Abstract
Epigenetic marks, such as DNA methylation, histone posttranslational modifications and microRNAs, are induced in B cells by the same stimuli that drive the antibody response. They play major roles in regulating somatic hypermutation (SHM), class switch DNA recombination (CSR) and differentiation to plasma cells or long-lived memory B cells. Histone modifications target the CSR and, possibly, SHM machinery to the immunoglobulin locus; they together with DNA methylation and microRNAs modulate the expression of critical elements of that machinery, such as AID, as well as factors central to plasma cell differentiation, such as Blimp-1. These inducible B cell-intrinsic epigenetic marks instruct the maturation of antibody responses. Their dysregulation plays an important role in aberrant antibody responses to foreign antigens, such as those of microbial pathogens, and self-antigens, such those targeted in autoimmunity, and B cell neoplasias.
Epigenetics and B cells
Epigenetics is the study of heritable changes in gene expression that are independent from the DNA sequence. Epigenetic marks include DNA methylation [1], histone posttranslational modifications [2] and non-coding RNAs [3], particularly microRNAs (Box 1). Together, these epigenetic marks comprise the “epigenome”. The epigenome of a cell is dynamic, exhibiting distinct patterns as the cell develops and differentiates in response to complex stimuli, and regulates gene activity for specification of cell differentiation fate.
Box 1. Features and molecular basis of epigenetics.
DNA methylation at the 5-position of the cytosine pyrimidine ring has been the first recognized and most investigated form of epigenetic information in the DNA. As recently discovered, DNA also carries cytosine hydroxylmethylation [1]. DNA methylation, which frequently occurs in CpG dinucleotide motifs in the promoter and the body of genes as well as intergenic regions, is the result of a balance between DNA methyltransferases (DNMTs)-mediated DNA methylation and DNA demethylation, as resulting from the passive loss of methylation during DNA replication and/or active methylation erasure mechanisms that remain to be defined and possibly involve cytidine deaminases and DNA repair pathways [87]. As a highly conserved transcriptional silencing mechanism, hypermethylated promoter DNA recruits methyl CpG binding protein 2 (MeCP2), which hinders the access of RNA polymerase II [1]. Conversely, demethylated CpG motifs recruit CXXC finger protein 1 (CFP1), a component of the Set1/MLL family, for H3K4me3 marking, thereby increasing chromatin accessibility [1, 114].
Histones H2A, H2B, H3 and H4 are major components of nucleosomes in chromatin and can be marked at their N-terminal tails by a variety of post-translational modifications, such as methylation, acetylation, phosphorylation, ubiquitination and sumolyation [2]. These modifications are introduced and catalyzed (“written”) or removed (“erased”) by their specific histone-modifying enzymes. They modulate interaction between histone and DNA as well as contacts between nucleosomes, leading to an open or closed chromatin state and, eventually, activation or repression of transcription [2]. Histone modifications also encrypt “histone codes” to recruit “histone code readers”, which are either specialized effector proteins or adaptors serving as docking sites for downstream effector proteins, thereby “transducing” epigenetic information to specify biological outcomes [115].
Non-coding RNAs include microRNAs, PIWI-interacting RNAs, small nucleolar RNAs and large intergenic non-coding RNAs [3]. Most of the work on the role of non-coding RNAs in the antibody response has focused on microRNAs (a main subject in this review). MicroRNAs regulate gene expression through targeting the 3’ untranslated region (3’ UTR) of mRNA transcripts and subsequent transcripts degradation or inhibiting their translation [3]. Generation of these short (average 22 nts) microRNA molecules depends on transcription of their host genes (e.g., Bic and Rtl1 as the host gene of miR-155 and miR-127, respectively [116, 117]) and processing of the long pri-microRNAs by the RNase III enzymes Drosha, followed by processing of the emerging pre-microRNAs by Dicer [3]. MicroRNAs also cross-regulate with DNA methylation and histone modifications [118].
Epigenetics changes regulate B cell development, which occurs in the bone-morrow, as timed by immunoglobulin (Ig) V(D)J (V, variable; D, diversity; J, joining) recombination, and eventually gives rise to naïve mature B cells (reviewed elsewhere [4]). Only recently has it become apparent that epigenetic marks also underpin peripheral B cell differentiation after antigen encounter. The purpose of this review article is to provide a conceptual framework for understanding how epigenetic marks contribute to Ig somatic hypermutation (SHM) and class switch DNA recombination (CSR), and generation of plasma cells and memory B cells. Here, we will be concerned with B cell epigenetic marks that underpin the generation of effective antibody responses; these are critical for the host defense against microbial pathogens and tumoral cells. We will also discuss the dysregulation of epigenetic processes, which is associated with autoimmunity and B cell neoplasias.
Epigenetic induction and functions in peripheral B cell differentiation
Once resting naive mature B cells encounter antigen in secondary lymphoid organs, they proliferate and differentiate into germinal center B cells, ultimately giving rise to long-lived plasma cells and memory B cells (Figure 1). Resting naïve B cells display genome-wide DNA hypermethylation, low amounts of overall histone acetylation [5, 6] and, consequently, low expression levels of a large number of genes, including β-actin and Gapdh housekeeping genes (G. Li et al., unpublished). In contrast, genes that are crucial for maintenance of B cell identity, such as Pax5, Spib and Ebf1, and B cell marker genes, such as Cd19, display an active epigenetic state [7, 8]. In resting B cells, most regions within the Ig heavy chain (Igh) locus exist in a closed chromatin state, devoid of activating histone modifications and enriched in repressive histone modifications (e.g., H3K9me3 and H3K27me3) [9, 10]. Resting, naïve B cells undergo VHDJH-Cμ transcription, which initiates at the VH promoter and runs through the intronic switch (S)μ region and Cμ/Cδ exon clusters. This encodes the surface B cell receptor (BCR), which comprises Igμ and Igδ heavy chain genes (Figure 2). Chromatin in the transcribed Igh VHDJH regions, the intronic μ enhancer (iEμ) and the Igh 3’ locus control region (3’ LCR) contains hypomethylated DNA and activating histone modifications [11–18] (Table 1). The epigenetic marks in the VHDJH region and, possibly, VλJλ and VκJκ regions are likely introduced during B cell development, as the open chromatin state of these regions is required for V(D)J recombination [4].
Figure 1.
Expression of selected microRNAs and their regulation of genes critical for different stages of peripheral B cell differentiation. Naïve mature B cells in peripheral lymphoid organs are activated by primary stimuli to undergo SHM and CSR (e.g., to IgG1, depicted) and to differentiate into plasma cells and memory B cells. In addition to DNA demethylation and histone modifications, which instruct genetic programs to specify expression of genes that drive distinct B cell differentiation stages, microRNAs play an important role in gene regulation through direct targeting of the 3’ UTR of transcripts of those genes, thereby regulating sequential B cell differentiation stages. Multiple microRNA molecules can cooperatively target one gene; an individual microRNA molecule can target multiple genes. Upregulation (red arrow) or downregulation (black arrow) of microRNAs results in alterations in the expression of key genes.
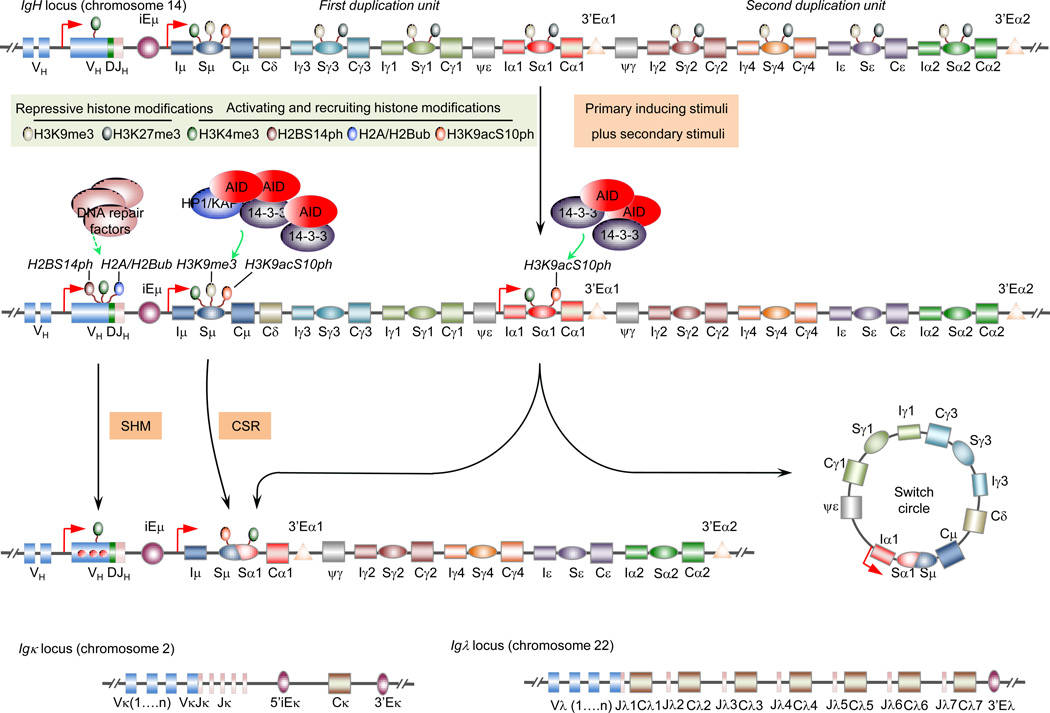
Figure 2.
Epigenetic targeting of SHM and CSR. The SHM and CSR machinery are targeted to the Igh locus VDJ and S regions, respectively (the human IgH locus is depicted; the SHM machinery also targets VκJκ or VλJλ region DNA in the Igκ and Igλ loci, as depicted). In resting naïve B cells as well as activated B cells, recombined VHDJH region chromatin displays DNA hypomethylation and activating histone marks, e.g., H3K4me3 (depicted here), H3K9ac/K14ac and H4K8ac. In B cells induced to undergo SHM, VHDJH region is enriched in H2AK119ub, H2BK120ub and H2B14ph, which probably recruit DNA repair factors to these regions for SHM. The Sμ region in the Igh locus is constitutively transcribed and marked by activating histone modifications, e.g., H3K4me3 (depicted here), H3K9ac/K14ac, H3K27ac, H3K36me3, H4K8ac and H2BK5ac, and combinatorial histone H3K9acS10ph modification (depicted here), starting in resting naïve B cells, in which all downstream S regions are marked by repressive histone modifications (e.g., H3K27me3, as depicted). In B cell undergoing CSR, the cytokine-selected acceptor S region(s) undergo high levels of germline IH-S-CH transcription, lose repressive histone modifications, acquire activating histone modifications, including H3K4me3 and combinatorial histone H3K9acS10ph modification (depicted here). H3K9acS10ph directly interacts with 14-3-3 proteins and stabilizes these adaptors and, therefore, AID on the donor Sμ and acceptor S region DNA for CSR to unfold. Also, H3K9me3, a repressive histone mark in general, occurs at low levels in Sμ and recruit the KAP1-HP1γ complex to stabilize AID in Sμ, but not downstream S regions.
Table 1.
Epigenetic marks and elements, and their functions in the antibody response.
| Target(s) | Epigenetic mark(s) | (Putative) functions of epigenetic marks | Refs. |
|---|---|---|---|
| DNA methylation | |||
| V(D)J | DNA hypomethylation | Increases V(D)J region accessibility | [14] |
| Igh 3’-LCR | DNA hypomethylation | Mediates germline VDJ and IH-S-CH transcription | [15] |
| Histone modifications in Ig loci* | |||
| V(D)J | H3K4me2/3, H3K9ac/K14ac, H4K8ac | Increase V(D)J region accessibility and transcription | [11, 18, 45] |
| iEμ | H3K4me3, H3K9ac/K14ac | Activate iEμ and enhance germline VDJ transcription and IH-S-CH transcription | [16, 17] |
| Igh 3’-LCR | H3K4me1/2 H3K9ac, H3K27ac, H4K8ac, H2BK5ac | Mediate VDJ and germline transcription | [12] |
| [17] | |||
| S region(s) | H3K27me3 | Decreases S region(s) accessibility | [9] |
| H3K4me3, H3K9ac/K14ac, H3K27ac, H4K8ac | Increase S region(s) accessibility | [9, 10, 13, 16, 17, 49–52] | |
| H3K9me3 | Recruits the HP1γ-KAP1 complex and AID to Sμ region | [10] | |
| H3K9acS10ph | Recruits 14-3-3 adaptors and AID to S region(s) | Li et al., (unpublished) | |
| H4K20me2 | Recruits 53BP1 to S region(s) in the DNA repair stage | [61] | |
| Histone modifications in B cell genes other than Ig loci | |||
| Aicda | H3K4me3, H3K9ac/K14ac, H3K36me3 | Increase transcription of Aicda | [17, 36] |
| Bcl6 | H3K4me3, H3K9ac | Increase transcription of Bcl6 | [19] |
| Pax5 | H3K4me3, H3K9ac | Increase transcription of Pax5 | [8, 73] |
| Cd19 | H3K4me3 | Increases transcription of Cd19 | [119] |
| Ciita | H3K4me3, H3K9ac, H3K18ac, H3K27ac | Increase transcription of Ciita | [73, 120] |
| c-Myc | H3 acetylation | Increases transcription of c-Myc | [72] |
| Spib | H3K4me2/3 H3 acetylation | Increase transcription of Spib | [73] |
| MicroRNAs | |||
| Aicda | miR-93* | Decrease expression of AID | [121] |
| miR-155** | [27, 28, 121] | ||
| miR-181b** | [122] | ||
| miR-361** | [123] | ||
| Prdm1 | miR-9** | Decrease Blimp-1 expression and/or inhibit plasma cell differentiation | [22] |
| miR-30** | [22] | ||
| miR-125b** | [21, 25] | ||
| miR-127** | [70] | ||
| let-7** | [68, 69] | ||
| Xbp1 | miR-127* | Decreases Xbp-1 expression and antibody secretion | [70] |
| Irf4 | miR-125b** | Decreases Irf-4 expression and inhibits plasma cell differentiation | [21, 25] |
| Bcl2 | miR-15a** | Decrease Bcl-2 expression | [21] |
| miR-16** | |||
| Pu.1 | miR-155** | Decreases Pu.1 and CSR to IgG1 | [26] |
| E2f1 | miR-223** | Decreases E2f-1 | [79] |
Repressive histone methylation: H3K9me3, H3K27me3.
Activating and recruiting histone modifications include histone acetylation: H3K9ac, H3K14ac, H3K27ac, H4K8ac, H2BK5ac and H3K9ac; histone methylations: H3K4me2/3, H4K20me2 and H3K36me3; combinatorial histone H3K9acS10ph modification; histone phosphorylation: H2BS14ph; histone ubiquitination: H2AK119ub and H2BK120ub.
MicroRNAs that target only the relevant human mRNA transcript.
MicroRNAs that target the relevant both human and mice mRNA transcript.
Active epigenetic marks in the Igh locus and in the Pax5, Spib, Ebf1 and Cd19 loci persist during naïve B cell activation [6, 11, 13] (Table 1). Proliferation and differentiation into hyper-mutating/class switching B cells is associated with genome-wide DNA hypomethylation and an overall increase in the amount of histone acetylation [5, 6]. Proto-oncogene B cell lymphoma 6 (Bcl-6) is induced in those somatically hyper-mutating/class-switching B cells with concomitant enrichment of activating histone marks H3K9ac and H3K4me3 in the Bcl6 promoter [19]. Bcl-6 inhibits B cell differentiation into plasma cells through transcription repressor functions that likely modify chromatin accessibility [20].
Naïve B cells, germinal center B cells, plasma cells and memory B cells display distinct microRNA expression profiles [21, 22]. Mice with a B cell-specific deletion in Dicer showed defects in generation of germinal center B cells, plasma cells and memory B cells [23]. Dicer-deficient B cells displayed defective biogenesis of multiple microRNAs, including miR-155 and miR-125b, which regulate expression of genes that fine-tune B cell germinal center differentiation [24, 25] (Table 1). A single microRNA molecule, such as miR-155, can target multiple genes, including AICDA/Aicda, which encodes activation-induced cytidine deaminase (AID), and Pu.1, thereby regulating sequential B cell differentiation stages [24, 26–28]. On the other hand, multiple microRNA molecules, such as miR-15a and miR-16, can cooperatively target one gene, such as Bcl2 in germinal center B cells [21] (Figure 1).
Thus, upon activation by antigens, B cells undergo DNA demethylation and histone modifications, and express a specific set of microRNAs. These epigenetic marks articulate transcription programs that specify distinct B cell differentiation stages and underpin the molecular changes, which are necessary for the maturation of the antibody response.
Epigenetics of SHM and CSR
SHM and CSR are critical for the maturation of the antibody response. SHM inserts point-mutations in Ig V(D)J DNA, thereby providing the structural substrate for selection of higher affinity antibody mutants by antigen. By deleting intervening DNA between an upstream (donor) and a downstream (acceptor) S region, CSR juxtaposes the expressed VHDJH DNA segment to a new CH exon cluster, thereby changing the CH chain of an antibody and endowing it with different biological effector functions (Figure 2). SHM and CSR are initiated by transcription through V(D)J and the donor/acceptor S regions that will undergo recombination, respectively, are mediated by AID, and share an array of DNA polymerases and repair enzymes [29, 30]. AID deaminates deoxycytidines (dCs) to yield deoxyuracils (dUs), thereby initiating the DNA lesions that will lead to insertions of point-mutations for SHM or double strand DNA breaks (DSBs) for CSR. The dU and dU:dG mispair DNA lesions are dealt with by uracil DNA glycosylase (Ung) and/or mismatch repair proteins (Msh2 and Msh6), leading to unfolding of a complex DNA repair process [31–34]. Epigenetic marks regulate expression of genes encoding AID and other SHM/CSR factors; they also target the SHM/CSR machinery, in a manner independent of V(D)J or S region transcription, by inducing an open chromatin state and directly recruiting critical adaptor proteins.
Epigenetic regulation of AID induction
AID is expressed at very low levels (mostly undetectable) in naïve B cells, highly induced in B cells undergoing SHM/CSR, and then repressed in plasma cells and memory B cells to preserve the specificity, affinity and isotype of a BCR/antibody [35–37]. AID is tightly regulated at the transcription, post-transcription and post-translation stages [38–40] (Table 1). Repression of the Aicda gene in naïve B cells was mediated by promoter hypermethylation [37]. During B cell activation, Aicda DNA was demethylated and the locus became enriched in H3K9ac/K14ac and H3K4me3 [17, 36]. These epigenetic changes, together with induction of HoxC4, NF-κB, STAT6 and other transcription factors [30, 41–43], activate Aicda transcription. Transcription elongation depends on induction of H3K36me3, an intragenic mark of gene activation, in the Aicda gene body, as suggested by AID downregulation following depletion of the H3K36me3 methyltransferase Setd2 [18]. Post-SHM/CSR downregulation of AID transcription probably results from re-methylation of the Aicda locus [37]. At the post-transcriptional stage, miR-155, miR-181b, miR-93 and miR-361 target different sites in the 3’ untranslated region (3’ UTR) of Aicda transcripts to downregulate AID [40]. These microRNAs likely repress AID in naïve B cells and repress it in B cells that complete SHM/CSR (Figure 1). Deleting the miR-155-targeting site in Aicda transcripts increased AID expression, enhanced CSR and promoted Myc/Igh translocations [27, 28]. Thus, microRNAs work in concert with DNA methylation and histone modifications to control the induction of AID for SHM/CSR.
Epigenetic targeting of the SHM machinery
The SHM machinery is targeted to V(D)J DNA through transcription, unique targeting sequences, and likely DNA demethylation and histone modifications [29, 44] (Figure 2). DNA hypomethylation seems to play a role in SHM, as in B cells carrying two nearly identical pre-rearranged transgenic Igκ alleles, only the hypomethylated allele was hypermutated despite comparable transcription of both alleles [14]. DNA demethylation probably facilitates SHM targeting by promoting H3K9ac/K14ac, H4K8ac and H3K4me3 histone modifications that are associated with an open chromatin state and are enriched in the V(D)J region [11, 18, 45]. Both H4K8ac and H3K4me3 play a role in SHM. H4K8ac increased concomitantly with VHDJH mutations in human BL2 cells upon treatment with trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor [45]. Sustained H4K8ac in VλJλ region required E2A, whose inactivation led to decreased mutations [29, 46]. Decreased H3K4me3 in VHDJH regions in BL2 cells upon knock-down of histone chaperone suppressor of Ty6 (Spt6) correlated with reduction in VHDJH mutations [18].
Histone modifications likely mediate recruitment of error-prone DNA polymerases at the DNA repair stage of SHM. H2AK119 ubiquitination (ub) and H2BK120ub were enriched in VHDJH but not CH regions and co-localized with error-prone translesion DNA synthesis (TLS) polymerase η (pol η) in AID-containing foci [47]. Pol η is likely recruited to those loci by H2AK119ub and H2BK120ub (as well as by monoubiquitinated PCNA) through its ubiquitin-binding domain [48]. H2BS14 phosphorylation (ph) marked V(D)J region in an AID-dependent manner [11], and probably also contributes to recruitment of DNA repair factors. Thus, DNA hypomethylation and activating histone modifications increase V(D)J region chromatin accessibility to the SHM machinery, including AID and error-prone DNA repair factors, which, in turn, are likely stabilized by modified histones.
Epigenetic targeting of the CSR machinery
Histone modifications target the CSR machinery to the upstream donor (e.g., Sμ) and the downstream acceptor S regions (e.g., Sγ3, Sγ1, Sα1, Sγ2, Sγ4, Sε or Sα2 in the human; Sγ3, Sγ1, Sγ2b, Sγ2a, Sε or Sα in the mouse) that are “primed” for CSR (Table 1). Activating histone marks, such as H2BK5ac, H3K9ac/K14ac, H3K27ac, H4K8ac, H3K4me3 and H3K36me3, together with germline Iμ-Sμ-Cμ transcription, were found in the donor Sμ region, even in naïve resting B cells [9, 13, 16, 17, 49–52] (Figure 2), indicating that Sμ is in a constitutively open state and poised for switching. Induction of CSR requires both “primary” and “secondary” (CSR-inducing) stimuli [30] (Figure 3). Primary stimuli comprise a T-dependent stimulus, i.e., engagement of CD40, which is constitutively expressed on B cells, by CD154 expressed on activated T cells, and T-independent stimuli, mainly, dual Toll-like receptor (TLR) and B cell receptor (BCR) engagement by MAMPs and antigenic epitopes, respectively, on bacteria or viruses. These stimuli induce B cell activation, proliferation and differentiation. Secondary stimuli consist of cytokine interleukin-4 (IL-4), transforming growth factor-β (TGF-β) and interferon-γ (IFN-γ, in the mouse, but not the human). In conjunction with primary stimuli, they direct class switching to IgG, IgA or IgE by selecting the acceptor S region, such as IL-4 in CSR to Sγ1 and Sε, TGF-β to Sγ2b and Sα, and IFN-γ to Sγ2a (mouse), through induction of specific germline IH-S-CH transcription and activating S region histone modifications [16, 17, 50, 51]. They also synergize with primary stimuli to induce maximal AID expression. When enabled by primary stimuli, which induce histone-modifying enzymes (Li et al., unpublished) and trigger chromatin decondensation in downstream S regions, e.g., by removing of repressive H3K9me3 and H3K27me3 [9, 10], cytokines activate transcription factors that bind to selected IH promoters and initiate germline IH-S-CH transcription [30] (Figure 2). These allow histone-modifying enzymes to ride on the RNA polymerase (pol) II, on which AID is also hitched through direct interaction with Spt5, to reach S regions during elongation of germline IH-S-CH transcription [16, 17, 53–55]. Upon RNA pol II stalling, likely due to complex secondary DNA structures (e.g., R-loop and cruciform-like structures) in S regions [30, 56], histone-modifying enzymes catalyze histone modifications in these regions. Modified histones together with DNA transcription open the chromatin in S regions, thereby allowing for access of CSR factors. The importance of histone-modifying enzymes in CSR is suggested by the defective S region histone modifications and CSR following inhibition of H3K4 methyltransferases MLL/SET1 [51] or H3K9 acetyltransferases Gcn5 and Pcaf (Li et al., unpublished).
Figure 3.
Epigenetic changes in B cells in response to primary and secondary (CSR-inducing) stimuli. Primary stimuli include CD40 engagement by CD154, dual TLR/BCR engagement by MAMPs and antigenic epitopes, respectively, on bacteria or viruses (a bacterium is depicted here; bacterial LPS engages both TLR4 and BCR through its monophosphoryl lipid A moiety and polysaccharidic moiety, respectively), and dual engagement of TACI (by BAFF or APRIL) and TLR, BCR or CD40. They induce histone-modifying enzymes, which catalyze activating histone modifications (e.g., H3K4me3 and H3K9ac/K14ac) in the promoter of microRNA host genes and genes encoding AID and other SHM/CSR factors, thereby creating an open chromatin state. This together with transcription factors activated by primary stimuli induce the expression of those genes. Induced microRNAs regulate expression of SHM/CSR factors. Histone-modifying enzymes also catalyze histone modifications, such as H2B14ph, in the Igh, Igλ and Igκ V(D)J regions for SHM targeting (depicted are VHDJH and VκJκ). When enabled by primary stimuli, secondary stimuli, e.g., the cytokines IL-4, TGF-β or IFN-γ, activate transcription factors that specifically bind selected Igh intervening (IH) promoters to initiate germline IH-S-CH transcription in the acceptor S region (depicted is recombination from Sμ to Sγ1). This allows primary stimuli-induced histone-modifying enzymes to ride on RNA polymerase II to reach Sγ1 region and catalyze activating histone modifications in this S region for CSR targeting. Inset: writing and erasure of histone acetylation by histone acetyltransferases (HATs) and histone deacetylases (HDACs, which are inhibited by HDAC inhibitors, HDIs), histone methylation by histone methyltransferases (HMTs) and histone demetylases (HDMs), histone phosphorylation by kinase and phosphatases, and histone ubiquitination by Ub ligases and deubiquitinases (DUBs). DNA methylation is catalyzed by DNA methyltransferase (DNMTs).
Activating histone modifications, including H3K9ac and H3K4me3, were enriched in the (intronic) donor and acceptor S regions at levels much higher than those in the related IH promoter and CH regions [16, 17]. These activating marks are “read” by CSR factors, including 14-3-3 adaptors, which, like AID and histone-modifying enzymes, were induced by primary stimuli (Mai et al., unpublished). 14-3-3 adaptors specifically bind to the combinatorial histone H3K9acS10ph modification in S regions (Li et al., unpublished) and 5’-AGCT-3’ repeats (Figure 2), which are characteristic motifs of all Igh locus S regions, but not IH promoters, CH regions or other genome areas [57]. Due to their high affinity for 5’-AGCT-3’ repeats, 14-3-3 can potentially bind all S regions. Nevertheless, these adaptors are recruited only to the S regions that undergo recombination [57]. This is due to the open chromatin state of these regions, as well as specific binding of 14-3-3 to H3K9acS10ph. The specificity of 14-3-3 adaptors for H3K9acS10ph and 5’-AGCT-3’ repeats are evocative of RAG1-RAG2 complex specificity for H3K4me3 and V, D and J gene RSS (recombination signal sequences) [4].
14-3-3 adaptors directly interact with AID and target it to the upstream and downstream S regions that undergo recombination [57], thereby “transducing” the epigenetic code (Figure 2). H3K9me3 was also found, albeit in low amounts, in Sμ and recruited the KAP1-HP1γ complex to stabilize AID on Sμ, but not on downstream S regions [10]. Accordingly, abrogation of the H3K9 methyltransferase Suv39 impaired CSR to IgA [58]. Histone readers with scaffold functions, e.g., 14-3-3 adaptors, together with scaffold proteins, such as RPA [52] and Rev1 [59], nucleate the assembly of macromolecular complexes on S region DNA, thereby stabilizing AID and/or Ung for generation of DNA lesions. The RNA exosome may also be recruited to the same complex and allow AID to deaminate dCs in both transcribed and non-transcribed DNA strands [60]. Histone modifications are also read by DNA repair factors, such as p53-binding protein 1 (53BP1). 53BP1 is recruited to DSBs where it is stabilized by H4K20me2, thereby playing an important role in CSR, as suggested by the findings that abrogation of histone methyltransferase MMSET expression impaired H4K20me2 enrichment and 53BP1 recruitment in/to the Igh locus S regions, resulting in defective CSR [61]. 53BP1 may functions as a scaffold to recruit/stabilize additional DNA repair factors for CSR.
Thus, activating histone modifications are induced in S regions and create an open chromatin state, which allows for the access of the CSR machinery. Epigenetic specification of CSR targeting entails reading of histone codes by scaffold proteins, which orchestrate the assembly of macromolecular complexes in the sequential DNA lesion and repair stages.
Epigenetics of plasma cell and memory B cell differentiation
Plasma cells are terminally differentiated effector cells that generally have undergone SHM and CSR. These cells do not proliferate, but secrete clonospecific antibodies at high rates (107 molecules/hour) [20]. While some memory B cells are Igμ+, most memory B cells are class-switched (Igγ+, Igε+ or Igα+) and express somatically hypermutated V(D)J gene segments. Upon reactivation, memory B cells differentiate into plasma cells to mediate an anamnestic response [62]. Reactivated memory B cells can undergo further SHM and/or CSR before differentiating into plasma cells or converting back to memory B cells [63]. The stimuli, signals and processes underlying differentiation into plasma cells and memory B cells are not well understood, but epigenetic mechanisms contribute to these processes.
Epigenetics of plasma cells
Upregulation of Blimp-1 (encoded by Prdm1) is central to B cell differentiation into plasma cells, [20, 64]. Downregulation of Pax-5 and Pax-5-driven Bcl-6 led to de-repression of the Prdm1 promoter from Bcl-6-mediated epigenetic silencing. This was associated with increased histone acetylation in the Prdm1 promoter, likely resulting from release of Bcl-6-bound HDACs [65, 66]. Blimp-1 expression was enhanced by HDAC inhibitor TSA, which also upregulated Syndecan-1 (CD138, a plasma cell hallmark) [67]. The hyperacetylated state of the Prdm1 locus likely remains intact in long-lived plasma cells in the bone marrow, thereby allowing for sustained expression of Blimp-1 to maintain cellular identity. Downregulation of Blimp-1 in a plasmacytoid cell line by enforced expression of Bcl-6 resulted in re-expression of B cell markers, including CD19 [66]. Expression of Blimp-1 was inhibited by microRNAs, including miR-9, miR-30, miR-125b, miR-127 and let-7 [21, 22, 25, 68–70] (Table 1). Overexpression of miR-125b in B cells inhibited Blimp-1 expression and differentiation into plasma cells [25]. Also, miR-125b downregulated Irf-4, which is also critical for plasma cell differentiation and reciprocally regulates Blimp-1 [21, 25, 71], and miR-127 downregulated Xbp-1, a CREB/ATF transcription factor that activates genes encoding proteins in secretory pathways for antibody secretion [20, 70].
Blimp-1 is a transcriptional repressor that binds to the promoters of Bcl6, Pax5 and Spib (all of which inhibit B cell differentiation into plasma cells) and downregulates the expression of these genes [20], possibly through deacetylation of their promoters. Indeed, Blimp-1 has been shown to bind HDACs [72]; Pax5, Spib and perhaps Bcl6 promoters showed decreased histone acetylation in plasma cells [72, 73]. In addition, Blimp-1, through a similar epigenetic mechanism, downregulates c-Myc in plasma cells [72], thereby maintaining the terminal differentiation state of these cells [20]. Finally, Blimp-1 was shown to interact with G9a, an H3K9 methyltransferase, and likely recruits this enzyme to the Pax5 and Spib promoters, thereby increasing H3K9me3 and repressing activation of these promoters [73, 74]. Thus, epigenetic induction of Blimp-1 and Blimp-1-mediated epigenetic inhibition of target genes drives plasma cell differentiation, and possibly maintains plasma cell identity.
Epigenetics of memory B cells
Memory B cells inherit epigenetic information from their precursor and acquire new epigenetic marks, which make these resting B cells prone to respond to antigen. Expression of memory B cell hallmark genes, e.g., Cd38 (in the mouse) and CD27 (in the human), likely results from de novo activating histone modifications, as induced during B cell activation. Genes that maintain B cell identity, such as Pax5 and Spib, are also expressed in memory B cells [7], possibly as a result of the activating epigenetic state that originated in naive B cells. Post-recombined Sμ-Sγ, Sμ-Sε or Sμ-Sα regions in the Igh locus probably display constitutive (Sμ portion) or induced (Sγ, Sε or Sα portion) histone modifications, which are likely transmitted to memory B cells, leading to comparable epigenetic landscapes in Igγ+, Igε+ and Igα+ memory B cells.
In quiescent memory B cells, Aicda and Prdm1 are silenced to preserve BCR integrity and prevent differentiation into plasma cells, respectively [75, 76]. Many genes are likely silenced by DNA methylation as catalyzed by DNMTs, such as DNMT3a, which is highly expressed in memory B cells [75], and upregulation of selected microRNAs, including miR-125b and let-7, which negatively regulate Prdm1 [21, 22, 68, 69]. Downregulation of miR-15a and miR-16, which target Bcl2, likely contributes to memory B cell survival [21, 77] and re-expression of Krüppel-like factor (KLFs), which can bind HDACs, mediates memory B cell quiescence [78].
Memory B cells are likely in a "poised" state, marked by active and repressive histone modifications in a balance, therefore displaying the potential for rapid activation upon antigen re-encounter. Poised genes include those needed for proliferation, such as Ki67, Ccnd2, Ccne1 and E2f1 [75] (E2f-1 is also downregulated by miR-223 [79]), or re-initiation of SHM or CSR, e.g., Bcl-6 [75, 80]. The switch from the poised state to the silenced state of those genes is likely mediated by KLFs, as ectopic expression of KLF-4 or KLF-9, converted memory B cells into cells with a “naive” B cell phenotype [81]. The poised state of genes, together with a high level of the BCR signaling molecule Malt1 [76] and the moderate level of KLFs [75], allow memory B cells to respond to antigen much more rapidly than naive B cells with the same BCR, showing a proliferative burst upon re-encounter with antigen. Thus, DNA methylation, histone modifications and microRNAs regulate gene expression programs that underpin B cell differentiation into plasma cells and memory B cells, and also play important roles in maintaining the identity of these differentiated cells.
Aberrant B cell epigenetic status and disease
Several studies have addressed the role of epigenetic changes in T cells, including regulatory T cells (Tregs), in health and autoimmune conditions [82, 83]. Aberrant patterns of DNA methylation, histone modifications and microRNA expression in B cells, however, have received less attention; only recently did they begin to be recognized in autoimmune conditions as well as B cell neoplasias [83–85].
Epigenetics in B cell (autoantibody)-mediated autoimmunity
Systemic autoimmune diseases such as systemic lupus erythematosus, rheumatoid arthritis, systemic sclerosis and dermatomyositis are associated with overall DNA hypomethylation, although the genomic regions that are hypomethylated were not well defined [82, 83, 85]. The role of DNA methylation in autoimmune diseases is epitomized by the discordance of systemic lupus erythematosus in monozygotic twins. B cells isolated from lupus patients displayed profound changes in DNA methylation, as compared to those from healthy monozygotic siblings [86]. The decreased DNA methylation in autoreactive B cells likely resulted from reduced DNMT1 and DNMT3b expression; it could also result from an active DNA demethylation dependent on AID-mediated cytosine deamination [87], consistent with AID upregulation in B cells of lupus patients or lupus-prone mice [88]. The role of B cell DNA hypomethylation in systemic autoimmunity is emphasized by development of lupus-like disease after prolonged treatment with hydralazine, procainamide or isoniazid (inhibitors of DNA methylation) [84]. Accordingly, adoptive transferring of mouse B cells treated with hydralazine led to productions of antinuclear autoantibodies in recipient mice [89]. The involvement of aberrant histone modifications in the development of lupus is indicated by the increased histone acetylation and reduced lupus symptoms in lupus-prone mice treated with HDAC inhibitors [90]. The amelioration of lupus symptoms results in part from decreased production of inflammatory cytokines by T cells, upregulation of Tregs [90] and, perhaps most importantly, inhibition of autoantibody secretion by B cells (H. Zan et al., unpublished).
MicroRNAs are aberrantly expressed in different cells types and tissues in patients with autoimmune disease [91, 92] and dysregulation of microRNAs has been associated with autoimmune traits. A B cell-specific Dicer deletion, which abolished mature microRNAs, led to a skewed BCR repertoire with hallmarks of autoreactivity, suggesting that selected microRNAs prevent the generation of self-reactive antibodies [93]. Expression of miR-150 was downregulated in spleen B cells from lupus-prone MRL/lpr mice, as compared to MRL/lpr−/− mice [94]. This likely stemmed from decreased histone acetyltransferase activity, resulting in impaired acetylation and decreased transcription of miR-150 host gene. Indeed, a B cell-specific conditional knock-in dominant negative p300 mutant resulted in production of class-switched anti-dsDNA autoantibodies and development of lupus-like traits in C57BL/6 × 129/sv F1 mice [95]. On the other hand, expression of certain microRNAs may contribute to the development of autoimmunity, as suggested by amelioration of autoimmune splenomegaly in lupus mice upon silencing of miR-21 [96] and increased numbers of germinal center B cells in the spleen and peripheral lymph nodes in miR-17-92 transgenic mice [97]. Thus, dysregulated expression of microRNAs can contribute to autoimmunity.
Epigenetics in B lymphomagenesis
The epigenome of tumoral cells of different origin is characterized by global changes in DNA methylation and altered histone modification patterns [98]. Distorted epigenetic marks interact with genetic alterations at all stages of cancer development to promote neoplastic transformation and tumor cell progression [99].
The abnormal DNA methylation in B cell lymphomas varies with chromosomal regions, local gene density and methylation status of neighboring genes [100]. Aberrant DNA hypomethylation of promoters can lead to increased transcription of genes with oncogenic potential and aberrant DNA hypermethylation lead to decreased transcription of genes with tumor suppressor function. For example, mantle B cell lymphomas display aberrant hypomethylation in promoter of genes that are involved in pathways controlling cell cycle or apoptosis such as Cdk5, and aberrant hypermethylation in the promoter of tumor suppressor genes such as Cdkn2b [101]. Aberrant hypomethylation or demethylation could involves AID, which can mediate active DNA demethylation [102] and is upregulated in human B cell non-Hodgkin lymphomas [103]. Hypermethylation is likely related to upregulation of DNMTs, such as DNMT3b, which is overexpressed in diffuse lager B cell lymphomas [104]. Accordingly, transgenic mice expressing Dnmt3b developed mediastinal B cell lymphomas displaying significantly altered methylation patterns [104].
Altered expression or mutation of the histone-modifying enzymes promotes aberrant gene expression patterns, which give rise to many cancer characteristics [105]. Overexpression of EZH2, a histone methyltransferase component of polycomb repression complex 2 (PRC2) that catalyzes H3K27me3 and stimulates tumor growth, has been implicated in B lymphomagenesis [106]. Somatic mutations of Y641 and A677 residues within the catalytic domain of EZH2 occur in diffuse large B cell lymphoma and follicular lymphoma [107]. These mutations promoted EZH2 activity and increased H3K27me3 levels in those cells [106]. Further, other histone methyltransferases, such as MLL2, histone demethylases UTX and JMJD2C and histone acetyltransferases CBP and p300, are frequently mutated in B cell lymphomas [104]. Mutations in these histone-modifying enzymes are likely mediated by AID, which is highly expressed in those neoplastic B cells, and result in aberrant patterns of histone modifications and disruption of chromatin structure, ultimately dysregulated gene transcription programs.
Aberrant expression of microRNAs contributes to B lymphomagenesis [22, 108]. Expression of miR-15 and miR-16-1, both of which target Bcl-2, was found to be downregulated in chronic lymphocytic leukemia B cells [109, 110]. Deletion of miR-15a and miR-16-1 in B cells resulted in clonal lymphoproliferative disorders [111]. Conversely, the miR-17-92 cluster, which targets the tumor repressor genes Bim and Pten, is often amplified in human lymphomas [97, 112]. Accordingly, ectopic expression of the miR-17–92 cluster in lymphocytes led to development of lymphoproliferative disease in these mice [97, 112]. B lymphomas often express substantial amounts of mRNA isoforms with shorter 3’ UTRs, which lack microRNA-binding sites, thereby escaping microRNA-directed silencing [113]. For example, a short cyclin D1 mRNA isoform that lacks part of 3’ UTR is expressed in a subset of mantle cell lymphomas [113]. These cells have increased cyclin D1 expression and B cell proliferation. Thus, by allowing selected oncogenes to elude the regulation by their modulatory microRNAs, the shortening of 3’ UTRs may be provide an important mechanism in B cell neoplastic transformation.
Concluding remarks
During an antibody response, primary stimuli induce epigenetic marks/changes in B cells, thereby regulating expression of genes crucial for SHM, CSR and/or B lymphocyte differentiation into plasma cells or memory B cells. This notion is supported by increasing new findings, but also calls for more and better mechanistic insights into how a B lymphocyte acquires specific epigenetic modifications and/or elements in selected gene loci, in response to a variety of stimuli that differentially activate transcription factors. The cross-regulation and feedback-regulation of different epigenetic modifications/elements further increase the complexity of the B cell epigenome. The epigenome “interacts” with complex genetic information for precise control of gene expression, a paradigm that is well illustrated by the highly specific AID induction in B cells undergoing SHM/CSR through epigenetic modifications and activation of key transcription factors.
Histone modifications in Ig V(D)J and S regions are critical for targeting of the SHM and CSR machinery, respectively. As histone modifications are generally coordinated, it is difficult to envision that an individual modification provide unique targeting signals. More likely, combinatorial histone modifications provide complex multi-step targeting signals recognized by the SHM and CSR machinery. Thus, the dynamic composition of combinatorial histone modifications needs a better definition to better understand the targeting of SHM and CSR.
Epigenetic dysregulation in B cells can result in aberrant antibody responses and, by compounding genetic susceptibility, mediate autoimmunity and neoplasia. The reversible nature of epigenetic marks, however, makes them attractive targets of small molecule or biologic therapeutics. A better understanding of the altered epigenetic processes in autoimmune diseases and B cell cancers will open perspectives for the development of new therapeutics.
Epigenetic marks are induced in B cells by the stimuli that drive the antibody response
Histone modifications target the CSR and, possibly, SHM machinery to the Ig locus
Histone modifications, DNA methylation and microRNAs modulate AID and Blimp-1 expression
Epigenetic dysregulation plays an important role in autoimmunity and B cell neoplasias
Acknowledgements
We apologize that owing to space limitations only a fraction of the relevant literature was cited in this review article. This work was supported by U.S. National Institutes of Health grants AI 079705, AI 045011 and AI 060573 to Paolo Casali.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 4.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 5.Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, Ballon G, Yang SN, Weinhold N, Reimers M, Clozel T, Luttrop K, Ekstrom TJ, Frank J, Vasanthakumar A, Godley LA, Michor F, Elemento O, Melnick A. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;118:3559–3569. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z, Mai A, Sun J. Lysine acetylation regulates Bruton's tyrosine kinase in B cell activation. J. Immunol. 2010;184:244–254. doi: 10.4049/jimmunol.0902324. [DOI] [PubMed] [Google Scholar]
- 7.Schmidlin H, Diehl SA, Blom B. New insights into the regulation of human B-cell differentiation. Trends Immunol. 2009;30:277–285. doi: 10.1016/j.it.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury M, Forouhi O, Dayal S, McCloskey N, Gould HJ, Felsenfeld G, Fear DJ. Analysis of intergenic transcription and histone modification across the human immunoglobulin heavy-chain locus. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15872–15877. doi: 10.1073/pnas.0808462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeevan-Raj BP, Robert I, Heyer V, Page A, Wang JH, Cammas F, Alt FW, Losson R, Reina-San-Martin B. Epigenetic tethering of AID to the donor switch region during immunoglobulin class switch recombination. J. Exp. Med. 2011;208:1649–1660. doi: 10.1084/jem.20110118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odegard VH, Kim ST, Anderson SM, Shlomchik MJ, Schatz DG. Histone modifications associated with somatic hypermutation. Immunity. 2005;23:101–110. doi: 10.1016/j.immuni.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3' end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol. Cell. Biol. 2005;25:1511–1125. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Whang N, Wuerffel R, Kenter AL. AID-dependent histone acetylation is detected in immunoglobulin S regions. J. Exp. Med. 2006;203:215–226. doi: 10.1084/jem.20051774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraenkel S, Mostoslavsky R, Novobrantseva TI, Pelanda R, Chaudhuri J, Esposito G, Jung S, Alt FW, Rajewsky K, Cedar H, Bergman Y. Allelic 'choice' governs somatic hypermutation in vivo at the immunoglobulin kappa-chain locus. Nat. Immunol. 2007;8:715–722. doi: 10.1038/ni1476. [DOI] [PubMed] [Google Scholar]
- 15.Giambra V, Volpi S, Emelyanov AV, Pflugh D, Bothwell AL, Norio P, Fan Y, Ju Z, Skoultchi AI, Hardy RR, Frezza D, Birshtein BK. Pax5 and linker histone H1 coordinate DNA methylation and histone modifications in the 3' regulatory region of the immunoglobulin heavy chain locus. Mol. Cell. Biol. 2008;28:6123–6133. doi: 10.1128/MCB.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329:917–923. doi: 10.1126/science.1187942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. J. Biol. Chem. 2012;287:32415–32429. doi: 10.1074/jbc.M112.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandrareddy H, Bouska A, Shen Y, Ji M, Rizzino A, Chan WC, McKeithan TW. BCL6 promoter interacts with far upstream sequences with greatly enhanced activating histone modifications in germinal center B cells. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11930–11935. doi: 10.1073/pnas.1004962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat. Rev. Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 21.Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 24.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 25.Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, Reina San-Martin B, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat. Rev. Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 30.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casali P, Pal Z, Xu Z, Zan H. DNA repair in antibody somatic hypermutation. Trends Immunol. 2006;27:313–321. doi: 10.1016/j.it.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casali P. Somatic recombination and hypermutation in the immune system. In: Krebs JE, et al., editors. Lewin's Genes X. 10th edn. Jones & Bartlett: 2009. pp. 570–623. [Google Scholar]
- 34.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat. Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasqualucci L, Guglielmino R, Houldsworth J, Mohr J, Aoufouchi S, Polakiewicz R, Chaganti RS, Dalla-Favera R. Expression of the AID protein in normal and neoplastic B cells. Blood. 2004;104:3318–3325. doi: 10.1182/blood-2004-04-1558. [DOI] [PubMed] [Google Scholar]
- 36.Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montano C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J. Exp. Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujimura S, Matsui T, Kuwahara K, Maeda K, Sakaguchi N. Germinal center B-cell-associated DNA hypomethylation at transcriptional regions of the AID gene. Mol. Immunol. 2008;45:1712–1719. doi: 10.1016/j.molimm.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Stavnezer J. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 2011;32:194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li G, Pone EJ, Tran DC, Patel PJ, Dao L, Xu Z, Casali P. Iron inhibits activation-induced cytidine deaminase enzymatic activity and modulates immunoglobulin class switch DNA recombination. J. Biol. Chem. 2012;287:21520–21529. doi: 10.1074/jbc.M112.366732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:81–99. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dedeoglu F, Horwitz B, Chaudhuri J, Alt FW, Geha RS. Induction of activation-induced cytidine deaminase gene expression by IL-4 and CD40 ligation is dependent on STAT6 and NF-κB. Int. Immunol. 2004;16:395–404. doi: 10.1093/intimm/dxh042. [DOI] [PubMed] [Google Scholar]
- 42.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat. Commun. 2012;3:767. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv. Immunol. 2010;105:159–191. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo CJ, Martin A, Scharff MD. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 46.Kitao H, Kimura M, Yamamoto K, Seo H, Namikoshi K, Agata Y, Ohta K, Takata M. Regulation of histone H4 acetylation by transcription factor E2A in Ig gene conversion. Int. Immunol. 2008;20:277–284. doi: 10.1093/intimm/dxm140. [DOI] [PubMed] [Google Scholar]
- 47.Borchert GM, Holton NW, Edwards KA, Vogel LA, Larson ED. Histone H2A and H2B are monoubiquitinated at AID-targeted loci. PLoS One. 2010;5:e11641. doi: 10.1371/journal.pone.0011641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 49.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 50.Kuang FL, Luo Z, Scharff MD. H3 trimethyl K9 and H3 acetyl K9 chromatin modifications are associated with class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5288–5293. doi: 10.1073/pnas.0901368106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22190–22195. doi: 10.1073/pnas.1016923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat. Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J. Exp. Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storck S, Aoufouchi S, Weill JC, Reynaud CA. AID and partners: for better and (not) for worse. Curr. Opin. Immunol. 2011;23:337–344. doi: 10.1016/j.coi.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Kenter AL. AID targeting is dependent on RNA polymerase II pausing. Semin. Immunol. 2012;24:281–286. doi: 10.1016/j.smim.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, Steinacker P, Li Z, Yates J, 3rd, Herron B, Otto M, Zan H, Fu H, Casali P. 14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradley SP, Kaminski DA, Peters AH, Jenuwein T, Stavnezer J. The histone methyltransferase Suv39h1 increases class switch recombination specifically to IgA. J. Immunol. 2006;177:1179–1188. doi: 10.4049/jimmunol.177.2.1179. [DOI] [PubMed] [Google Scholar]
- 59.Zan H, White CA, Thomas LM, Mai T, Li G, Xu Z, Zhang J, Casali P. Rev1 recruits Ung to switch regions and enhances dU glycosylation for immunoglobulin class switch DNA recombination. Cell Rep. 2012;2:1220–1232. doi: 10.1016/j.celrep.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Basu U, Meng FL, Keim C, Grinstein V, Pefanis E, Eccleston J, Zhang T, Myers D, Wasserman CR, Wesemann DR, Januszyk K, Gregory RI, Deng H, Lima CD, Alt FW. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pei H, Wu X, Liu T, Yu K, Jelinek DF, Lou Z. The histone methyltransferase MMSET regulates class switch recombination. J. Immunol. 2013;190:756–763. doi: 10.4049/jimmunol.1201811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat. Rev. Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, Reynaud CA, Weill JC. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 64.Nutt SL, Taubenheim N, Hasbold J, Corcoran LM, Hodgkin PD. The genetic network controlling plasma cell differentiation. Semin. Immunol. 2011;23:341–349. doi: 10.1016/j.smim.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O, Khochbin S. Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 66.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 67.Lee SC, Bottaro A, Insel RA. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol. Immunol. 2003;39:923–932. doi: 10.1016/s0161-5890(03)00029-4. [DOI] [PubMed] [Google Scholar]
- 68.West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nie K, Zhang T, Allawi H, Gomez M, Liu Y, Chadburn A, Wang YL, Knowles DM, Tam W. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: a potential role of the microRNA let-7. Am. J. Pathol. 2010;177:1470–1479. doi: 10.2353/ajpath.2010.091291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leucci E, Onnis A, Cocco M, De Falco G, Imperatore F, Giuseppina A, Costanzo V, Cerino G, Mannucci S, Cantisani R, Nyagol J, Mwanda W, Iriso R, Owang M, Schurfeld K, Bellan C, Lazzi S, Leoncini L. B-cell differentiation in EBV-positive Burkitt lymphoma is impaired at posttranscriptional level by miRNA-altered expression. Int. J. Cancer. 2010;126:1316–1326. doi: 10.1002/ijc.24655. [DOI] [PubMed] [Google Scholar]
- 71.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 72.Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 2000;20:2592–2603. doi: 10.1128/mcb.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su ST, Ying HY, Chiu YK, Lin FR, Chen MY, Lin KI. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell. Biol. 2009;29:1421–1431. doi: 10.1128/MCB.01158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 75.Luckey CJ, Bhattacharya D, Goldrath AW, Weissman IL, Benoist C, Mathis D. Memory T and memory B cells share a transcriptional program of self-renewal with long-term hematopoietic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3304–3309. doi: 10.1073/pnas.0511137103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhattacharya D, Cheah MT, Franco CB, Hosen N, Pin CL, Sha WC, Weissman IL. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 2007;179:6808–6819. doi: 10.4049/jimmunol.179.10.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klein U, Tu Y, Stolovitzky GA, Keller JL, Haddad J, Jr, Miljkovic V, Cattoretti G, Califano A, Dalla-Favera R. Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao Z, Sun X, Icli B, Wara AK, Feinberg MW. Role of Kruppel-like factors in leukocyte development, function, and disease. Blood. 2010;116:4404–4414. doi: 10.1182/blood-2010-05-285353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pulikkan JA, Dengler V, Peramangalam PS, Peer Zada AA, Müller-Tidow C, Bohlander SK, Tenen DG, Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuo TC, Shaffer AL, Haddad J, Jr, Choi YS, Staudt LM, Calame K. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J. Exp. Med. 2007;204:819–830. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Good KL, Tangye SG. Decreased expression of Kruppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13420–13425. doi: 10.1073/pnas.0703872104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 2011;7:263–271. doi: 10.1038/nrrheum.2011.16. [DOI] [PubMed] [Google Scholar]
- 83.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol. Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strickland FM, Richardson BC. Epigenetics in human autoimmunity. Autoimmunity. 2008;41:278–286. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Portela A, Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 86.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell. Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zan H, Zhang J, Ardeshna S, Xu Z, Park SR, Casali P. Lupus-prone MRL/faslpr/lpr mice display increased AID expression and extensive DNA lesions, comprising deletions and insertions, in the immunoglobulin locus: concurrent upregulation of somatic hypermutation and class switch DNA recombination. Autoimmunity. 2009;42:89–103. doi: 10.1080/08916930802629554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mazari L, Ouarzane M, Zouali M. Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6317–6322. doi: 10.1073/pnas.0610434104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reilly CM, Regna N, Mishra N. HDAC inhibition in lupus models. Mol. Med. 2011;17:417–425. doi: 10.2119/molmed.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ceribelli A, Nahid MA, Satoh M, Chan EK. MicroRNAs in rheumatoid arthritis. FEBS Lett. 2011;585:3667–3674. doi: 10.1016/j.febslet.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang D, Shen N. MicroRNA involvement in lupus: the beginning of a new tale. Curr. Opin. Rheumatol. 2012;24:489–498. doi: 10.1097/BOR.0b013e3283563363. [DOI] [PubMed] [Google Scholar]
- 93.Belver L, de Yébenes VG, Ramiro AR. MicroRNAs prevent the generation of autoreactive antibodies. Immunity. 2010;33:713–722. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dai R, Zhang Y, Khan D, Heid B, Caudell D, Crasta O, Ahmed SA. Identification of a common lupus disease-associated microRNA expression pattern in three different murine models of lupus. PLoS One. 2010;5:e14302. doi: 10.1371/journal.pone.0014302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Forster N, Gallinat S, Jablonska J, Weiss S, Elsässer HP, Lutz W. p300 protein acetyltransferase activity suppresses systemic lupus erythematosus-like autoimmune disease in mice. J. Immunol. 2007;178:6941–6948. doi: 10.4049/jimmunol.178.11.6941. [DOI] [PubMed] [Google Scholar]
- 96.Garchow BG, Bartulos Encinas O, Leung YT, Tsao PY, Eisenberg RA, Caricchio R, Obad S, Petri A, Kauppinen S, Kiriakidou M. Silencing of microRNA-21 in vivo ameliorates autoimmune splenomegaly in lupus mice. EMBO Mol. Med. 2011;3:605–615. doi: 10.1002/emmm.201100171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 100.De S, Shaknovich R, Riester M, Elemento O, Geng H, Kormaksson M, Jiang Y, Woolcock B, Johnson N, Polo JM, Cerchietti L, Gascoyne RD, Melnick A, Michor F. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013;9:e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, Yu Y, Remache Y, Weniger MA, Rafiq S, Suh KS, Goy A, Wilson W, Verma A, Braunschweig I, Muthusamy N, Kahl BS, Byrd JC, Wiestner A, Melnick A, Parekh S. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116:1025–1034. doi: 10.1182/blood-2009-12-257485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010;24:2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Greeve J, Philipsen A, Krause K, Klapper W, Heidorn K, Castle BE, Janda J, Marcu KB, Parwaresch R. Expression of activation-induced cytidine deaminase in human B-cell non-Hodgkin lymphomas. Blood. 2003;101:3574–3580. doi: 10.1182/blood-2002-08-2424. [DOI] [PubMed] [Google Scholar]
- 104.Shaknovich R, Melnick A. Epigenetics and B-cell lymphoma. Curr. Opin. Hematol. 2011;18:293–299. doi: 10.1097/MOH.0b013e32834788cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 106.Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM, Elemento O. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood. 2010;116:5247–5255. doi: 10.1182/blood-2010-04-280149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, Marquez VE, Marra MA, Gascoyne RD, Humphries RK, Arrowsmith CH, Morin GB, Aparicio SA. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 109.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 110.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 111.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 112.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 113.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 114.Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 117.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 118.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS. J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 119.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Green MR, Yoon H, Boss JM. Epigenetic regulation during B cell differentiation controls CIITA promoter accessibility. J. Immunol. 2006;177:3865–3873. doi: 10.4049/jimmunol.177.6.3865. [DOI] [PubMed] [Google Scholar]
- 121.Borchert GM, Holton NW, Larson ED. Repression of human activation induced cytidine deaminase by miR-93 and miR-155. BMC cancer. 2011;11:347. doi: 10.1186/1471-2407-11-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, Dalla-Favera R. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J. Exp. Med. 2012;209:2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]