.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 14.Ohnemus S, Bobola N, Kanzler B, Mallo M. Different levels of Hoxa2 are required for particular developmental processes. Mechanisms of development. 2001;108:135–47. doi: 10.1016/s0925-4773(01)00502-0. [DOI] [PubMed] [Google Scholar]
- 15.Mallo M, Kanzler B, Ohnemus S. Reversible gene inactivation in the mouse. Genomics. 2003;81:356–60. doi: 10.1016/s0888-7543(03)00032-6. [DOI] [PubMed] [Google Scholar]
- 16.Richardson-Jones JW, Craige CP, Guiard BP, Stephen A, Metzger KL, Kung HF, Gardier AM, Dranovsky A, David DJ, Beck SG, Hen R, Leonardo ED. 5-HT1A autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron. 2010;65:40–52. doi: 10.1016/j.neuron.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson-Jones JW, Craige CP, Nguyen TH, Kung HF, Gardier AM, Dranovsky A, David DJ, Guiard BP, Beck SG, Hen R, Leonardo ED. Serotonin-1A autoreceptors are necessary and sufficient for the normal formation of circuits underlying innate anxiety. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:6008–18. doi: 10.1523/JNEUROSCI.5836-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka KF, Ahmari SE, Leonardo ED, Richardson-Jones JW, Budreck EC, Scheiffele P, Sugio S, Inamura N, Ikenaka K, Hen R. Flexible Accelerated STOP Tetracycline Operator-knockin (FAST): a versatile and efficient new gene modulating system. Biological psychiatry. 2010;67:770–3. doi: 10.1016/j.biopsych.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Boehmer H, Aifantis I, Azogui O, Feinberg J, Saint-Ruf C, Zober C, Garcia C, Buer J. Crucial function of the pre-T-cell receptor (TCR) in TCR beta selection, TCR beta allelic exclusion and alpha beta versus gamma delta lineage commitment. Immunol Rev. 1998;165:111–9. doi: 10.1111/j.1600-065x.1998.tb01234.x. [DOI] [PubMed] [Google Scholar]
- 20.Shortman K, Egerton M, Spangrude GJ, Scollay R. The generation and fate of thymocytes. Semin Immunol. 1990;2:3–12. [PubMed] [Google Scholar]
- 21.Guidos CJ. Positive selection of CD4+ and CD8+ T cells. Curr Opin Immunol. 1996;8:225–32. doi: 10.1016/s0952-7915(96)80061-6. [DOI] [PubMed] [Google Scholar]
- 22.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellmeier W, Sawada S, Littman DR. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu Rev Immunol. 1999;17:523–54. doi: 10.1146/annurev.immunol.17.1.523. [DOI] [PubMed] [Google Scholar]
- 24.Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. doi: 10.1016/S0065-2776(04)83002-5. [DOI] [PubMed] [Google Scholar]
- 25.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–33. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 26.Taniuchi I, Sunshine MJ, Festenstein R, Littman DR. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol Cell. 2002;10:1083–96. doi: 10.1016/s1097-2765(02)00735-9. [DOI] [PubMed] [Google Scholar]
- 27.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, Littman DR. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–6. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 28.Yu M, Wan M, Zhang J, Wu J, Khatri R, Chi T. Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development. Proc Natl Acad Sci U S A. 2008;105:3873–8. doi: 10.1073/pnas.0800810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–8. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 30.de Luca C, Kowalski TJ, Zhang Y, Elmquist JK, Lee C, Kilimann MW, Ludwig T, Liu SM, Chua SC., Jr. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. The Journal of clinical investigation. 2005;115:3484–93. doi: 10.1172/JCI24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo K, McMinn JE, Ludwig T, Yu YH, Yang G, Chen L, Loh D, Li C, Chua S, Jr., Zhang Y. Disruption of peripheral leptin signaling in mice results in hyperleptinemia without associated metabolic abnormalities. Endocrinology. 2007;148:3987–97. doi: 10.1210/en.2007-0261. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med. 2005;202:1471–6. doi: 10.1084/jem.20051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–9. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 34.Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–82. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 35.Jani A, Wan M, Zhang J, Cui K, Wu J, Preston-Hurlburt P, Khatri R, Zhao K, Chi T. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J Exp Med. 2008;205:2813–25. doi: 10.1084/jem.20080938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang H, Peterlin BM. Differential chromatin looping regulates CD4 expression in immature thymocytes. Mol Cell Biol. 2008;28:907–12. doi: 10.1128/MCB.00909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahemtulla A, Fung-Leung WP, Schilham MW, Kundig TM, Sambhara SR, Narendran A, Arabian A, Wakeham A, Paige CJ, Zinkernagel RM, et al. Normal development and function of CD8+ cells but markedly decreased helper cell activity in mice lacking CD4. Nature. 1991;353:180–4. doi: 10.1038/353180a0. [DOI] [PubMed] [Google Scholar]
- 38.Brugnera E, Bhandoola A, Cibotti R, Yu Q, Guinter TI, Yamashita Y, Sharrow SO, Singer A. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- 39.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, Bujard H. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10933–8. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long MA, Rossi FM. Silencing inhibits Cre-mediated recombination of the Z/AP and Z/EG reporters in adult cells. PloS one. 2009;4:e5435. doi: 10.1371/journal.pone.0005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS genetics. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson A, Perea J, Tolmachova T, Thomas PK, Huxley C. Effects of mouse strain, position of integration and tetracycline analogue on the tetracycline conditional system in transgenic mice. Gene. 2002;282:65–74. doi: 10.1016/s0378-1119(01)00793-4. [DOI] [PubMed] [Google Scholar]
- 43.Mansuy IM, Winder DG, Moallem TM, Osman M, Mayford M, Hawkins RD, Kandel ER. Inducible and reversible gene expression with the rtTA system for the study of memory. Neuron. 1998;21:257–65. doi: 10.1016/s0896-6273(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 44.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–98. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol. 1999;145:1341–54. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Memedula S, Belmont AS. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr Biol. 2003;13:241–6. doi: 10.1016/s0960-9822(03)00048-4. [DOI] [PubMed] [Google Scholar]
- 47.Deuschle U, Meyer WK, Thiesen HJ. Tetracycline-reversible silencing of eukaryotic promoters. Mol Cell Biol. 1995;15:1907–14. doi: 10.1128/mcb.15.4.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. Journal of virology. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3:109–16. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 50.Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J Biol Chem. 2007;282:34535–41. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.