Abstract
Enteroaggregative Escherichia coli (EAEC) has been acknowledged as an emerging cause of gastroenteritis worldwide for over two decades. Epidemiologists are revealing the role of EAEC in diarrheal outbreaks as a more common occurrence than ever suggested before. EAEC induced diarrhea is most commonly associated with travelers, children and immunocompromised individuals however its afflictions are not limited to any particular demographic. Many attributes have been discovered and characterized surrounding the capability of EAEC to provoke a potent pro-inflammatory immune response, however cellular and molecular mechanisms underlying initiation, progression and outcomes are largely unknown. This limited understanding can be attributed to heterogeneity in strains and the lack of adequate animal models. This review aims to summarize current knowledge about EAEC etiology, pathogenesis and clinical manifestation. Additionally, current animal models and their limitations will be discussed along with the value of applying systems-wide approaches such as computational modeling to study host-EAEC interactions.
Keywords: enteroaggregative E. coli, animal model, computational modeling, EAEC pathogenesis, Th17
Introduction
Escherichia coli are classified as motile, rod-shaped, non-spore forming, Gram-negative Enterobacteriaceae. The majority of E. coli strains co-exist in the gastrointestinal tract as harmless commensal symbionts. Commensal E. coli strains colonize the gastrointestinal tract within hours of life and remain the most predominant facultative anaerobe within the colonic microflora of humans.1 However, disease-causing pathogenic E. coli strains have the ability to induce life-threatening illnesses that often require hospitalization and can result in death.2 Pathotypes known to induce enteric disease have been categorized into six groups: enteropathogenic E. coli (EPEC), enteroheamorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), diffusely adherent E. coli (DAEC), enteroinvasive E. coli (EIEC) and enteroaggregative E. coli (EAEC).3
Enteroaggregative Escherichia coli (EAEC) was first identified in the late 1980s as an enteric pathogen that causes diarrhea.4 Since its discovery, scientists have been studying host response to EAEC with aims to identify pathognomonic factors implicated in lesion formation in the gut and enteric disease. Increasing attention to EAEC has given rise to improved diagnostic techniques prompting more comprehensive epidemiological studies. For instance, a meta-analysis was conducted using all published literature about EAEC infections from 1987 through 2006 and revealed EAEC as a causative agent of diarrheal illnesses among many different subpopulations in both developing and industrialized regions worldwide.5 Etiological efforts have uncovered striking numbers of infectious cases identifying EAEC as the causative agent of diarrhea in travelers, children (especially malnourished populations) and immunocompromised individuals (specifically HIV-infected patients).6-8 Also, EAEC has been identified as a common cause of acute diarrheal illness in children and adults in inpatient and emergency units throughout the United States.9 The alarming rise in attention to EAEC led to its inclusion on the National Institutes of Health category B list of infectious organisms of potential importance as a bioterrorism weapon in 2002.10 In May 2011 an outbreak of E. coli O104:H4 occurred in Germany where more than 4,000 people became victims to infection and 54 of these cases resulted in death; the highest frequency of deaths ever recorded for an E. coli outbreak.11 Nucleotide analysis of the genome sequence classified E. coli O104:H4 within the EAEC pathotype though it was Shiga-toxin (Stx2) producing. This hybrid strain acquired the phage-borne gene encoding Stx2, a characteristic associated with EHEC strains, likely through lateral gene transfer providing clear evidence for enhanced virulence and detrimental effects caused by emerging heterogeneity among strains.12
Transmission of EAEC is most commonly associated with contaminated food and water. In Mexico, EAEC is the most common bacterial pathogen isolated from food.13 Poor sanitation and crowded living conditions increase the propensity for EAEC to spread.14 Recent research has identified food handlers, especially those working in tourist hotels, as primary carriers of EAEC. Over 65% of the isolates from these individuals are multidrug resistant thus posing a significant public health threat.15 Furthermore, the prevalence of EAEC induced travelers’ diarrhea throughout winter and summer seasons remains constant unlike other diarrheagenic E. coli strains such as ETEC, EPEC and EIEC whose rate of infection significantly decreases in lower temperatures.16 Genetic predisposition has also been alluded to in EAEC susceptibility. Single-nucleotide polymorphisms (SNPs) in the IL-8 gene promoter have proven to be associated with increased incidence of EAEC-associated diarrhea, and individuals with lactoferrin SNPs have higher susceptibility to traveler’s diarrhea.17,18
According to the CDC’s 2011 estimates, diarrheal episodes and enteric infections caused by foodborne illness affect an estimated 47.8 million people annually in the United States alone, from which approximately 130,000 people seek hospitalization and 3,000 cases result in death (www.cdc.gov). EAEC is one of the primary, if not most common, bacterial instigator of diarrheal illness in people from industrialized and developing countries around the globe including the United States, especially children.19-24 Yet despite EAEC outbreaks and many years of high-level research, the disease pathogenesis remains widely unknown. This review will highlight known pathogenicity factors, describe host responses to disease and discuss current animal models. Lastly, an emphasis on the necessity for an integrated immunoinformatics approach that combines computational immunology and animal experimentation will be discussed. This review aims to prompt future perspectives and advancements for safe, effective, preventative and therapeutic treatments toward EAEC.
Host-EAEC Interactions at the Intestinal Epithelium
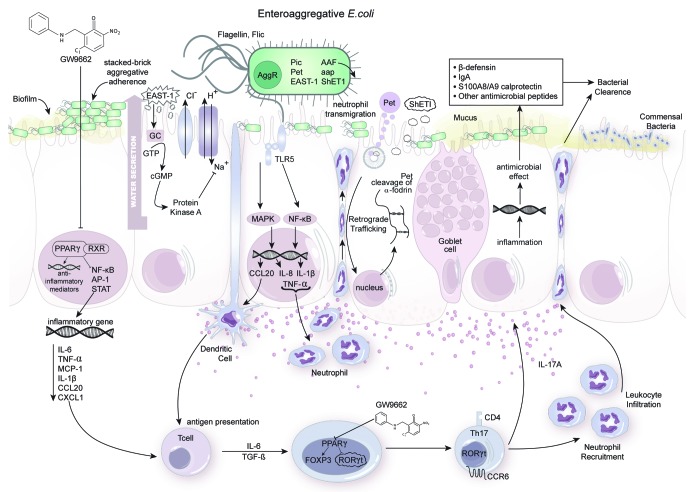
Understanding the complex interactions between host and bacterium is crucial for revealing disease pathogenesis of infectious diseases. The intestinal epithelium is constantly exposed to trillions of microorganisms and faces the challenge to peacefully coexist with harmless commensal bacteria while swiftly responding to pathogens.25 The ability for a host to resist bacterial colonization or clear infection is determined by carefully arranged cellular and molecular interactions between the host and pathogen at the mucosal interface. A single layer of epithelial cells, the epithelial barrier, provides the first line of defense against pathogenic microorganisms. The epithelial barrier integrity is formed by “tight-junctions” between cells and the protective mucus-gel that coats the cells.26 If an enteric pathogen passes through the mucus layers, evolutionarily conserved pathogen-associated molecular patterns (PAMPs) expressed on the microbial surfaces are recognized by pattern recognition receptors (PRRs) expressed on epithelial cell surfaces such as toll-like receptors (TLRs). TLRs activate potent innate responses by triggering signaling pathways that regulate gene transcription, such as NFκB and MAPK and activate the production of a large repertoire of pro-inflammatory mediators to orchestrate the influx of leukocytes.27 More specifically, secretion of IL-8 and CXCL1 by enterocytes generates a chemotactic gradient promoting the recruitment of neutrophils to facilitate clearance of bacteria through phagocytosis.28 Epithelial cells also secrete CCL20 in response to enteric pathogen to enhance infiltration of cells expressing CCR6. Dendritic cells expressing CCR6 are brought to the underlying lamina propria to hasten antigen presentation and activation of the adaptive immune system.29 Th17 cells are CCR6+ and implicated as primary contributors to defense against extracellular bacterial infections. In addition to the secretion of cytokines to mediate cellular trafficking, epithelial cells produce potent antimicrobial proteins such as β-defensins, cathelicidins and calprotectin in response to stimulation from enteric pathogens or proinflammatory cytokines for further defense against infection.28 Importantly, a great amount of attention has recently shifted away from the host response and toward understanding the protective barricade created by commensal microbiota during infection.30 The combined efforts of innate and adaptive immune responses with the beneficial influence of the gastrointestinal microbiome generally contribute to successful eradication of disease in healthy individuals.
Pathogenic bacteria such as EAEC have developed strategic mechanisms to conceal recognition and/or enhance survivability during interaction with its host predominantly driven by genetically encoded virulence factors. EAEC strains harbor a 60- to 65-MDa virulence plasmid (pAA) that encodes many of the known virulence factors including the aggregative adherence fimbriae (AAF), Pet toxin, the transcriptional regulator AggR and the secretory protein dispersin.31 The detection of pAA by probe, also known as the CVD432 probe, was initially trusted as a common broad-spectrum analysis used to identify the prevalence of EAEC isolates, however studies using this methodology have since exposed a large variation in accurate sensitivity toward EAEC ranging between 15% and 90% in separate cases.32,33 The golden standard for EAEC identification remains the highly specialized HEp-2 cell-adherence culture but, due to the assay’s extensive requirements, the more common alternative is multiplex PCR though no molecular assays have been described with 100% specificity. Multiplex PCR evaluation of EAEC detects aggR, asp and aatA, three EAEC plasmid-borne genes and proven a suitable diagnostic test.34 A key virulence factor harbored by pAA is the transcriptional activator AggR which is considered the master regulator of virulence due to its capability to activate a large cluster of virulence genes in EAEC permitting adherence while also promoting the production of cytotoxins and enterotoxins.35 In fact, combined DNA microarray and real-time quantitative RT-PCR data confirm that AggR activates the expression of at least 44 genes in the EAEC prototype strain 042.36 To mediate protein secretion, EAEC possess a type VI secretion system (T6SS) that is chromosomally encoded on the pathogenicity island pheU and transcriptionally regulated by AggR. Sci-1 and sci-2 are two gene clusters present on pheU responsible for encoding T6S machines.37 Additionally, the ETT2 gene cluster has been identified in the EAEC O42 genome sequence providing evidence for T3SS mechanism prevalence as well.38 These secretion systems may play a key role in EAEC virulence due to expulsion of toxic proteins and association with biofilm formation39; their roles in pathogenesis remain widely unknown. Heterogeneity among EAEC strains remains an overarching issue that complicates elucidating pathogenic mechanisms underlying infection. Many virulence factors are not consistently expressed throughout various EAEC strains and the clinical manifestation of disease ranges significantly in severity. Moreover, successful immunoregulatory responses by the host that potentiate EAEC clearance are limited in the literature. Nevertheless, numerous studies suggest that infection can be summarized in three general stages: (1) adherence and colonization, (2) increased mucus production and (3) toxin release and host response.40
Stage I of pathogenesis
During the first step of pathogenesis, EAEC abundantly adhere to the intestinal mucosa in a stacked brick pattern termed aggregative adherence (AA) (Fig. 1). The AA phenotype was first described using a biological co-culture of EAEC with HEp-2 cells. Biopsies from pediatric intestinal mucosa cultured with EAEC strains 17-2 and 221 portrayed the ability for EAEC to adhere to jejunal, ileal and colonic mucosa.41 Another early study provided evidence that fimbria mediate EAEC adherence to HEp-2 cells.42 Four AA fimbriae (AAF) have since been described. Characteristics of AAF vary between EAEC strains both in morphology and genetic code however all mediate the essential role of bacterial attachment to epithelial cells. Prototype strains EAEC17-2, 042, 55989 and C1010–00 express AAF-I, II, III and IV respectively and all four strains develop the observed AA phenotype.43,44 Evidence from an in vivo intestinal cell model of EAEC infection shows that disruption between intestinal epithelial cells induced by strains 042 and JM221 is due to an AAF-dependent delocalization of tight junction proteins, claudin-1 and occludin. AafA, the major pilin protein of AAF fimbria, is directly linked to diminished transepithelial resistance.45 The expression of AAF-I, -III or –IV is sufficient for the induction of polymorphonuclear cell transmigration in vitro. More pertinently, human fetal intestinal xenografts implanted into SCID mice and inoculated with EAEC 042 and mutants verify an AAF-dependent inflammatory response.46 AAF are highly hydrophobic thus favoring agglutination in an aqueous environment. In order to promote the spreading of EAEC for efficient colonization EAEC secretes a low molecular weight protein known as dispersin (aap). Dispersin is a positively charged hydrophobic surface protein that maintains electrostatic interactions with the outer lipopolysaccharide layer of the bacteria preventing the positively charged AAF from clinging to bacterial membrane.47,48 AAF fimbriae actually collapse in the absence of dispersin and lack functionality critical for adherence.49
Figure 1. Enteroaggregative Escherichia coli (EAEC) pathogenesis and host response at the colonic mucosa. The clinical manifestation of EAEC infection is the outcome resulting from complex host-pathogen-microbiota interactions regulated at a molecular level. EAEC attach and aggregate on colonic epithelial cells in a stacked brick pattern by means of AAF fimbria and the secreted protein encoded by aap known as dispersin. EAEC form a thick biofilm enabling protection against host or interventional antimicrobial responses. FliC surface flagella are then recognized by TLR5 receptors expressed on the apical surface of enterocytes. Bacterial-epithelial cell contact triggers a cascade of events activating NFκB and MAPK pathways that result in the upregulation of proinflammatory cytokines IL-8, TNFα and CCL20 responsible for recruiting dendritic cells and neutrophils to the site of infection. Small red spheres underneath the colonic epithelial layer portray the chemokine gradient indicative of inflammation. EAEC harbors the transcriptional regulator AggR responsible for the expression of virulence factors including Pic, Pet, EAST-1, aap and ShET1 portrayed in the amplified image of the bacteria. EAST-1 toxin binds to extracellular guanylate cyclase (GC) on enterocytes and stimulates overproduction of intracellular cyclicGMP (cGMP) ultimately impairing Na/Cl transport. This causes water to be secreted from the enterocyte and contributes to the watery diarrhea seen in infected individuals. ShET1 is also proposed to affect intracellular cGMP levels however much of the biochemistry surrounding this enterotoxin remains unknown. Pet enters the cell via clathrin-mediated endocytosis and is translocated into the cytosol after being transferred from the Golgi complex to the endoplasmic reticulum through retrograde trafficking. In the cytosol, Pet cleaves the actin-binding protein α-Fodrin inducing cytotoxic disruption of the cytoskeleton. Systemic administration of PPAR γ antagonist GW9662 to malnourished EAEC infected hosts enhances an upregulation of inflammatory gene expression and potentiates a beneficial early T helper 17 (Th17) response that successfully facilitates neutrophil recruitment and antimicrobial production that clears the infection and ameliorates disease. A healthy enterocyte is depicted on the far right cohabitating peacefully with the beneficial microflora.
Other accessory molecules have been discovered and associated with EAEC colonization to include a serine protease autotransporter, Pic. Pic is encoded on the chromosomes of EAEC strain 042 and is suggested to mount a pivotal role in the colonization and growth of EAEC. Having hemagglutinin and mucinolytic activity, Pic is able to penetrate the intestinal mucus layer and possibly promote EAEC growth by enhancing the use of nutrients from mucin.49,50 Notably, Pic causes hypersecretion of intestinal mucus in EAEC infected rat ileal loops while also significantly increasing the number of mucus-containing goblet cells in intestinal villi.51 Moreover, Pic efficiently cleaves extracellular glycoproteins on human leukocytes like CD43, a highly expressed surface marker found on almost all cells from a hematopoietic lineage. Interestingly, Pic protein is a key virulence factor in other enteropathogens including uropathogenic E. coli and Shigella flexneri, underscoring its importance in EAEC pathogenicity. Human neutrophils treated with purified Pic protein experience impaired chemotaxis and transmigration but increased activation of the neutrophil oxidative burst while activated T cells experience Pic-induced apoptosis.52
Stage II of pathogenesis
Once EAEC successfully adhere, epithelial cells are stimulated to produce a thick mucus layer above the enterocytes forming a biofilm (Fig. 1, first enterocyte). The presence of AAF is critical in biofilm formation though other unidentified factors including those regulated by fis and AggR gene expression are likely contributors as well.53 The ability to form biofilm is closely associated with bacterial persistence and many chronic bacterial infections are linked to biofilm production.54 To enhance colonization, EAEC surround themselves with biofilm and recruit cells forming micro-colonies that are interspersed within fluid-filled channels. The biofilm then protects colonies by restricting antimicrobial penetration.55
Stage III of pathogenesis
During the third stage of pathogenesis EAEC secretes putative enterotoxins and cytotoxins that elicit a host inflammatory response. Mucosal toxicity can occur causing morphological changes in the architecture of the mucosa characterized by microvillus vesiculation, enlarged crypt openings and increased epithelial cell extrusion.44 Three primary enterotoxins have been discovered; namely EAEC heat-stable enterotoxin-1 (EAST1), Plasmid-encoded enterotoxin (Pet) and Shigella enterotoxin 1 (ShET1). EAST1 is a 4.1 kDa toxin first detected in EAEC strain 17–2 that has now been associated with other diarrheagenic strains of E. coli providing evidence for its relationship to enteropathogenic induced diarrhea.56 The role of EAST1 in the molecular pathogenesis remains incompletely understood, however, scientists hypothesize that the toxin promotes the initial phase of watery diarrhea seen in many patients.57 EAST1 binds to the extracellular domain of guanylate cyclase (GC) on the apical membrane of enterocytes (Fig. 1, second enterocyte). EAST1 then induces high production levels of cGMP inside cells inhibiting the Na/Cl transport system. This significantly reduces the absorption of electrolytes and water from the intestine at the villus tips resulting in elevated secretion of water in crypt cells.54 Pet is a serine protease autotransporter enterotoxin that generates high toxicity in human epithelial cells resulting in structural damage to the cell. After internalization via receptor-mediated endocytosis, Pet is delivered to the cytoplasm by means of retrograde trafficking (Fig. 1, fourth enterocyte). This is then accompanied by cleavage of spectrin, also known as the actin-binding protein fodrin, within microvilli cytoskeleton leading to cell elongation, rounding and ultimately the release of cells from the substratum.58-60 ShET1 appears to induce intestinal secretion via cAMP and cGMP however much of the biochemistry and mechanism of action surrounding this toxin remain elusive.61
Most EAEC strains harbor genes encoding class I and class II serine protease autotransporter toxins (SPATEs). Class I SPATEs are cytotoxic to epithelial cells and include Pet, Sat, EspP and SigA while the non-cytotoxic class II category includes pic and sepA.62,63 Sat, originally discovered in uropathogenic and diffusely adhering E. coli, has been described as the most commonly detected SPATE among EAEC strains. Sat, like its homolog Pet, is believed to cleave the intracellular protein spectrin and cause cytoskeletal damage to tight junctions between intestinal epithelial cells.64 Likewise, SigA, a SPATE largely associated with Shigella flexneri pathogenesis, is capable of inducing fodrin degradation causing catastrophic morphological changes in cells.65 Interestingly, although only moderately prevalent in EAEC strains, SepA is the SPATE most strongly associated with severe diarrheal illness63 though its role in EAEC pathogenesis remains largely uncharacterized.
Shiga toxin producing EAEC strains
E. coli O104:H4, reported as a causative agent of diarrhea since 2001 and the disease causing strain in the 2011 German outbreak, is an EAEC strain that has adopted the ability to produce Shiga-toxin (Stx2),66 a chromosomally encoded cytotoxic verotoxin that targets globotriaosylceramide (Gb3) receptors located on host intestinal and kidney cells. Death from infection with Stx2-producing EAEC strains is strongly linked to the development of hemolytic uremic syndrome (HUS), a life-threatening disease induced by Stx2 shortly after the onset of diarrhea. Stx2 undergoes retrograde transport to induce endothelial cell apoptosis causing significant gastrointestinal damage. Additionally, Stx2 is able to enter systemic circulation and induce glomerular occlusion as blood is filtered through the capillary arrangement in the kidney. The resulting hemolytic anemia and acute renal failure are complications that most commonly affect children and contribute to increased mortality rates.67,68 The 2011 EAEC O104:H4 outbreak an unusually high proportion of adult patients (especially women) and significantly increased incidence of HUS (25% of patients).66 Interestingly, death occurred in patients who had not developed HUS; these cases most commonly occurred in elderly females.69
Whole genome-phylogenesis confirmed strain O104:H4 as an EAEC strain. Acquisition of a Stx2 bacteriophage is the leading factor for hypervirulence. This phenomenon could have occurred in mammalian intestines or an environment where both human and ruminant feces were present.70 Alignment of an EAEC O104:H4 isolate TY2482 against the prototype EAEC strain 55989 chromosome ultimately revealed the presence of the large conjugative plasmid pAA which resembled the AAF gene-coding cluster from strain 55989. Interestingly, pAA TY2482 encoded for AAF/I rather than the more common AAF/III. The isolate lacked the locus of enterocyte effacement (LEE; responsible for bacterial adherence), intimin adherence factor and a type III secretion system normally identified in enterohemmorhagic E. coli (EHEC) strains.66 Since EAEC virulence factors are encoded on plasmids, bacteriophages and genetic pathogenicity islands, the traits are easily transferred to new emerging strains.71
Survivability and Shiga toxin production alone are not likely the sole causes of HUS in EAEC infected patients. EAEC O104:H4 adherence to the intestinal mucosa is mediated by AAF/I and potentially more aggressive than EHEC LEE mediated adherence. Additionally, EAEC infections induce proinflammatory responses and epithelial barrier disruption possibly enhancing systemic dissemination of shiga-toxin and HUS induction providing an explanation for the strain’s hypervirulent activity. In addition to Stx2 gaining systemic accessibility, severe epithelial damage induced by the toxin could have allowed bacterial components to enter peripheral blood exaggerating inflammation systemically leading to death by sepsis in non-HUS patients. Most importantly, the genome sequence of TY2492 illuminates the ability for Shiga toxin-producing E. coli to produce various adhesion mechanisms portraying the ability for pathotypes to overlap and evolve into more virulent strains. Rapid responses in sequencing efforts during the EAEC O104:H4 outbreak suggest that genomic epidemiology will become a standard molecular strategy to elucidate infectious disease outbreaks.72
Host response to colonization and virulence factors of EAEC
Measured immune responses in infected subjects represent the result of a delicate balance between host–microbial interactions. Additionally, responses are specific and dependent on variables found among hosts and EAEC strains. For instance, genetic variability seen in both host and EAEC strains can significantly impact the susceptibility and outcome of EAEC infection. The capacity for specific EAEC strains to produce Stx2 and cause HUS-induced mortality demonstrates enhanced virulence. In other instances, host age dictates disease severity portrayed when children are more susceptible to persistent EAEC infection compared with healthy travelers. Regardless, studies have proven that bacterial-epithelial contact is a key determinant of host response to EAEC.73 The EAEC bacterial surface protein flagellin (FliC) has been shown to mediate NFκB and p38 MAP kinase activation in epithelial cells by cellular signaling through Toll-like receptor 5 resulting in Interleukin 8 (IL-8) production (Fig. 1, third enterocyte).74,75 FliC is the major inducer of IL-8 release however other AggR regulated factors contribute and AAF adhesion is required for full IL-8 induction.76 IL-8 is a cytokine associated with infection with EAEC and other enteric pathogens. IL-8 production is involved in recruitment of neutrophils and the transmigration of these cells into the intestinal mucosa disrupts epithelial tight junctions ultimately inducing colitis: a mechanism of action common among diarrhea-inducing pathogens. Some research suggests that the induction of IL-8 and subsequent disruption of the epithelial barrier is beneficial for EAEC pathogenicity enhancing toxic effects on the host though in vivo models are yet to validate this theory. Elevated levels of fecal IL-1β, another cytokine that can induce neutrophil migration,77 have also been reported in adults diagnosed with EAEC induced traveler’s diarrhea.78 Lactoferrin, an iron-binding antimicrobial glycoprotein,79 has been a target in other studies that demonstrate significantly increased levels of this protein alongside fecal leucocytes in EAEC infected patients.80 Not surprisingly, CCL20, a dendritic cell recruiter, is also known to be upregulated after persistent EAEC stimulus.73
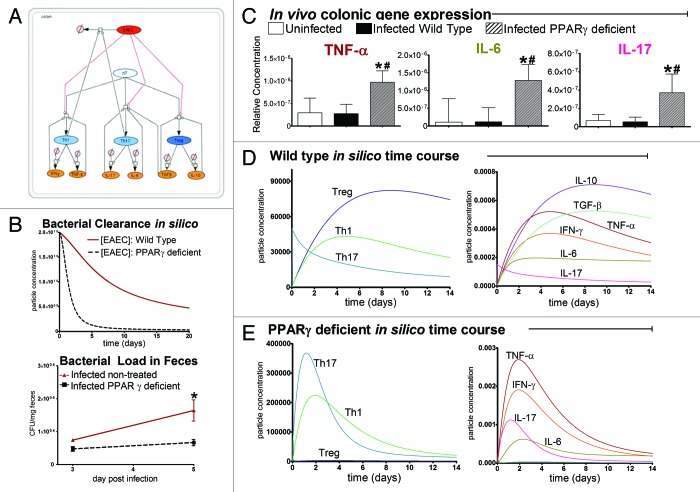
Recently, our group published in vivo data reporting for the first time the importance of T helper (Th)17 cells in host responses to EAEC and EAEC clearance.81 By pharmacologically and genetically disrupting the activity of the transcription factor peroxisome proliferator-activated receptor (PPAR) γ in malnourished mice, we modulated mucosal inflammation that resulted in enhanced Th17 phenotypes and disease amelioration (Fig. 1, first and fifth enterocytes, underlying mucosa). PPAR γ regulates anti-inflammatory responses through its ability to inhibit signaling pathways such as NFκB, AP-1 and STAT in epithelial cells, macrophages and T and B lymphocytes.82,83 Mice infected with EAEC strain JM221 that were treated with a potent PPAR γ antagonist, GW9662, or those that lacked functional PPAR γ in T-cells (PPAR γ fl/fl CD4-cre+ mice) cleared EAEC significantly faster than untreated wild type mice. Colonic gene expression for inflammatory cytokines and chemokines, including TNFα, IL-6, MCP-1, CCL20, CXCL1 and IL-1β was significantly upregulated early during infection in PPAR γ deficient mice when compared with wild-type counterparts. During the chronic phase of infection, PPAR γ deficiency significantly enhanced IL-6, TGF-β and IL-17A expression in the colon suggesting the presence and importance of CD4+Th17 cells during EAEC infection. Flow cytometry validated higher percentages of Th17 cells in colonic lamina propria of PPAR γ deficient mice. Histopathological analysis of colons also provided consistent evidence that PPAR γ blockade enhanced the inflammatory response without causing collateral tissue damage at the gut mucosa. Th17 responses are known to enhance antimicrobial inflammatory responses by increasing the expression of antimicrobial peptides and effectively recruiting and activating neutrophils that contribute to destroying invading extracellular pathogens.84,85 Notably, EAEC clearance was directly correlated with the upregulation of colonic calprotectin in GW9662 treated mice. The beneficial role of Th17 cells to enhance effector mucosal responses during EAEC infections is a pivotal finding and future studies should focus on how EAEC induces these responses.
EAEC appears to strategically orchestrate an inflammatory response in the host’s intestinal mucosa regardless of the presence or absence of diarrhea.7,86 Infiltration of innate immune cells, disruption of the epithelial barrier and increased mucus production best explain the most commonly reported symptoms including watery diarrhea, with or without blood and mucus, abdominal pain, nausea and fever.10,58 However, this inflammatory response is sometimes not sufficient to facilitate pathogen clearance, thereby resulting in extended host tissue damage. New data suggests that EAEC may also induce a Th17 response and that enhancing this response in a malnourished host early during the infection process is beneficial to overcome disease. Of note, EAEC can persist subclinically facilitating a chronic inflammatory state that can impair nutrient absorption and developmental processes at the intestinal wall.87 Over the past few decades, a few key animal models have been developed in order to help gain a better understanding of how EAEC modulates mucosal immune responses.
Current Animal Models Used in Examining EAEC Infection
The first reported animal model used to study EAEC infection was published in 1988.88 Specifically, a team of scientists commenced the preliminary infectious trials using ligated intestinal loops in NZB rabbits and Fischer 344 rats. EAEC infection resulted in intestinal lesions, limb paralysis and even death in some animals. These studies provided ample evidence that EAEC exhibited sufficient distinct characteristics in comparison to other diarrheagenic E. coli (DEC) strains to become its own discrete category. These studies also proposed EAEC virulence was likely accompanied by toxins.88 Currently there is a substantial deficiency in the development of reliable and reproducible animal models that effectively portray immunological responses toward EAEC at the gut mucosa. The developing animal models can be subdivided into two general categories. The first group includes models that use young animals while the second uses adults. Known animal models used to date will be described in more detail below.
Animal models investigating infantile and childhood EAEC induced diarrhea
A global analysis of child mortality in 2008 estimated that infectious diseases cause the majority (68%) of deaths in children younger than 5 y of age worldwide. Moreover, diarrhea is the second leading cause of death under the infectious disease category, thus generating a demand for impactful research in this area.89
Gnotobiotic piglets
One approach to study the pathogenicity of EAEC in neonates uses 24-h-old germ-free piglets. This model clearly demonstrates detrimental effects caused by EAEC at the colonic mucosa and most closely resembles pathology observed in humans. Infected animals suffer from severe diarrhea and sometimes mortality. Importantly, in this model EAEC adhere to the mucosal epithelium in the described “stacked-brick” pattern verified during in vitro studies and elicit edema and lesions in the intestinal lamina propria. The gnotobiotic piglet appears to be one of the best whole-animal models reported however confines to using the model are high.90 Piglet models have low scalability and are extremely labor intensive in comparison to rodent models. Also, large animal biosafety level 2 (ABSL2) facilities are not as readily available as mouse ABLS2 facilities. Lastly there is a large deficiency in the production of swine carrying targeted gene mutations (conditional knockout animals) restricting studies to “wild type” animals. More than a decade has passed since advances in a swine model of EAEC infection have been reported. Despite boundaries, swine remain an ideal model for studying human diseases, especially those affecting the gastrointestinal tract. Pigs are monogastrics and leverage the gastrointestinal, nutritional, metabolic and immunologic similarities of humans.91-93
Neonatal and weaned C57BL/6 mouse model with or without malnutrition
Another neonatal model was developed more recently using six-day-old C57BL/6 mice to study EAEC induced infantile diarrhea. Mice are challenged orally with a bacterial inoculum of prototype EAEC strains 042 or JM221 and remain with their dam for regulated time periods until weaning. Additionally, to comparatively analyze vulnerability to EAEC in young children, a weaned mouse model was established in parallel with an emphasis on the effects of malnutrition. In these studies, weaned C57BL/6 mice are fed either regular (20% protein) or malnourished (2% protein) diet throughout the duration of infection. Both neonatal and weaned mice experience significant developmental stunting due to infection and malnutrition intensifies this effect, especially in mice infected with strain JM221. Mild histopathological changes in the colonic epithelium including localized inflammation and goblet cell hyperplasia are noticed as early as four days post infection. Due to protein malnutrition, the infectious burden can become chronic and bacterial shedding persists for over three weeks post infection. While these models have opened the door to potentially divulge novel characteristics of EAEC induced childhood infection, the model has limited translational value. Experimental limitations include the fact that symptoms are only mild in relation to reported human infections and mice do not develop overt diarrhea. Nonetheless, these experiments were successful in portraying heterogeneity between separate EAEC strains (042 and JM221) that remain novel and beneficial to the scientific community though research advancements using this model remain remarkably underreported.94
Antibiotic treatment of adult mice in combination with infection
Adult mice over the age of six weeks are treated with 5 g streptomycin for up to 48 h prior to infection and for the duration of the experiment. Some experimental designs also include treatment with sodium bicarbonate just prior to infection in order to neutralize gastric acid. This model has been successful at recapitulating in vitro studies demonstrating the ability for Pic, a serine protease autotransporter secreted by EAEC, to enhance EAEC growth in mouse colons by its utilization of nutrients from mucin.49 Another study using this model provides insight on EAEC promoter induction in vivo through luminescence imaging.95 Though advancements in understanding a key role of Pic in EAEC pathogenesis have resulted from using this approach, mice in this model do not develop clinical signs or histopathological abnormalities according to authors using this protocol, therefore gaining no expansion in the ability to study mechanisms of inflammation at the gut mucosa.
Immunodeficient mice and human intestinal xenograph implantation
Most recently, the need for an effective in vivo model prompted a unique investigation using severe-combined immunodeficient (SCID) mice and xenographs from fetal intestinal tissue to generate a model with intact and functional human tissue during infection. Fetal tissue was implanted into the subscapular region of SCID mice and then infected by direct intraluminal inoculation. Findings using this approach demonstrated the ability for AAF to trigger polymorphonuclear cell (PMN) migration across mucosal surfaces of the intestinal epithelial barrier.46 However, the availability of fetal tissue is rare and restricted, which will significantly constrict the broad acceptability and use of this model. Also, this approach does not allow scientists to address critical interaction between innate and adaptive immunity during EAEC infection leaving a significant portion of the pathogenesis story untold.
The development of animal models as a means of studying chronic and acute EAEC infections has provided insight into novel features related to EAEC pathogenesis, however all established animal models are underused and limited in their ability to fully characterize immunological responses to EAEC at the intestinal mucosa. This is likely due to many factors and scientists studying this particular infectious disease have proposed two probable causes. First, disease severity is mild at most during in vivo trials likely resulting from dampened immune responses compared with human studies. Second, it is likely that EAEC pathogenesis has primarily adapted to human intestinal tissue enhancing variability in clinical manifestation that does not imitate natural disease.4,46 Collectively, there is still a desperate need for a reproducible and comprehensive animal model that results in significant disease activity, weight loss and intestinal pathology.
Future Perspective in EAEC Animal Models
Systems biology is an emerging field that transcends biology, engineering and computer science and aims to elucidate mechanistic functions in complex systems through the integration of computational work and biological research using mathematical models.96,97 These models range in size, purpose and specificity to include infectious mechanisms at the cellular level, tissue level and individual level, to population-scale disease spread.96,98,99 The first step in creating a model consists of a comprehensive literature search and construction of a network portraying mechanisms of interest. A specific mathematical modeling approach is selected and applied to the network allowing calibration to begin. Model refinement continues in a cyclic process to incorporate experimental validation or add additional dynamics (Fig. S1).
Two common mathematical approaches used to implicitly model the kinetics of biological processes include stochastic and deterministic modeling systems.100,101 Stochastic models hold an advantage in accounting for randomness in a system and they produce results based on probability mimicking individual variation more realistically. Agent based models (ABM) are an example of a powerful stochastic modeling technique for predicting and simulating biological events. In ABM, each entity, or agent, in the system assesses its status and makes a decision based on the current environment. This constant sensing generates randomness in the data thus providing the most realistic approach for modeling systems that are nonlinear and discrete. However, stochastic models are extremely mathematically complex requiring extensive time for development, fitting and calibration, requiring mathematical expertise. This is explained by the fact that, as the name implies, ABM simulate individual agents and thus simulating the behavior of large systems with many entities as one unit is extremely computationally intense and often require high-performance computing solutions.102 In contrast, deterministic models require less data, are considered more user-friendly and multiple software programs currently exist to assist the user in construction.103,104 Deterministic models can be built around the law of mass action, a fundamental law that governs rates of reactions in biochemistry. This is performed by assigning rates of creation and degradation for each species. Ordinary differential equations (ODEs) then combine the set of functions assigned to each species and, through numerical integration using given data, express the rate of change of each molecule; a species’ concentration over time. Since deterministic models are equation-based systems, the evaluation and execution of a simulation will be consistent each time the task is performed unless the user manually changes the rates and parameters. This minimizes the complexity and time requirement for simulations when compared with the ABM systems. Deterministic models have been vastly useful for scientists inquiring mechanisms behind infection, especially using a series of ODEs.96,97,100
The Modeling Immunity to Enteric Pathogens (MIEP) program at Virginia Tech has developed a complex network representing host interactions with EAEC at the colonic mucosa and CD4+ T cell differentiation using the graphical software package, CellDesigner. Interactive annotated EAEC and CD4+ T cell differentiation models developed by MIEP are deposited and archived on the team website, www.modelingimmunity.org/models, available to download. The EAEC model is comprised of four compartments including the colonic lumen, epithelium, lamina propria and mesenteric lymph node (MLN). EAEC enters the system at the lumen and immediately contacts epithelial cells causing a cascade of reactions triggering cytokine secretion, neutrophil activation and macrophage differentiation in the lamina propria. Additionally, dendritic cells sample the EAEC present in the lumen and subsequently migrate to the MLN facilitating naïve T cell differentiation activating adaptive immunity. Phagocytic macrophages and neutrophils play primary roles in expediting EAEC death in the model. Th1 and Th17 CD4+ cell populations are known to possess antimicrobial properties and provide defense against extracellular bacteria.98,99 Thus after T cell migration into the LP, Th1 and Th17 cells also assist in bacterial clearance. Initial steps in EAEC model calibration involved isolating parameters regulating T cell differentiation and bacterial clearance to reconstruct a smaller network (Fig. 2A, Fig. S2). A calibration database was generated using in house gene expression, flow cytometry and bacterial shedding data from malnourished mice infected with EAEC strain JM221 (Fig. S3); parameters were estimated in COPASI. The model successfully portrays chronic bacterial burden in malnourished EAEC infected mice (Fig. 2B) and significantly reduced T cell populations, lack of effector responses and low concentrations of proinflammatory cytokines signifying immunodeficiency (Fig. 2C and D). To assess the model’s ability to predict immune responses to EAEC in PPARγ deficient mice, we reduced the ability for naïve T (nT) cells to differentiate into Treg and enhanced Th1 and Th17 differentiation. The model successfully predicts a dominant proinflammatory Th17 effector response correlated with successful bacterial clearance by day 5 post infection (Fig. 2B, C and E). The systems biology approach to predict the modulation of PPARγ on CD4+ T cell responses using this approach has been extensively developed and provided novel unintuitive characterizations of unforeseen mechanistic pathways.105 We are confident that this new approach will deliver favorable predictive results with new revelations surrounding innate and adaptive host responses toward EAEC.
Figure 2. Modeling immune responses to EAEC. The EAEC T cell differentiation model network created in CellDesigner using systems biology markup language (A) was linked to COPASI software for the calibration. A calibrated wild type system was created using in house data from malnourished EAEC strain JM221 infected mice (C).Asterisks (*) indicate statistical significance compared with uninfected mice and the number symbol (#) represents statistical difference compared with infected wild type mice (p < 0.05).81 Modulating the parameters regulating T cell differentiation into separate phenotypes simulated PPARγ deficiency: Th1 and Th17 cell differentiation was enhanced equally while Treg differentiation was decreased in an equal magnitude. Bacterial clearance predicted in silico mimicked EAEC quantification in feces from infected mice (B). In silico simulations of a time course infection over 14 d were performed using COPASI. The wild type system (D) portrays immunodeficiency while the PPARγ deficient system (E) predicted enhanced effector responses.
Conclusion
EAEC has been recognized as a causative agent of persistent diarrheal illness worldwide for over two decades. A better understanding of the cellular responses, particularly the adaptive immunity, involved in host response toward EAEC is critical for the development of treatments to ameliorate disease. The ability to validate that effector Th17 responses are induced by EAEC would have great value on targeting cellular responses and specific molecular mechanisms during therapeutic treatments in chronically ill or immunocompromised patients. To date, no such studies are presented and this is likely attributed to the lack of a reliable animal model. Limitations of animal models have hindered advancements in novel discoveries of pathophysiology and preclinical testing for therapeutics. Thus there is still a desperate need for highly reproducible animal models that provide an outlet to examine cellular responses at the intestinal mucosa during EAEC infection. Lastly, transdisciplinary immunoinformatics approaches that combine omics data and computational modeling to compile complex and heterogeneous data regarding host responses to EAEC hold great potential in unveiling dynamic commonalities in mechanisms of infection that have otherwise been undetected.
Supplementary Material
Glossary
Abbreviations:
- Escherichia coli
E. coli
- enteroaggregative E. coli
EAEC
- interleukin
IL
- chemokine C-X-C motif ligand
CXCL
- chemokine C-C motif ligand
CCL
- chemokine receptor
CCR
- pathogen-associated molecular pattern
PAMP
- pattern recognition receptor
PRR
- Toll-like receptor
TLR
- aggregative adherence fimbria
AAF
- mitogen-activated protein kinase
MAPK
- enteroaggregative heat-stable toxin 1
EAST-1
- Shigella enterotoxin 1
ShET1
- plasmid-encoded toxin
Pet
- Shiga toxin 2
Stx2
- hemolytic uremic syndrome
HUS
- severe combined immunodeficiency
SCID
- agent based modeling
ABM
- ordinary differential equation
ODE
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/24826
References
- 1.Donnenberg MS, Whittam TS. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J Clin Invest. 2001;107:539–48. doi: 10.1172/JCI12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton Behravesh C, Jones TF, Vugia DJ, Long C, Marcus R, Smith K, et al. FoodNet Working Group Deaths associated with bacterial pathogens transmitted commonly through food: foodborne diseases active surveillance network (FoodNet), 1996-2005. J Infect Dis. 2011;204:263–7. doi: 10.1093/infdis/jir263. [DOI] [PubMed] [Google Scholar]
- 3.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 4.Gomes TA, Blake PA, Trabulsi LR. Prevalence of Escherichia coli strains with localized, diffuse, and aggregative adherence to HeLa cells in infants with diarrhea and matched controls. J Clin Microbiol. 1989;27:266–9. doi: 10.1128/jcm.27.2.266-269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang DB, Nataro JP, DuPont HL, Kamat PP, Mhatre AD, Okhuysen PC, et al. Enteroaggregative Escherichia coli is a cause of acute diarrheal illness: a meta-analysis. Clin Infect Dis. 2006;43:556–63. doi: 10.1086/505869. [DOI] [PubMed] [Google Scholar]
- 6.Glandt M, Adachi JA, Mathewson JJ, Jiang ZD, DiCesare D, Ashley D, et al. Enteroaggregative Escherichia coli as a cause of traveler’s diarrhea: clinical response to ciprofloxacin. Clin Infect Dis. 1999;29:335–8. doi: 10.1086/520211. [DOI] [PubMed] [Google Scholar]
- 7.Steiner TS, Lima AA, Nataro JP, Guerrant RL. Enteroaggregative Escherichia coli produce intestinal inflammation and growth impairment and cause interleukin-8 release from intestinal epithelial cells. J Infect Dis. 1998;177:88–96. doi: 10.1086/513809. [DOI] [PubMed] [Google Scholar]
- 8.Wanke CA, Mayer H, Weber R, Zbinden R, Watson DA, Acheson D. Enteroaggregative Escherichia coli as a potential cause of diarrheal disease in adults infected with human immunodeficiency virus. J Infect Dis. 1998;178:185–90. doi: 10.1086/515595. [DOI] [PubMed] [Google Scholar]
- 9.Kaur P, Chakraborti A, Asea A. Enteroaggregative Escherichia coli: An Emerging Enteric Food Borne Pathogen. Interdiscip Perspect Infect Dis. 2010;2010:254159. doi: 10.1155/2010/254159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang DB, Okhuysen PC, Jiang ZD, DuPont HL. Enteroaggregative Escherichia coli: an emerging enteric pathogen. Am J Gastroenterol. 2004;99:383–9. doi: 10.1111/j.1572-0241.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 11.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al Safadi R, Abu-Ali GS, Sloup RE, Rudrik JT, Waters CM, Eaton KA, et al. Correlation between in vivo biofilm formation and virulence gene expression in Escherichia coli O104:H4. PLoS One. 2012;7:e41628. doi: 10.1371/journal.pone.0041628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koo HL, Jiang ZD, Brown E, Garcia C, Qi H, Dupont HL. Coliform contamination of vegetables obtained from popular restaurants in Guadalajara, Mexico, and Houston, Texas. Clin Infect Dis. 2008;47:218–21. doi: 10.1086/589249. [DOI] [PubMed] [Google Scholar]
- 14.Pawlowski SW, Warren CA, Guerrant R. Diagnosis and treatment of acute or persistent diarrhea. Gastroenterology. 2009;136:1874–86. doi: 10.1053/j.gastro.2009.02.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oundo JO, Kariuki SM, Boga HI, Muli FW, Iijima Y. High incidence of enteroaggregative Escherichia coli among food handlers in three areas of Kenya: a possible transmission route of travelers’ diarrhea. J Travel Med. 2008;15:31–8. doi: 10.1111/j.1708-8305.2007.00174.x. [DOI] [PubMed] [Google Scholar]
- 16.Paredes-Paredes M, Okhuysen PC, Flores J, Mohamed JA, Padda RS, Gonzalez-Estrada A, et al. Seasonality of diarrheagenic Escherichia coli pathotypes in the US students acquiring diarrhea in Mexico. J Travel Med. 2011;18:121–5. doi: 10.1111/j.1708-8305.2010.00488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang ZD, Okhuysen PC, Guo DC, He R, King TM, DuPont HL, et al. Genetic susceptibility to enteroaggregative Escherichia coli diarrhea: polymorphism in the interleukin-8 promotor region. J Infect Dis. 2003;188:506–11. doi: 10.1086/377102. [DOI] [PubMed] [Google Scholar]
- 18.Mohamed JA, DuPont HL, Jiang ZD, Belkind-Gerson J, Figueroa JF, Armitige LY, et al. A novel single-nucleotide polymorphism in the lactoferrin gene is associated with susceptibility to diarrhea in North American travelers to Mexico. Clin Infect Dis. 2007;44:945–52. doi: 10.1086/512199. [DOI] [PubMed] [Google Scholar]
- 19.Nataro JP, Mai V, Johnson J, Blackwelder WC, Heimer R, Tirrell S, et al. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin Infect Dis. 2006;43:402–7. doi: 10.1086/505867. [DOI] [PubMed] [Google Scholar]
- 20.Al-Gallas N, Bahri O, Bouratbeen A, Ben Haasen A, Ben Aissa R. Etiology of acute diarrhea in children and adults in Tunis, Tunisia, with emphasis on diarrheagenic Escherichia coli: prevalence, phenotyping, and molecular epidemiology. Am J Trop Med Hyg. 2007;77:571–82. [PubMed] [Google Scholar]
- 21.Sarantuya J, Nishi J, Wakimoto N, Erdene S, Nataro JP, Sheikh J, et al. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol. 2004;42:133–9. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pabst WL, Altwegg M, Kind C, Mirjanic S, Hardegger D, Nadal D. Prevalence of enteroaggregative Escherichia coli among children with and without diarrhea in Switzerland. J Clin Microbiol. 2003;41:2289–93. doi: 10.1128/JCM.41.6.2289-2293.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyo SJ, Maselle SY, Matee MI, Langeland N, Mylvaganam H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect Dis. 2007;7:92. doi: 10.1186/1471-2334-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alikhani MY, Sedighi I, Zamani A, Aslani MM, Sadrosadat T. Incidence of diarrhoeagenic Escherichia coli isolated from young children with diarrhoea in the west of Iran. Acta Microbiol Immunol Hung. 2012;59:367–74. doi: 10.1556/AMicr.59.2012.3.7. [DOI] [PubMed] [Google Scholar]
- 25.Vossenkämper A, Macdonald TT, Marchès O. Always one step ahead: How pathogenic bacteria use the type III secretion system to manipulate the intestinal mucosal immune system. J Inflamm (Lond) 2011;8:11. doi: 10.1186/1476-9255-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gouyer V, Gottrand F, Desseyn JL. The extraordinarily complex but highly structured organization of intestinal mucus-gel unveiled in multicolor images. PLoS One. 2011;6:e18761. doi: 10.1371/journal.pone.0018761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eddy SF, Storey KB. p38 MAPK regulation of transcription factor targets in muscle and heart of the hibernating bat, Myotis lucifugus. Cell Biochem Funct. 2007;25:759–65. doi: 10.1002/cbf.1416. [DOI] [PubMed] [Google Scholar]
- 28.Eckmann L, Kagnoff MF. Intestinal mucosal responses to microbial infection. Springer Semin Immunopathol. 2005;27:181–96. doi: 10.1007/s00281-005-0207-5. [DOI] [PubMed] [Google Scholar]
- 29.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4607–14. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–23. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, et al. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One. 2010;5:e8801. doi: 10.1371/journal.pone.0008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernier C, Gounon P, Le Bouguénec C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun. 2002;70:4302–11. doi: 10.1128/IAI.70.8.4302-4311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snelling AM, Macfarlane-Smith LR, Fletcher JN, Okeke IN. The commonly-used DNA probe for diffusely-adherent Escherichia coli cross-reacts with a subset of enteroaggregative E. coli. BMC Microbiol. 2009;9:269. doi: 10.1186/1471-2180-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cordeiro F, da Silva Gomes Pereira D, Rocha M, Asensi MD, Elias WP, Campos LC. Evaluation of a multiplex PCR for identification of enteroaggregative Escherichia coli. J Clin Microbiol. 2008;46:828–9. doi: 10.1128/JCM.01865-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aslani MM, Alikhani MY, Zavari A, Yousefi R, Zamani AR. Characterization of enteroaggregative Escherichia coli (EAEC) clinical isolates and their antibiotic resistance pattern. Int J Infect Dis. 2011;15:e136–9. doi: 10.1016/j.ijid.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Morin N, Santiago AE, Ernst RK, Guillot SJ, Nataro JP. Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect Immun. 2013;81:122–32. doi: 10.1128/IAI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–82. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 38.Ren CP, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, Barnes WM, et al. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J Bacteriol. 2004;186:3547–60. doi: 10.1128/JB.186.11.3547-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aschtgen MS, Bernard CS, De Bentzmann S, Lloubès R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol. 2008;190:7523–31. doi: 10.1128/JB.00945-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrada-Garcia T, Navarro-Garcia F. Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol Med Microbiol. 2012;66:281–98. doi: 10.1111/j.1574-695X.2012.01008.x. [DOI] [PubMed] [Google Scholar]
- 41.Hicks S, Candy DC, Phillips AD. Adhesion of enteroaggregative Escherichia coli to pediatric intestinal mucosa in vitro. Infect Immun. 1996;64:4751–60. doi: 10.1128/iai.64.11.4751-4760.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nataro JP, Deng Y, Maneval DR, German AL, Martin WC, Levine MM. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun. 1992;60:2297–304. doi: 10.1128/iai.60.6.2297-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76:3281–92. doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrington SM, Dudley EG, Nataro JP. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol Lett. 2006;254:12–8. doi: 10.1111/j.1574-6968.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 45.Strauman MC, Harper JM, Harrington SM, Boll EJ, Nataro JP. Enteroaggregative Escherichia coli disrupts epithelial cell tight junctions. Infect Immun. 2010;78:4958–64. doi: 10.1128/IAI.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boll EJ, Struve C, Sander A, Demma Z, Nataro JP, McCormick BA, et al. The fimbriae of enteroaggregative Escherichia coli induce epithelial inflammation in vitro and in a human intestinal xenograft model. J Infect Dis. 2012;206:714–22. doi: 10.1093/infdis/jis417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, Le Bouguénec C, et al. A novel dispersin protein in enteroaggregative Escherichia coli. J Clin Invest. 2002;110:1329–37. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortensen NP, Fowlkes JD, Maggart M, Doktycz MJ, Nataro JP, Drusano G, et al. Effects of sub-minimum inhibitory concentrations of ciprofloxacin on enteroaggregative Escherichia coli and the role of the surface protein dispersin. Int J Antimicrob Agents. 2011;38:27–34. doi: 10.1016/j.ijantimicag.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Harrington SM, Sheikh J, Henderson IR, Ruiz-Perez F, Cohen PS, Nataro JP. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect Immun. 2009;77:2465–73. doi: 10.1128/IAI.01494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun. 1999;67:5587–96. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarro-Garcia F, Gutierrez-Jimenez J, Garcia-Tovar C, Castro LA, Salazar-Gonzalez H, Cordova V. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect Immun. 2010;78:4101–9. doi: 10.1128/IAI.00523-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz-Perez F, Wahid R, Faherty CS, Kolappaswamy K, Rodriguez L, Santiago A, et al. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc Natl Acad Sci U S A. 2011;108:12881–6. doi: 10.1073/pnas.1101006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheikh J, Hicks S, Dall’Agnol M, Phillips AD, Nataro JP. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol. 2001;41:983–97. doi: 10.1046/j.1365-2958.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- 54.Telli M, Guiral E, Martínez JA, Almela M, Bosch J, Vila J, et al. Prevalence of enterotoxins among Escherichia coli isolates causing bacteraemia. FEMS Microbiol Lett. 2010;306:117–21. doi: 10.1111/j.1574-6968.2010.01945.x. [DOI] [PubMed] [Google Scholar]
- 55.Mohamed JA, Huang DB, Jiang ZD, DuPont HL, Nataro JP, Belkind-Gerson J, et al. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol. 2007;45:121–6. doi: 10.1128/JCM.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ménard LP, Dubreuil JD. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit Rev Microbiol. 2002;28:43–60. doi: 10.1080/1040-840291046687. [DOI] [PubMed] [Google Scholar]
- 57.Savarino SJ, McVeigh A, Watson J, Cravioto A, Molina J, Echeverria P, et al. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J Infect Dis. 1996;173:1019–22. doi: 10.1093/infdis/173.4.1019. [DOI] [PubMed] [Google Scholar]
- 58.Huang DB, Mohanty A, DuPont HL, Okhuysen PC, Chiang T. A review of an emerging enteric pathogen: enteroaggregative Escherichia coli. J Med Microbiol. 2006;55:1303–11. doi: 10.1099/jmm.0.46674-0. [DOI] [PubMed] [Google Scholar]
- 59.Navarro-García F, Sears C, Eslava C, Cravioto A, Nataro JP. Cytoskeletal effects induced by pet, the serine protease enterotoxin of enteroaggregative Escherichia coli. Infect Immun. 1999;67:2184–92. doi: 10.1128/iai.67.5.2184-2192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Navarro-Garcia F, Sonnested M, Teter K. Host-Toxin Interactions Involving EspC and Pet, Two Serine Protease Autotransporters of the Enterobacteriaceae. Toxins (Basel) 2010;2:1134–47. doi: 10.3390/toxins2051134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Navarro-Garcia F, Elias WP. Autotransporters and virulence of enteroaggregative E. coli. Gut Microbes. 2011;2:13–24. doi: 10.4161/gmic.2.1.14933. [DOI] [PubMed] [Google Scholar]
- 62.Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg. 2009;80:294–301. [PMC free article] [PubMed] [Google Scholar]
- 63.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, Simon J, et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis. 2012;205:431–44. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guignot J, Chaplais C, Coconnier-Polter MH, Servin AL. The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell Microbiol. 2007;9:204–21. doi: 10.1111/j.1462-5822.2006.00782.x. [DOI] [PubMed] [Google Scholar]
- 65.Al-Hasani K, Navarro-Garcia F, Huerta J, Sakellaris H, Adler B. The immunogenic SigA enterotoxin of Shigella flexneri 2a binds to HEp-2 cells and induces fodrin redistribution in intoxicated epithelial cells. PLoS One. 2009;4:e8223. doi: 10.1371/journal.pone.0008223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohde H, Qin J, Cui Y, Li D, Loman NJ, Hentschke M, et al. E. coli O104:H4 Genome Analysis Crowd-Sourcing Consortium Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365:718–24. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 67.Pacheco AR, Sperandio V. Shiga toxin in enterohemorrhagic E.coli: regulation and novel anti-virulence strategies. Front Cell Infect Microbiol. 2012;2:81. doi: 10.3389/fcimb.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lingwood CA, Binnington B, Manis A, Branch DR. Globotriaosyl ceramide receptor function - where membrane structure and pathology intersect. FEBS Lett. 2010;584:1879–86. doi: 10.1016/j.febslet.2009.11.089. [DOI] [PubMed] [Google Scholar]
- 69.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, et al. HUS Investigation Team Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–80. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 70.Laing CR, Zhang Y, Gilmour MW, Allen V, Johnson R, Thomas JE, et al. A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104:H4 outbreak strain. PLoS One. 2012;7:e37362. doi: 10.1371/journal.pone.0037362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brzuszkiewicz E, Thürmer A, Schuldes J, Leimbach A, Liesegang H, Meyer FD, et al. Genome sequence analyses of two isolates from the recent Escherichia coli outbreak in Germany reveal the emergence of a new pathotype: Entero-Aggregative-Haemorrhagic Escherichia coli (EAHEC) Arch Microbiol. 2011;193:883–91. doi: 10.1007/s00203-011-0725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M, et al. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci U S A. 2012;109:3065–70. doi: 10.1073/pnas.1121491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edwards LA, Bajaj-Elliott M, Klein NJ, Murch SH, Phillips AD. Bacterial-epithelial contact is a key determinant of host innate immune responses to enteropathogenic and enteroaggregative Escherichia coli. PLoS One. 2011;6:e27030. doi: 10.1371/journal.pone.0027030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steiner TS, Nataro JP, Poteet-Smith CE, Smith JA, Guerrant RL. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. J Clin Invest. 2000;105:1769–77. doi: 10.1172/JCI8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khan MA, Kang J, Steiner TS. Enteroaggregative Escherichia coli flagellin-induced interleukin-8 secretion requires Toll-like receptor 5-dependent p38 MAP kinase activation. Immunology. 2004;112:651–60. doi: 10.1111/j.1365-2567.2004.01923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harrington SM, Strauman MC, Abe CM, Nataro JP. Aggregative adherence fimbriae contribute to the inflammatory response of epithelial cells infected with enteroaggregative Escherichia coli. Cell Microbiol. 2005;7:1565–78. doi: 10.1111/j.1462-5822.2005.00588.x. [DOI] [PubMed] [Google Scholar]
- 77.Patton LM, Saggart BS, Ahmed NK, Leff JA, Repine JE. Interleukin-1 beta-induced neutrophil recruitment and acute lung injury in hamsters. Inflammation. 1995;19:23–9. doi: 10.1007/BF01534377. [DOI] [PubMed] [Google Scholar]
- 78.Greenberg DE, Jiang ZD, Steffen R, Verenker MP, DuPont HL. Markers of inflammation in bacterial diarrhea among travelers, with a focus on enteroaggregative Escherichia coli pathogenicity. J Infect Dis. 2002;185:944–9. doi: 10.1086/339617. [DOI] [PubMed] [Google Scholar]
- 79.Yen CC, Shen CJ, Hsu WH, Chang YH, Lin HT, Chen HL, et al. Lactoferrin: an iron-binding antimicrobial protein against Escherichia coli infection. Biometals. 2011;24:585–94. doi: 10.1007/s10534-011-9423-8. [DOI] [PubMed] [Google Scholar]
- 80.Bouckenooghe AR, Dupont HL, Jiang ZD, Adachi J, Mathewson JJ, Verenkar MP, et al. Markers of enteric inflammation in enteroaggregative Escherichia coli diarrhea in travelers. Am J Trop Med Hyg. 2000;62:711–3. doi: 10.4269/ajtmh.2000.62.711. [DOI] [PubMed] [Google Scholar]
- 81.Philipson CW, Bassaganya-Riera J, Viladomiu M, Pedragosa M, Guerrant RL, Roche JK, et al. The role of peroxisome proliferator-activated receptor γ in immune responses to enteroaggregative Escherichia coli infection. PLoS One. 2013;8:e57812. doi: 10.1371/journal.pone.0057812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res. 2000;49:497–505. doi: 10.1007/s000110050622. [DOI] [PubMed] [Google Scholar]
- 83.Adachi M, Kurotani R, Morimura K, Shah Y, Sanford M, Madison BB, et al. Peroxisome proliferator activated receptor gamma in colonic epithelial cells protects against experimental inflammatory bowel disease. Gut. 2006;55:1104–13. doi: 10.1136/gut.2005.081745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peck A, Mellins ED. Precarious balance: Th17 cells in host defense. Infect Immun. 2010;78:32–8. doi: 10.1128/IAI.00929-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006;203:2271–9. doi: 10.1084/jem.20061308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adachi JA, Ericsson CD, Jiang ZD, DuPont MW, Pallegar SR, DuPont HL. Natural history of enteroaggregative and enterotoxigenic Escherichia coli infection among US travelers to Guadalajara, Mexico. J Infect Dis. 2002;185:1681–3. doi: 10.1086/340419. [DOI] [PubMed] [Google Scholar]
- 87.Okhuysen PC, Dupont HL. Enteroaggregative Escherichia coli (EAEC): a cause of acute and persistent diarrhea of worldwide importance. J Infect Dis. 2010;202:503–5. doi: 10.1086/654895. [DOI] [PubMed] [Google Scholar]
- 88.Vial PA, Robins-Browne R, Lior H, Prado V, Kaper JB, Nataro JP, et al. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158:70–9. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 89.Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Child Health Epidemiology Reference Group of WHO and UNICEF Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–87. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 90.Tzipori S, Montanaro J, Robins-Browne RM, Vial P, Gibson R, Levine MM. Studies with enteroaggregative Escherichia coli in the gnotobiotic piglet gastroenteritis model. Infect Immun. 1992;60:5302–6. doi: 10.1128/iai.60.12.5302-5306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saif LJ, Ward LA, Yuan L, Rosen BI, To TL. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Arch Virol Suppl. 1996;12:153–61. doi: 10.1007/978-3-7091-6553-9_17. [DOI] [PubMed] [Google Scholar]
- 92.Birchall MA, Bailey M, Barker EV, Rothkötter HJ, Otto K, Macchiarini P. Model for experimental revascularized laryngeal allotransplantation. Br J Surg. 2002;89:1470–5. doi: 10.1046/j.1365-2168.2002.02234.x. [DOI] [PubMed] [Google Scholar]
- 93.Rothkötter HJ, Sowa E, Pabst R. The pig as a model of developmental immunology. Hum Exp Toxicol. 2002;21:533–6. doi: 10.1191/0960327102ht293oa. [DOI] [PubMed] [Google Scholar]
- 94.Roche JK, Cabel A, Sevilleja J, Nataro J, Guerrant RL. Enteroaggregative Escherichia coli (EAEC) impairs growth while malnutrition worsens EAEC infection: a novel murine model of the infection malnutrition cycle. J Infect Dis. 2010;202:506–14. doi: 10.1086/654894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morin N, Tirling C, Ivison SM, Kaur AP, Nataro JP, Steiner TS. Autoactivation of the AggR regulator of enteroaggregative Escherichia coli in vitro and in vivo. FEMS Immunol Med Microbiol. 2010;58:344–55. doi: 10.1111/j.1574-695X.2010.00645.x. [DOI] [PubMed] [Google Scholar]
- 96.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–72. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 97.Kitano H. Computational systems biology. Nature. 2002;420:206–10. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 98.Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–82. doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liévin-Le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and microbiota. Clin Microbiol Rev. 2006;19:315–37. doi: 10.1128/CMR.19.2.315-337.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goss PJ, Peccoud J. Quantitative modeling of stochastic systems in molecular biology by using stochastic Petri nets. Proc Natl Acad Sci U S A. 1998;95:6750–5. doi: 10.1073/pnas.95.12.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alves R, Antunes F, Salvador A. Tools for kinetic modeling of biochemical networks. Nat Biotechnol. 2006;24:667–72. doi: 10.1038/nbt0606-667. [DOI] [PubMed] [Google Scholar]
- 102.Bonabeau E. Agent-based modeling: methods and techniques for simulating human systems. Proc Natl Acad Sci U S A. 2002;99(Suppl 3):7280–7. doi: 10.1073/pnas.082080899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, et al. COPASI--a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–74. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 104.H. Trottier PP. Deterministic Modeling Of Infectious Diseases: Theory and Methods. The Internet Journal of Infectious Diseases 2001; 1.
- 105.Carbo A, Hontecillas R, Kronsteiner B, Viladomiu M, Pedragosa M, Lu P, et al. Systems modeling of molecular mechanisms controlling cytokine-driven CD4+ T cell differentiation and phenotype plasticity. PLoS Computational. doi: 10.1371/journal.pcbi.1003027. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.