Abstract
Internal tandem duplication (ITD) mutations of the FLT3 gene have been associated with a poor prognosis in acute myeloid leukemia (AML). Detection of ITD-positive minor clones at the initial diagnosis and during the minimal residual disease (MRD) stage may be essential. We previously designed a delta-PCR strategy to improve the sensitivity to 0.1% ITD-positive leukemia cells and showed that minor mutants with an allele burden of less than 1% can be clinically significant. In this study, we report on tandem duplication PCR (TD-PCR), a modified inverse PCR assay, and demonstrate a limit of detection of a few molecules of ITD mutants. The TD-PCR was initially designed to confirm ITD mutation of an amplicon which was undetectable by capillary electrophoresis and was incidentally isolated by a molecular fraction collecting tool. Subsequently, TD-PCR detected ITD mutation in 2 of 77 patients previously reported as negative for ITD mutation by a standard PCR assay. TD-PCR can also potentially be applied to monitor MRD with high analytic sensitivity in a portion of ITD-positive AML patients. Further studies using TD-PCR to detect ITD mutants at diagnosis may clarify the clinical significance of those ITD mutants with extremely low allele burden.
Keywords: FLT3, Internal Tandem Duplication, acute myeloid leukemia, minimal residual disease, Tandem duplication PCR
Introduction
FMS-like tyrosine kinase (FLT3), a member of the class III receptor tyrosine kinase family,1 plays a key role in survival of hematopoietic progenitor cells.2 Internal tandem duplication (ITD) mutations of the FLT3 gene occur in 20-30% of acute myeloid leukemia (AML) and have been associated with an inferior prognosis.3-6 Currently, most AML patients with ITD mutations need hematopoietic stem cell transplantation. As a corollary, identification of FLT3 mutations has become a routine assay to provide optimal treatment for AML patients. ITD mutations also provide a potentially useful molecular marker for monitoring minimal residual disease (MRD).7,8
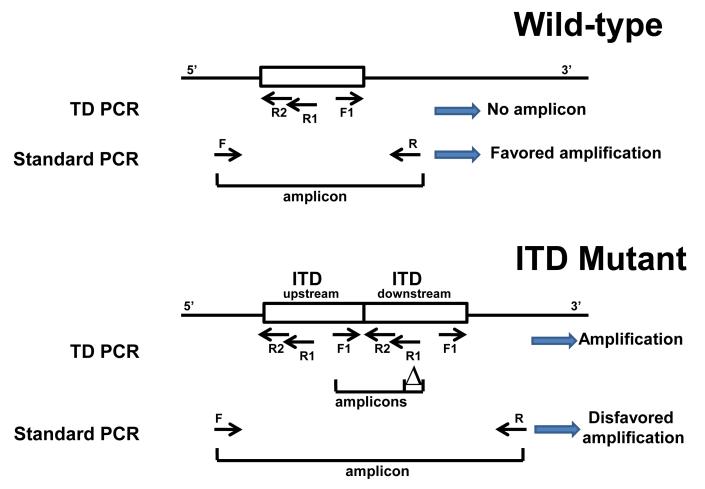
ITD mutations of the FLT3 gene typically result from head-to-tail insertion of a duplicated portion of the juxtamembrane region (Fig. 1).9 A standard PCR assay using primers that straddle the ITD mutation has been developed to detect length-altering mutations of ITD.10 Amplicons with a size greater than that of wild-type and labeled with both 6-FAM and HEX are interpreted as positive for ITD mutation. The limit of detection is generally reported as about 10% leukemia cells (5% mutant alleles) since amplicons with low peak heights, even though labeled with both fluorochromes, may represent non-specific amplification.
Fig. 1.
Design for TD-PCR. In standard PCR for detecting ITDs, template-specific PCR products are amplified by forward primer F and reverse primer R from both wild-type (top) and mutant (bottom) alleles with preferential amplification of the smaller sized wild-type allele. In the TD-PCR, template-specific PCR products are only amplified from the mutant allele by forward primer F1 and reverse external and internal primers R1 and R2 . “△” indicates the defined size difference of the paired amplicons (19+1 bases in this study). “→” indicates primers.
We recently developed a triple-primer strategy, delta-PCR, to ensure PCR specificity and improve the limit of detection to 0.1% leukemia cells.11 We demonstrated that minor leukemia mutants with an ITD mutant allelic burden of less than 1% detected by delta-PCR at the initial diagnosis could be clinically significant. An assay with even higher sensitivity, however, is needed for monitoring MRD. However, both standard PCR assays and our delta-PCR approach are intrinsically flawed for detecting ITD: the amplicon from the wild-type allele is always shorter than the mutant allele and thus favored in amplification (Fig. 1). Attempts have been made to reduce this intrinsic bias by reducing PCR cycle number, but this of course reduces the ability to detect rare mutants, i.e., MRD.5 In the current study, we report a generic assay for tandem duplication detection with greatly improved sensitivity and specificity, to a limit of detection of only a few molecules of an ITD. This assay can be applied to monitor MRD.
Materials and Methods
Design of tandem duplication PCR (TD-PCR), a modified inverse PCR
Inverse PCR is generally conducted with restriction enzyme digested DNA that is circularized by ligation, which allows adjacent PCR primers that face away from each other to amplify a product across the restriction site. The modification we use has been described previously to detect de novo duplication of the human alpha globin gene.12 There is no digestion or ligation; the arrangement of tandem duplicates permits PCR primers facing away from each other to produce an amplicon across the duplication junction. The reaction in TD-PCR includes an upstream reverse primer and a downstream forward primer, in contrast to an upstream forward primer and a downstream reverse primer for standard PCR (Fig. 1). When both primers are located within the tandem duplicated sequences, amplification occurs across the insertion junction using the forward primer located within the upstream duplicated sequence and the reverse primer located within the downstream duplicated sequence. Since the length-altering mutation varies for each ITD mutant, we have also added a third primer to the reaction to improve the specificity of the reaction, analogous to our delta-PCR.13 No template-specific amplification occurs from the wild-type allele or if one or both primers lie outside the duplicated segment. By contrast, in standard PCR, template specific products are amplified from both the smaller wild-type allele and the larger mutant allele, resulting in competition for amplification and reduced mutant signal (Fig. 1).
Polymerase chain reaction (standard PCR, delta-PCR and TD-PCR)
The standard PCR for simultaneously detecting an ITD mutation and D835 point mutation was performed as described.10 Amplicons with a size greater than that of wild-type (328+1 bases on ABI 3130xL capillary electrophoresis using the previously described oligonucleotides, Fig. 2a) and which were labeled with both 6-FAM and HEX fluorochromes were interpreted as positive for ITD mutation. Delta-PCR assay was performed using a forward primer and 2 reverse primers (external primer labeled with 6-FAM and internal primer labeled with HEX) as described previously.11 The presence of a pair of amplicons with a delta of 19+1 bases and with sizes larger than the pair of amplicons from the wild-type allele (160-base green peak and 179-base blue peak) was interpreted as positive for ITD mutation.
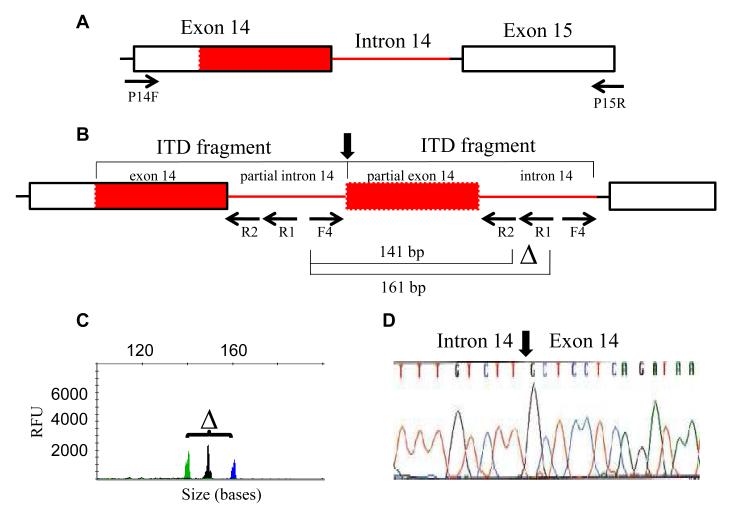
Fig. 2.
Design of TD-PCR according to duplication sequences of an ITD mutant undetectable by both delta-PCR13 and standard PCR.10 The ITD fragment extends from exon 14 to near the 3′end of intron 14 (A). A pair of amplicons migrating at 141 bases and 161 bases was detected by TD-PCR using forward primer INT14F4 (F4) and a pair of reverse primers, external INT14R1 (R1) primer and internal INT14R2 (R2) primer (B). The internal primer served as a “probe” to ensure PCR specificity. A pair of amplicons with a delta (“△”) of 19+1 bases confirms an ITD mutation (C). The black peak in (C) is an internal control amplicon unrelated to the ITD (see text). Direct sequencing of the 161-base amplicon confirmed the abnormal junction (vertical arrow) between intron 14 and exon 14 (D). Red box indicates the duplicated part of exon 14 and the red line indicates the duplicated part of intron 14. P14F and P15R are the primers used for standard PCR for detection of ITD mutations.10 RFU: relative fluorescence units.
TD-PCR was conducted using a forward primer located at the 3′end of intron 14 of the FLT3 gene (primer INT14F4: 5′-aat gca cgt act cac cat ttg tc-3′) and a reverse primer located at the middle portion of intron 14 (primer INT14R1: 5′-caa tgg aaa aga aat gct gca g-3′, labeled with 6- FAM) (Figure 2b). The additional “internal” reverse primer (primer INT14R2: 5′-cag aaa cat ttg gca cat tcc a-3′, labeled with HEX) was located at the 5′end of intron 14. These two reverse primers are the same as those used for delta-PCR.11 As described previously for delta-PCR,11,13 the internal primer was used as a probe to ensure PCR specificity. For both delta-PCR and the new TD-PCR, a pair of amplicons with a delta of 19+1 bases was interpreted as positive for ITD mutation.
TD-PCR was performed in a 20 μL volume containing up to 250 ng DNA, 10 pmol of forward primer, 5 pmol of reverse external primer, 5 pmol of reverse internal primer, 1.5 mM MgCl2, 0.2 mM each deoxyribonucleotide, 1.0 units AmpliTaq Gold DNA polymerase and 2 μL buffer (Applied Biosystems, Foster City, CA). A PCR quality control reaction (to the activation loop of FLT3) was included in every run.10 Samples were subjected to 40 cycles of denaturation (95°C, 30 sec), annealing (57°C, 30 sec) and extension (72°C, 60 sec).
Materials
DNA was isolated from the peripheral blood or bone marrow of 40 normal donors and 117 consecutive AML patients at initial diagnosis or relapse using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA). Only cases reported to contain greater than 20% blasts in the sample were included. Detection of the ITD using standard PCR had been previously performed: 40 were ITD-positive and 77 were ITD-negative. An additional 6 ITD-positive AML patients with a larger duplication (more than 480 bases as compared to the wild-type peak migrating at 328+1 bases) were also selected for testing by TD-PCR. The Johns Hopkins Medicine institutional review board granted approval to this study.
Capillary Electrophoresis
One μL PCR products, 0.5 μL ROX size standard and 8.5 μL deionized formamide were mixed according to the manufacturer’s protocol (Applied Biosystems), heated at 95°C for 2 minutes and placed on ice for at least 1 minute before electrokinetic injection (10 and 25 sec for TD-PCR) on the ABI 3130xL Genetic Analyzer (Applied Biosystems) as described previously.14 The size and positions of peaks on the electropherogram were analyzed using GeneMapper® analysis software (Applied Biosystems).
DNA sequencing
Primers INT14R1M13R and INT14F4M13F were their parent primers tagged with M13 sequences for direct sequencing of amplicons. PCR products were purified using USB ExoSapit (GE Healthcare, Uppsala, Sweden) and cycle sequenced using the BigDye Terminator version 3.1 cycle sequencing kit according to the manufacturer’s protocol and resolved on an ABI 3500xL sequencer (Applied Biosystems). Sequences were analyzed using Sequencher software (Gene Codes Corp, Inc., Ann Arbor, MI).
RESULTS
We provide comparative results for three assays (standard PCR, delta-PCR and TD-PCR) in the setting of FLT3 ITD and show the benefits of TD-PCR.
Rationale for designing TD-PCR
We previously reported the delta-PCR assay to detect minor ITD-positive mutants representing as little as approximately 0.1% of all cells. However, this is insufficient to measure true minimal residual leukemia. An assay with higher sensitivity is needed. TD-PCR was initially designed to confirm an ITD mutant which was undetectable by standard PCR or delta-PCR. This mutant, along with other 4 mutants detectable by delta-PCR, was isolated incidentally by a molecular fraction collecting tool.13,14 A pair of amplicons, demonstrating the expected delta of 19+1 bases and migrating at 141 bases and 161 bases, was repeatedly detected by TD-PCR in the bone marrow specimen in which this mutant was isolated (Fig. 2c), but not the concurrent peripheral blood specimen (data not shown), suggesting that this minor clone was present in the bone marrow at a very low level. Sanger sequencing of the TD-PCR amplicons confirmed the presence of the same ITD sequences (c.1748_1838-5dup) identified from the amplicons isolated by our molecular fraction collection tool (Fig. 2d).
Limit of detection of TD-PCR
In a previous study, we demonstrated that a minor mutant with an ITD mutation may be clinically significant.11 In a patient with at least 3 ITD-positive mutants at initial diagnosis, a minor mutant with an allelic burden of 0.5%, which migrated at a position slightly more than 500 bases and was barely detected by standard PCR and delta-PCR, became the only mutant, with an allelic burden of 44% at the time of relapse. By contrast, in the initial diagnosis sample TD-PCR easily detected this minor mutant with a peak height over 100-fold higher than standard PCR (data not shown). Sanger sequencing confirmed the presence of an ITD of 186 bases extending from exon 14 into exon 15 (c.1754_1840 dup) that is consistent with the ITD mutant detected by standard PCR and delta-PCR.
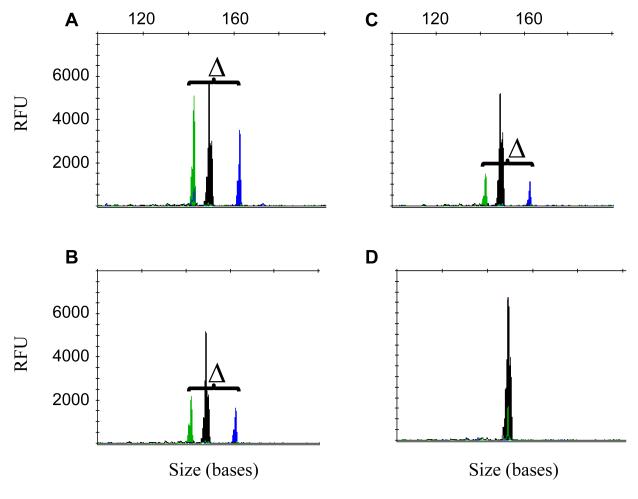
The relapsed specimen of this patient was serially diluted with an ITD negative specimen at 1:1,000 (10−3), 1:2,000 (5×10−4), 1:10,000 (10−4), and 1:20,000 (5×10−5). TD-PCR was conducted in replicates with 250 ng DNA (equivalent to approximately 40,000 cells) in each reaction. The expected pair of amplicons was detected in 4 of 4 replicates at a dilution of 10−3, 8 of 8 replicates at a dilution of 5×10−4 (equivalent to 20 mutant cells), 6 of 8 replicates at a dilution of 10−4 (equivalent to 4 mutant cells), and 5 of 8 replicates at a dilution of 5×10−5 (equivalent to 2 mutant cells), assuming all the leukemia cells contain this ITD mutant and each cell contains 6-7 pg DNA (Fig. 3). TD-PCR was also tested with a reduced amount of DNA (50 ng) in each reaction. The expected product was also detected in 15/16 replicates at a dilution of 5×10−4 (equivalent to 4 mutant cells) and in 2 of 16 replicates at a dilution of 10−4 (equivalent to 0.8 mutant cells). The results indicate that TD-PCR is a very sensitive assay for ITD, likely capable of detecting less than 5 mutant copies per reaction. In all cases including the 40 normal donor specimens, background was negligible.
Fig. 3.
Limit of detection of TD-PCR. DNA samples from case A115 with ITD mutation were serially diluted with normal DNA samples (1 in 2,000 in 3A, 1 in 10,000 in 3B, 1 in 20,000 in 3C and negative control in 3D). “△” indicates the defined size difference of the paired amplicons at 19+1 bases. The 150-base NED-labeled black peak is an internal control amplicon for PCR. RFU: relative fluorescence unit.
Detection of ITD by TD-PCR in patients with AML
TD-PCR was used to detect ITD in 117 cases of newly diagnosed AML or frank relapsed AML. The assay was positive in 8 of 40 cases previously diagnosed as ITD-positive AML by standard PCR. Sanger sequencing confirmed the presence of ITD with duplications extending from exon 14 into intron 14 or exon 15 of the FLT3 gene and confirms that the current design of primers detected only ITD of larger size with duplication extending from exon 14 into intron 14. The cases not detected by TD-PCR had smaller ITDs by standard PCR or had ITDs restricted within exon 14 and would not be expected to yield a positive result. An additional 6 selected cases with duplication sequences of more than 150 bases were also positive. Again, the duplication sequences were confirmed by Sanger sequencing of the amplicons from TD-PCR.
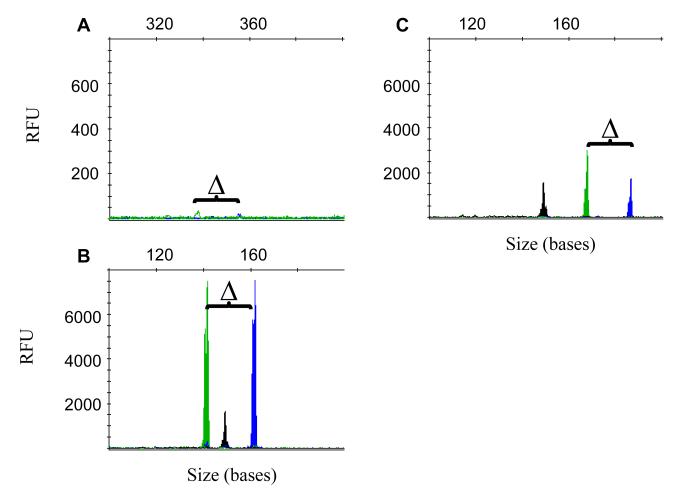
As a test of the clinical sensitivity of TD-PCR, we assayed 77 cases of AML that were previously reported as negative for ITD mutation by standard PCR. TD-PCR was conducted using 250 ng DNA. Two cases missed by standard PCR (cases A23 and A185) were identified as mutated by TD-PCR. The presence of ITD sequences was confirmed by Sanger sequencing in both cases. In case A185, a pair of amplicons with peak height less than 50 relative fluorescence units (RFU) (90 second injection) which was not easy to distinguish from background noise was observed in 2 of 5 reactions by delta-PCR (Fig. 4a). However, TD-PCR can easily detect this ITD mutation with a pair of peaks of more than 7,000 RFU (25 second injection) (Fig. 4b). TD-PCR consistently shows this ITD mutant even with as little as 50 ng input DNA. In case A23, a pair of amplicons was detected in TD-PCR reactions (Fig. 4c), but not in the corresponding delta-PCR reactions. This mutant was seen by TD-PCR in 6 of 16 additional replicates containing 50 ng DNA, suggesting a very low level of ITD mutant in case A23.
Fig. 4.
TD-PCR was used to detect ITD mutations which were undetectable by standard PCR. Delta-PCR showed a pairs of amplicons with extremely low peak heights at 90-second injection in case A185 (A). A pair of amplicons with peak heights of several thousands at 25-second injection was detected by TD-PCR in case A185 (B) and case A23 (C). “△” indicates the defined size difference of the paired amplicons at 19+1 bases. The 150-base NED-labeled black peak is an internal control amplicon for PCR. RFU: relative fluorescence unit.
Discussion
ITD mutations of the FLT3 gene have been associated with a poor prognosis in AML. Detection of ITD mutations at diagnosis is now routine clinical practice. Although detection of minimal residual AML with ITD mutations is theoretically important, the current standard PCR assay for simultaneous detection of ITD and D835 mutations for newly diagnosed AML is not sensitive enough for monitoring MRD. In this proof-of-principle study, we show that TD-PCR is a sensitive strategy to detect ITD mutation at a level of only a few molecules. In contrast to the current commercially available standard PCR assay, no amplification and, thus, no competition from the wild-type allele results in higher sensitivity in the TD-PCR assay.
Improved sensitivity can also be obtained by real-time PCR using mutant-specific primers or probes.8 However, each mutant needs a mutant-specific primer/probe designed from the junctional sequence; due to the constraints of the sequence at the junction, this may not be possible for every case. An added complication is that relatively low allelic burden mutants (as identified by standard PCR) may not be obtainable for direct sequencing since the wild type DNA is present in great excess. We have successfully isolated ITD mutants of less than 1% allelic frequency by using the molecular fraction collecting tool that we constructed.13,14 The procedure, however, is cumbersome. In addition, it is laborious but mandatory to test the analytic sensitivity and specificity for each clone-specific PCR assay to have a valid clinical test. The single TD-PCR assay need only be validated once for clinical testing. ITD mutants at relapse occasionally differ from the mutant at diagnosis.3,6,15 These newly emerging ITD-positive mutants will not be detected by a real-time assay using mutant-specific primers/probes designed for MRD detection of a specific ITD mutant.
Although TD-PCR is ultrasensitive as compared to the standard PCR or delta-PCR assays, short ITD mutations (less than 40-50 bases) cannot be detected by TD-PCR since approximately 20 bases are needed for each primer. The TD-PCR assay as currently constructed can potentially be used to monitor MRD in approximately 20% of AML patients with ITD mutations. Delta-PCR, although not as sensitive as the real-time PCR assay or the TD-PCR assay, can provide broader screening of ITD mutations and monitoring of the emergence of a new ITD mutant. Combining delta-PCR and TD-PCR may provide the broadest and most sensitive monitor of MRD.
Our current TD-PCR assay detected ITD mutants with extremely low allelic burden in two cases that were reported as ITD-negative by standard PCR. Like most cases with ITD-negative AML cases, there was no serial follow-up of the FLT3 gene mutation in these two cases. Therefore, the clinical significance of such low allelic burden in these two cases is not known. In our previous report, we have shown that minor ITD clones with an allele burden of less than 1% at diagnosis became the dominant clone later in the disease course.11 It may be worthwhile to test all AML cases with TD-PCR serially and follow up those cases with initial extremely low mutant allelic burden.
In summary, we have shown a proof-of-principle design of TD-PCR with a high analytic sensitivity, likely less than 5 mutant molecules per reaction. This approach can be generalized and should be useful in any cancer involving tandem duplications, particularly for MRD monitoring. The TD-PCR assay may identify AML patients with an extremely low allelic burden of ITD mutations at initial diagnosis. Further clinical studies with serial follow-up of such patients may clarify the significance of ITD mutations with extremely low allelic burden.
Acknowledgments
Supported in part by R21HG004315 and R21HG005745 from National Institutes of Health.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 2.Turner AM, Lin NL, Issarachai S, et al. FLT3 receptor expression on the surface of normal and malignant human hematopoietic cells. Blood. 1996;88:3383–3390. [PubMed] [Google Scholar]
- 3.Kottaridis PD, Gale RE, Frew ME, et al. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 4.Meshinchi S, Woods WG, Stirewalt DL, et al. Prevalence and prognostic significance of Flt3 internal tandem duplication in pediatric acute myeloid leukemia. Blood. 2001;97:89–94. doi: 10.1182/blood.v97.1.89. [DOI] [PubMed] [Google Scholar]
- 5.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 6.Schnittger S, Schoch C, Dugas M, et al. Analysis of FLT3 length mutations in 1003 patients with acute myeloid leukemia: correlation to cytogenetics, FAB subtype, and prognosis in the AMLCG study and usefulness as a marker for the detection of minimal residual disease. Blood. 2002;100:59–66. doi: 10.1182/blood.v100.1.59. [DOI] [PubMed] [Google Scholar]
- 7.Schnittger S, Schoch C, Kern W, et al. FLT3 length mutations as marker for follow-up studies in acute myeloid leukaemia. Acta Haematol. 2004;112:68–78. doi: 10.1159/000077561. [DOI] [PubMed] [Google Scholar]
- 8.Stirewalt DL, Willman CL, Radich JP. Quantitative, real-time polymerase chain reactions for FLT3 internal tandem duplications are highly sensitive and specific. Leuk Res. 2001;25:1085–1088. doi: 10.1016/s0145-2126(01)00087-x. [DOI] [PubMed] [Google Scholar]
- 9.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 10.Murphy KM, Levis M, Hafez MJ, et al. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5:96–102. doi: 10.1016/S1525-1578(10)60458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beierl K, Tseng LH, Beierl R, et al. Detection of minor clones with internal tandem duplication mutations of FLT3 gene in acute myeloid leukemia using delta-PCR. Diagn Mol Pathol. 2012 doi: 10.1097/PDM.0b013e31825d81f4. (in press) [DOI] [PubMed] [Google Scholar]
- 12.Lam KW, Jeffreys AJ. Processes of de novo duplication of human alpha-globin genes. Pros Natl Acad Sci USA. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin MT, Tseng LH, Rich RG, et al. Δ-PCR, a simple method to detect translocations and insertion/deletion mutations. J Mol Diagn. 2011;13:85–92. doi: 10.1016/j.jmoldx.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin MT, Rich RG, Shipley RF, et al. A molecular fraction collecting tool for the ABI 310 automated sequencer. J Mol Diagn. 2007;9:598–603. doi: 10.2353/jmoldx.2007.070022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shih LY, Huang CF, Wu JH, et al. Internal tandem duplication of FLT3 in relapsed acute myeloid leukemia: a comparative analysis of bone marrow samples from 108 adult patients at diagnosis and relapse. Blood. 2002;100:2387–2392. doi: 10.1182/blood-2002-01-0195. [DOI] [PubMed] [Google Scholar]