Abstract
The β and proliferating cell nuclear antigen (PCNA) sliding clamps were first identified as components of their respective replicases, and thus were assigned a role in chromosome replication. Further studies have shown that the eukaryotic clamp, PCNA, interacts with several other proteins that are involved in excision repair, mismatch repair, cellular regulation, and DNA processing, indicating a much wider role than replication alone. Indeed, the Escherichia coli β clamp is known to function with DNA polymerases II and V, indicating that β also interacts with more than just the chromosomal replicase, DNA polymerase III. This report demonstrates three previously undetected protein–protein interactions with the β clamp. Thus, β interacts with MutS, DNA ligase, and DNA polymerase I. Given the diverse use of these proteins in repair and other DNA transactions, this expanded list of β interactive proteins suggests that the prokaryotic β ring participates in a wide variety of reactions beyond its role in chromosomal replication.
Keywords: PCNA, mismatch repair
The Sliding Clamp in Replication
The highly processive DNA polymerases that replicate cellular genomes combine a tight grip on DNA with rapid mobility along the duplex during synthesis (reviewed in refs. 1 and 2). The apparent contradiction of binding to DNA tightly, yet rapidly moving on DNA, is resolved by a mechanism that on hindsight is quite elegant and simple. The solution lies in a protein molecule in the shape of a ring that, rather than binding to DNA by using direct chemical forces, binds to DNA by encircling it (i.e., topological binding). This mechanism was first discovered in the Escherichia coli system in which the β dimer, a subunit of DNA polymerase III holoenzyme (Pol III H.E.), was found to be able to bind tightly to circular DNA but to slide off the ends of linear DNA (3). The dependence of the beta-DNA interaction on the topological state of DNA indicated that the protein does not bind DNA through the usual chemical forces, but instead binds DNA by virtue of its shape (i.e., as a ring, or clamp). The β subunit therefore serves as a mobile sliding clamp to continuously tether Pol III H.E. to DNA during synthesis, thus assuring its processivity.
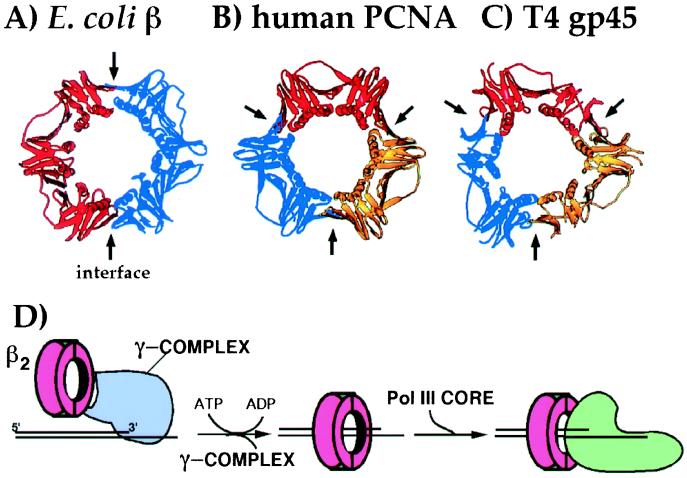
Crystal structure analysis of E. coli β and its functional homologues, eukaryotic proliferating cell nuclear antigen (PCNA) and phage T4 gp45, has revealed the ring shape of all of these “processivity factors” (Fig. 1 A–C; refs. 4–7). The fold of the polypeptide chain in β and PCNA is essentially the same, which is remarkable given the fact that no homology is observed between the two proteins. The chain fold of gp45 is similar, with the exception of the loss of two secondary structural elements. The overall structures of the homoligomeric β and PCNA rings can be described as being composed of six globular domains, each of the same shape, which pack together to form a six domain ring. The architecture of the ring includes a continuous layer of pleated sheet structure around the entire circumference, and a set of 12 α-helices that line the inside of the ring. A major difference between β, PCNA, and gp45 is their oligomeric composition. β is a homodimer in which each monomer consists of three domains; PCNA and gp45 monomers consist of only two domains and trimerize to form the ring.
Figure 1.
DNA sliding clamps from E. coli, human, and phage T4. Ribbon diagrams are shown for sliding clamps from different organisms: (A) E. coli β (4), (B) human PCNA (6), and (C) phage T4 gp45 (7). Each monomeric unit of the ring is shown in a different color. The arrows indicate the protomer interfaces. (D) The scheme illustrates assembly of a β ring on a primed template by γ complex in an ATP-driven reaction, followed by association of the core polymerase with β for processive DNA synthesis.
Sliding clamps require a clamp loader assembly for their topological association with DNA. Clamp loaders consist of multiple subunits and require ATP to perform their task as a “molecular matchmaker” between the sliding clamp and the DNA (reviewed in refs. 1 and 8). The workings of the E. coli clamp loader, called the γ complex, will be discussed later in this report. After assembly of the sliding clamp onto DNA, the polymerase associates with the same side of the ring used by the clamp loader (see scheme in Fig. 1D).
PCNA and gp45 Participate in Many DNA Metabolic Processes
In this report, we demonstrate that β interacts with other proteins besides those directly involved in chromosomal replication. First, however, it seems prudent to briefly review what is known in this regard in other systems, especially considering the broad scope of proteins that are known to interact with PCNA. Although the clamps were first discovered for their use in replication, it soon became apparent that they are also involved in many other DNA metabolic processes. The first example of this was in the T4 system in which Geiduschek and coworkers demonstrated that the T4 gp45 clamp can act as a “mobile enhancer” to activate transcription of phage late genes (9). This work demonstrated that T4 gp45 assembles onto DNA at a nicked site and interacts with phage-modified E. coli RNA polymerase, which is then active for specific transcription from the phage late-gene promoters.
PCNA is known to function with DNA polymerases δ and ɛ, enzymes responsible for chromosome replication (2). The first indication that PCNA may act with a protein other than a DNA polymerase came from an observation of PCNA in complex with p21C1P1/WAF1, the cyclin kinase inhibitor (10). This interaction is thought to play a regulatory role in replication because p21 blocks use of PCNA by Pol δ (11, 12). Since this observation was made, several other proteins have been shown to interact with PCNA (reviewed in ref. 10). These include XPG, MSH3, MSH6, MLH1, PMS2, and hMYH (13), implying roles for PCNA in nucleotide excision repair, mismatch repair, and base excision repair. PCNA also interacts with Fen1 endonuclease and DNA ligase I, suggesting a role in the maturation of Okazaki fragments or perhaps other processes involving the trimming of 5′ ends or filling of DNA gaps. PCNA also binds the chromatin assembly factor 1 (CAF-1; ref. 14), and several other proteins, including a DNA methyltransferase and gadd45.
Interaction of β with Pol III H.E., Pol II, and Pol V
The β clamp interacts not only with the α DNA polymerase subunit of DNA polymerase III H.E., but also with three subunits of the clamp loader. The clamp loader, γ complex, consists of five different subunits, γ, δ, δ′, χ, and ψ. The δ, δ′, χ, and ψ subunits are present in single copy in the complex (15, 16), and γ is present as a homotrimer (17). The β subunit interacts most strongly with the δ subunit (18), although it also shows a week interaction with the γ and χ subunits (A. Yuzhakov and M.O.D., unpublished results).
The γ, δ, and δ′ subunits are needed for efficient clamp loading; neither χ nor ψ are required (1). The δ subunit has been shown to be capable of opening the β ring by itself and can remove β from DNA, but cannot load β onto DNA without γ and δ′ (19). The γ and δ′ subunits presumably coordinate ring opening with the positioning of DNA inside the ring, and with ring closure. The γ subunit is the only subunit that binds and hydrolyzes ATP, and thus is the motor of the clamp loader. In the absence of ATP, the interaction of γ complex with β is decreased. ATP binding results in the interaction with β, and ring opening. ATP hydrolysis is promoted by primed DNA and results in dissociation of γ complex from β, leading to the closed ring on DNA (ref. 17, and reviewed in ref. 1). The exact roles of the γ-β and χ-β interactions are not clear at the present time.
The β subunit has also been shown to interact with DNA polymerase II (Pol II), increasing its processivity in synthesis (20, 21). The role of Pol II is unclear; it has been implicated in replication restart (22). The recently discovered DNA polymerase activity present in UmuC, renamed Pol V, requires the presence of γ complex and β [plus UmuD′, RecA, and ssDNA-binding protein (SSB)] for activity, suggesting that it too interacts with the β clamp (23).
Interaction of β with MutS
In vitro, E. coli mismatch repair depends specifically on DNA polymerase III H.E (24), suggesting that some component of this replicase contacts the mismatch repair machinery. Further, the fact that in eukaryotes PCNA binds MSH2 and MSH6 suggested that perhaps β interacts with MutS.
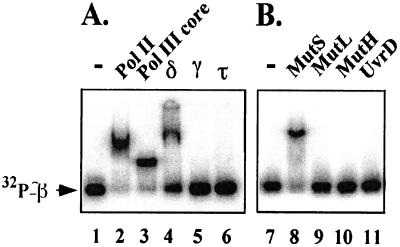
We initially examined MutS and β for interaction by gel filtration, a technique that detects fairly strong interactions (e.g., Kd = 300 nM or less), but did not observe a Muts–β complex (data not shown). Next, we examined β for a MutS interaction by using a native PAGE (Fig. 2). In this technique, we used β with a six-residue C-terminal extension that can be phosphorylated with protein kinase (25). Either MutS, or other proteins, were mixed with [32P]β, and then the mixture was analyzed in a native polyacrylamide gel followed by autoradiography. An interaction between [32P]β and the added protein results in a gel shift. In Fig. 2A, this technique was applied to proteins known to bind β. The [32P]β was gel shifted by Pol II, Pol III core, and the δ subunit of γ complex, all proteins that have previously been shown to bind to β. The γ and τ subunits did not produce a gel shift with [32P]β, yet we have recently detected a weak association between γ and β by surface plasmon resonance (A. Yuzhakov and M.O.D., unpublished results). [32P]β was also examined for interaction with MutL, MutH, and UvrD (helicase II), all proteins involved in mismatch repair. The results using this technique, in Fig. 2B, demonstrate an interaction between β and MutS but no interaction with MutL, MutH, or UvrD.
Figure 2.
A protein-shift assay reveals a MutS interaction with the β clamp. (A) Interaction of β with replication proteins. The β subunit, modified at the C terminus to incorporate a protein kinase motif, was labeled with [32P]ATP by protein kinase as described (25). Reactions (15 μl) contained 20 mM Tris⋅Cl (pH 7.5), 0.1 mM EDTA, 4% glycerol, 50 μg/ml BSA, 100 mM NaCl, 5 mM DTT, and 90 nM [32P]β. Lanes 1 and 7 are controls; lane 2, 0.5 μM DNA polymerase II; lane 3, 0.5 μM DNA polymerase III core; lane 4, 3 μM δ; lane 5, 4 μM γ; lane 6, 3 μM τ. (B) Analysis of β interaction with mismatch repair proteins. Reactions were as in A. Lanes 8–11 contained 3 μM of either MutS, MutL, MutH, or UvrD, respectively. Reactions were incubated at 37°C for 4 min before loading 5 μl on a native 4% polyacrylamide gel. Electrophoresis was performed by using TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) at 17 mA and 22°C. Gels were dried, and detection of radioactive β was performed by using a PhosphorImager (Molecular Dynamics).
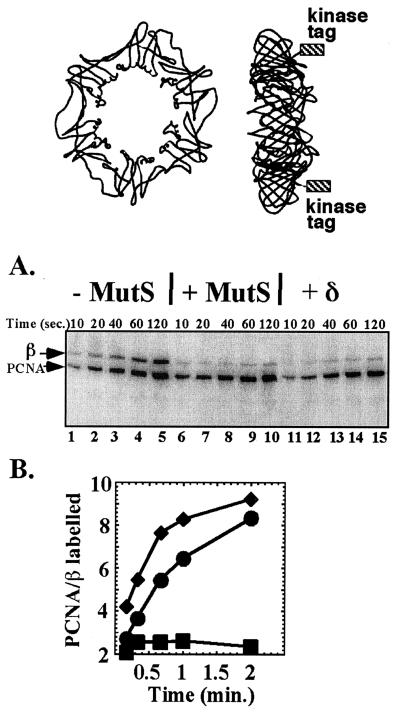
The interaction between β and MutS was also examined by the kinase protection technique (25). In this assay the rate of phosphorylation of the six-residue kinase tag at the C terminus of β is followed in the presence or absence of a protein that binds β. As illustrated in the diagram at the Top of Fig. 3, the C termini of the β dimer on which the kinase tags are placed protrude from one face of the ring. If the β binding protein interacts with the face of β from which the C-terminal kinase tags protrude, kinase action is blocked, and the phosphorylation rate of β is diminished. Previous studies have demonstrated that both Pol III core and δ subunit protect β from phosphorylation, indicating that they bind to the face of β from which the C termini protrude. The first five lanes of Fig. 3A show the time course of phosphorylation of β in the absence of MutS. As an internal control, kinase-tagged PCNA is also present in the reaction. The same experiment, but in the presence of MutS, is shown in lanes 6–10. The result shows protection of β, but no protection of the PCNA internal control, indicating that MutS binds the face of β from which the C-terminal tags protrude. A positive control using δ subunit is shown in lanes 11–15. The δ subunit acts like MutS, protecting β but not PCNA from phosphorylation by the kinase. These results are quantitated in the plot of Fig. 3B.
Figure 3.
Kinase protection analysis of the β–MutS interaction. The diagram at the Top illustrates the location of the six-residue kinase tags placed onto the C terminus of β. The diagram to the Left is a front view (no tags shown). The diagram to the Right shows a side view; the location of the kinase tags is indicated by the boxes. (A) Interaction of MutS with β is specific and occurs on the same face of the ring as δ and α. Kinase-tagged β and PCNA were labeled with [32P]ATP using protein kinase as described in Fig. 2. Lanes 1–5 show a time course of phosphorylation of kinase-tagged β or PCNA in the absence of additional protein. Addition of MutS (lanes 6–10) or δ (lanes 11 to 15) inhibits labeling of β but not of PCNA, indicating a specific interaction of MutS with β. Reaction conditions were as described (26). Concentrations of β and PCNA (as dimer or trimer, respectively) were 1 μM, and those of MutS and δ, when present, 10 μM. Reactions (50 μl) were incubated at 37°C, and aliquots (5 μl) were removed and quenched with 1% SDS and 40 mM EDTA at the times indicated. Quenched reactions were analyzed in a 12% SDS polyacrylamide gel, followed by autoradiography. Positions of β and PCNA are indicated to the left of the gel. (B) Quantitation of the gel in A. The plot indicates the ratio of labeled PCNA to labeled β in each time point. ■, no protein added; ♦, MutS added; and ●, δ added.
What Role May the β–MutS Interaction Play in Mismatch Repair?
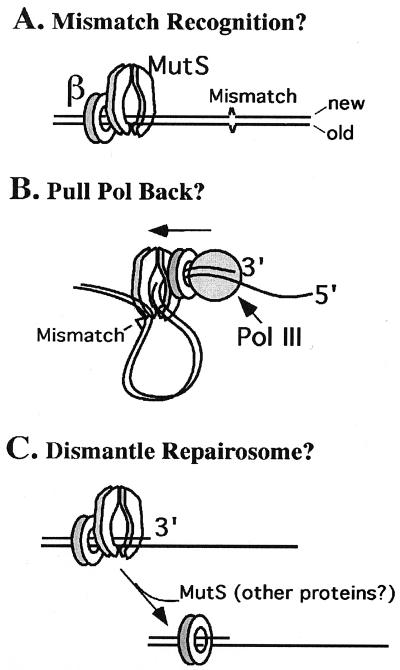
How may the β clamp participate in the mismatch repair process? Perhaps the β clamps that are deposited on DNA during lagging strand replication help localize MutS to regions of active DNA synthesis (Fig. 4A). In this scenario, MutS is mainly targeted to the lagging strand on which β clamps are deposited. However, it is possible that the β-MutS interaction simply increases the local concentration of MutS at regions of newly replicated DNA, thereby helping to target MutS to both daughter strands. Indeed, DNA replication has been shown to take place at discrete loci within the cell (27), where the β clamp could serve as the common interacting platform for the actions of PolIII, MutS, and other proteins involved in replication. The MutS–β interaction may also help orient MutS on DNA, providing it with information as to which strand is newly replicated. Mechanisms such as these have been hypothesized for roles of PCNA in mismatch repair (28–30). The crystal structures of both β and MutS (4, 31) show that they have structurally distinct faces (i.e., a front and a back) as they sit on DNA, consistent with the hypothesis that an interaction between β and MutS will orient one protein relative to the other on DNA. In E. coli, the status of the methylation state of DNA provides the signal to the mismatch repair machinery to discriminate the new strand from the template strand. However, methylation-dependent strand discrimination does not exclude the possibility that other mechanisms for strand discrimination may be available to the bacterium (e.g., such as β-directed orientation). Further, other bacteria such as Gram-positive organisms lack the high degree of DNA methylation observed in E. coli, and lack the Dam methylase and MutH endonuclease (32). Hence, many prokaryotes share in common the same problem that eukaryotic organisms face in solving strand discrimination and endonucleolytic nicking in mismatch repair.
Figure 4.
Possible uses of β in mismatch repair. (A) MutS may be oriented on DNA by β clamps deposited on DNA by the replication machinery. The fact that MutS has distinct front and back faces (like β) and interacts with the same face of β that the polymerase binds may direct MutS action to the newly replicated strand. (B) MutS interaction with β may exert a drag force on the replicase in the direction opposite to DNA synthesis. This force may result in activating the 3′-5′ exonuclease for excision past the mismatch. (C) The β clamp, on binding to MutS, may signal the dissociation of proteins from excised DNA, enabling polymerase to fill in the gap.
Another possible role for a β–MutS interaction is illustrated in Fig. 4B, in which MutS may “drag” the replicase backward via pulling on the β clamp, or may inhibit the DNA synthetic activity of Pol III H.E., resulting in excision of DNA past the mismatch by the proofreading nuclease in Pol III H.E. (also a proposed role for PCNA; refs. 28–30).
Yet another possible role for β–MutS interaction is shown in Fig. 4C. The excision step of mismatch repair is likely to be overseen by MutS and MutL, ensuring that excision proceeds past the mismatch. However, in the fill-in step, the DNA polymerase must recruit a 3′ primer terminus that, up to the time for fill-in, has been acted on by UvrD helicase, exonuclease, and possibly MutS/MutL. Perhaps the β–MutS interaction functions to recruit this 3′ terminus in a process in which repair proteins are dismantled from the DNA, clearing the excised DNA for fill-in by a DNA polymerase.
Despite these various hypotheses on possible roles of the β–MutS interaction, it remains quite possible that it functions in a very different fashion from any of the above speculations. Clearly, further studies of Pol III H.E. action in mismatch repair are required to delineate the role(s) of β and other proteins in this complicated reaction.
The β Clamp Interacts with Pol I and Ligase
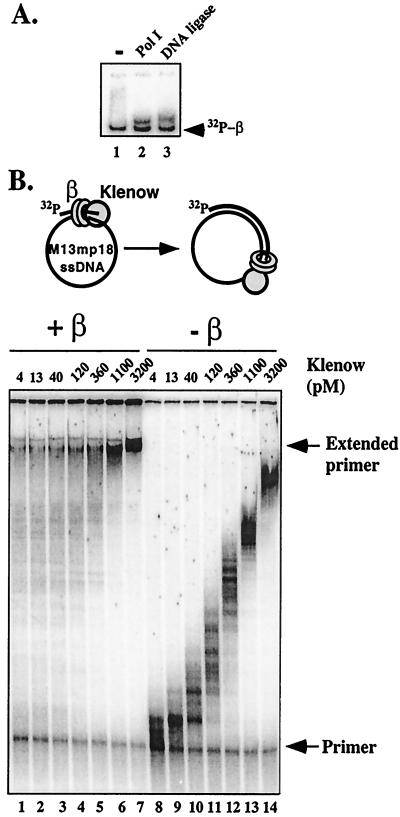
Early reports using accessory factors of Pol III indicated that they stimulated DNA synthesis by Pol II, and possibly Pol I (33, 34). On hindsight, these accessory factors were most likely the β clamp and the γ complex clamp loader. Follow-up studies using recombinant proteins demonstrated that β and γ complex do indeed function with Pol II (20, 21). We used the native gel mobility shift assay to determine whether β binds Pol I. We also examined β for an interaction with E. coli DNA ligase, because PCNA is known to interact with eukaryotic DNA ligase I (35). The results, shown in Fig. 5A, indicate the formation of a β–Pol I complex and a β–ligase complex.
Figure 5.
The β subunit interacts with DNA polymerase I and DNA ligase. (A) Analysis of [32P]β interaction with Pol I and ligase by gel shift in a native polyacrylamide gel was performed as described in the legend to Fig. 2. Lane 1, no addition ([32P]β only); lane 2, addition of 3.5 μM Pol I; and lane 3, addition of 3.5 μM DNA ligase. (B) The β clamp stimulates Pol I activity. DNA synthesis by Pol I was examined on a 32P-labeled primed M13mp18 ssDNA template in the presence of a β clamp (see diagram above the gel) and in the absence of β. Reactions (25 μl) contained 20 mM Tris⋅Cl (pH 7.5), 0.1 mM EDTA, 100 mM NaCl, 4% glycerol, 40 μg/ml BSA, 5 mM DTT, 8 mM MgCl2, 0.5 mM ATP, 60 μM each of dGTP, dATP, dTTP, and dCTP, 1.7 nM M13mp18 ssDNA primed with a 32P-5′-end-labeled 30-mer DNA, 0.42 μM ssDNA-binding protein (as tetramer), and 2 nM γ-complex (as γ3 δ1 δ′1 χ1 ψ1). Reactions in lanes 1–7 contained 30 nM β. The Klenow fragment of DNA polymerase I was added to each reaction as indicated above each lane. Reactions were incubated at 37°C for 30 min and then quenched on addition of 25 μl of 1% SDS and 90% formamide. Reactions were analyzed in a 8% polyacrylamide gel containing 6 M urea. The gel was dried, and radiolabeled DNA was visualized by using a PhosphorImager. Positions of primer and extended primer are indicated to the right of the gel.
Does the interaction of β with Pol I influence its action during DNA synthesis? The influence of β on Pol I was examined in the experiment of Fig. 5B by using an ssDNA-binding protein-coated M13mp18 ssDNA template primed with a unique 5′ 32P-labeled oligonucleotide. Reactions were performed for 30 min in either the presence or absence of β (γ complex is present in both experiments), using different amounts of Pol I as indicated in the figure. Reactions were quenched with SDS/EDTA and analyzed in a urea polyacrylamide gel. The autoradiogram shows a widely different pattern of synthesis by Pol I in the presence of β compared with Pol I in the absence of β. The pattern of Pol I in the absence of β is characteristic of a distributive enzyme that extends the entire population of primed template with a speed that depends on protein concentration. The reaction is not saturated at even the highest Pol I concentration. In the presence of β (and γ complex) a significant proportion of the substrate becomes fully replicated at even the lowest concentration of Pol I. This result is likely explained by an increase in processivity of Pol I, with some molecules perhaps remaining with the same template from beginning to end. In the presence of β/γ complex, the reaction would appear to be saturated at the highest Pol I concentration (assuming the template is completed, and that the small amount of starting material that remains is inactive primer). Further studies are needed to more fully understand the effects of β on Pol I and ligase.
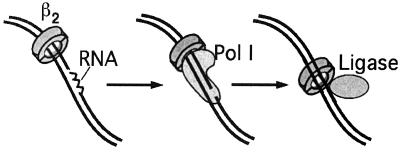
What roles may β play with Pol I and ligase in DNA metabolism? One obvious possibility could be the processing of the RNA primers at Okazaki fragments, where Pol I and ligase function at nearly the same time and place. On the completion of each lagging strand fragment, Pol III H.E. disengages from its β clamp and the completed DNA and cycles to a new upstream primed site near the replication fork (reviewed in refs. 8 and 36). The “used β clamp” remains on the completed Okazaki fragment. To complete the joining of Okazaki fragments, Pol I digests the RNA primer at the 5′ end of each fragment and fills this section in with DNA by using its nick translation capacity. After this, DNA ligase seals the nick. Combining the findings that β clamps are left by Pol III H.E. on each Okazaki fragment, and that β interacts with both Pol I and ligase, it seems possible that the β clamp that Pol III leaves behind on DNA may aid Pol I and ligase in Okazaki fragment maturation, as illustrated in Fig. 6. Similar events have been hypothesized to occur in eukaryotes based on interactions between PCNA and both the Fen1 nuclease and DNA ligase I (37). Clearly, further studies on the effects of β on Pol I and ligase are required to determine whether they may function as hypothesized, or perhaps act in other aspects of DNA metabolism. For example, the role of Pol I in various repair reactions may be aided by β (e.g., nucleotide excision repair). Ligase is also needed in a variety of DNA transactions involving repair and recombination (e.g., mismatch repair, a reaction in which Pol III H.E. and ligase are both involved).
Figure 6.
Possible role of β in Okazaki fragment maturation. During lagging strand synthesis, β clamps are left behind on lagging strand fragments. The remaining β clamps may serve to target Pol I to the lagging strand for removal of the RNA used to initiate each fragment. The β subunit interaction with ligase may serve a similar purpose in targeting, or otherwise aiding, DNA ligase in sealing lagging strand fragments after Pol I action.
The finding that Pol I can use the beta clamp may be exploited in modified versions of reactions important in biotechnology, such as PCR, because most of these involve polymerases of the Pol I family.
Conclusions
The DNA sliding clamp was originally identified as a key component of the chromosomal replicating machine. However, studies presented here and elsewhere demonstrate that sliding clamps, prokaryotic and eukaryotic, directly interact with numerous DNA metabolic enzymes including ligase, mismatch repair proteins, and several different types of DNA polymerases involved in replication, repair, and the bypass of DNA lesions. In fact, Pols I, II, III, and V all belong to different polymerase classes, making it all the more remarkable that β can function with all of them. This report increases the list of factors that interact with the E. coli β clamp to nine different proteins (Table 1). The emerging view of the sliding clamp is now more of a mobile platform upon which a variety of different proteins may attach, indicating that β and PCNA are amazingly versatile and may play many roles, not only as a sliding clamp in replication, but also in repair and recombination.
Table 1.
Proteins that interact with the E. coli β clamp
| Protein | Protein function | Reference |
|---|---|---|
| DNA polymerase I | Repair and replication | This work |
| DNA polymerase II | Replication restart | 18, 19 |
| DNA polymerase III (α) | Chromosomal replication | 3 |
| DNA polymerase V | Lesion bypass | 21 |
| γ complex (γδδ′χψ) | Clamp loader | 16 |
| γ | Motor protein | * |
| δ | Clamp opener | 16 |
| χ | Salt stabilization, primase switch | * |
| MutS | Mismatch recognition | This work |
| DNA ligase | Ligation | This work |
A. Yuzhakov and M.O.D., unpublished observations.
Acknowledgments
We are grateful to Dr. Myron Goodman for purified DNA polymerase II. This work was supported by the Howard Hughes Medical Institute.
Abbreviations
- Pol III H.E.
DNA polymerase III holoenzyme
- PCNA
proliferating cell nuclear antigen
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Kelman Z, O'Donnell M. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 2.Waga S, Stillman B. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 3.Stukenberg P T, Studwell-Vaughan P S, O'Donnell M. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- 4.Kong X P, Onrust R, O'Donnell M, Kuriyan J. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 5.Krishna T S, Kong X P, Gary S, Burgers P M, Kuriyan J. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 6.Gulbis J, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 7.Moarefi J, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- 8.Marians K J. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 9.Tinker R L, Williams K P, Kassavetis G A, Geiduschek E P. Cell. 1994;77:225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 10.Kelman Z, Hurwitz J. Trends Biochem Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]
- 11.Flores-Rozas H, Kelman Z, Dean F B, Pan Z-Q, Harper J W, Elledge S J, O'Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waga S, Hannon G J, Beach D, Stillman B. Nature (London) 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 13.Parker A, Gu Y, Mahoney W, Lee S-H, Singh K K, Lu A-L. J Biol Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 14.Shibahara K, Stillman B. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 15.Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O'Donnell M. J Biol Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 16.Dallmann H G, McHenry C S. J Biol Chem. 1995;270:29563–29569. [PubMed] [Google Scholar]
- 17.Pritchard A E, Dallmann H G, Glover B P, McHenry C S. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naktinis V, Onrust R, Fang L, O'Donnell M. J Biol Chem. 1995;270:13358–13365. [PubMed] [Google Scholar]
- 19.Turner J, Hingorani M M, Kelman Z, O'Donnell M. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes A J, Jr, Bryan S K, Chen H, Moses R E, McHenry C S. J Biol Chem. 1991;266:4568–4573. [PubMed] [Google Scholar]
- 21.Bonner C A, Stukenberg P T, Rajagopalan M, Eritja R, O'Donnell M, McEntee K, Echols H, Goodman M F. J Biol Chem. 1992;267:11431–11438. [PubMed] [Google Scholar]
- 22.Rangarajan S, Woodgate R, Goodman W F. Proc Natl Acad Sci USA. 1999;96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahue R S, Au K G, Modrich P. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 25.Stukenberg P T, Turner J, O'Donnell M. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- 26.Naktinis V, Turner J, O'Donnell M. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- 27.Lemon K P, Grossman A D. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 28.Clark A B, Valles F, Drotschmann K, Gary R K, Kunkel T A. J Biol Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 29.Flores-Rozas H, Clark D, Kolodner R D. Nat Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 30.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 31.Obmolova G, Ban C, Hsieh P, Yang W. Nature (London) 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 32.Malik H S, Henikoff S. Trends Biochem Sci. 2000;25:414–418. doi: 10.1016/s0968-0004(00)01623-6. [DOI] [PubMed] [Google Scholar]
- 33.Hurwitz J, Wickner S, Wright M. Biochem Biophys Res Commun. 1973;51:257–267. doi: 10.1016/0006-291x(73)91251-5. [DOI] [PubMed] [Google Scholar]
- 34.Wickner S, Hurwitz J. Proc Natl Acad Sci USA. 1976;73:1053–1057. doi: 10.1073/pnas.73.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin D S, Bai W, Yao N, O'Donnell M, Tomkinson A E. Proc Natl Acad Sci USA. 1997;94:12863–12868. doi: 10.1073/pnas.94.24.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hingorani M M, O'Donnell M. Curr Biol. 2000;10:R25–R29. doi: 10.1016/s0960-9822(99)00252-3. [DOI] [PubMed] [Google Scholar]
- 37.Levin D S, McKenna A E, Motycka T A, Matsumoto Y, Tomkinson A E. Curr Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]