Abstract
Crystal structures and biochemical analyses of PcrA helicase provide evidence for a model for processive DNA unwinding that involves coupling of single-stranded DNA (ssDNA) tracking to a duplex destabilization activity. The DNA tracking model invokes ATP-dependent flipping of bases between several pockets on the enzyme formed by conserved aromatic amino acid residues. We have used site-directed mutagenesis to confirm the requirement of all of these residues for helicase activity. We also demonstrate that the duplex unwinding defects correlate with an inability of certain mutant proteins to translocate effectively on ssDNA. Moreover, the results define an essential triad of residues within the ssDNA binding site that comprise the ATP-driven DNA motor itself.
Keywords: DNA–protein interactions, motor proteins, DNA translocation, mutagenesis
The DNA helicases are a large and diverse group of motor proteins that are involved in the modulation of DNA structure. Frequently, they are found to catalyze the separation of duplex DNA into its component single strands (for reviews see refs. 1 and 2) but also are involved in events such as processing stalled replication forks and Holliday junction migration (3). These reactions are essential during DNA replication, recombination, and repair. Consequently, DNA helicases are found to be ubiquitous in nature, and organisms encode multiple DNA helicases that play specific cellular roles (4).
PcrA is a superfamily I DNA helicase, found in Gram-positive bacteria, which is involved in DNA repair and plasmid rolling circle replication (5, 6). Its primary structure is very similar to the intensively studied Rep, UvrD and RecB(CD) helicases of Escherichia coli. PcrA from Bacillus stearothermophilus has been crystallized alone and in a variety of complexes with DNA and/or nucleotide analogues (7–9), and these structures have been used to devise a detailed molecular mechanism for PcrA-catalyzed helicase activity (8, 10). In this model, PcrA helicase is functional as a monomer that separates duplex DNA in a processive manner by coupling two distinct activities. First, PcrA translocates unidirectionally along single-stranded DNA (ssDNA) by virtue of a ssDNA tracking motor that is powered by ATP hydrolysis. Progression into duplex DNA is facilitated by a duplex destabilization activity acting ahead of the tracking motor, which also is modulated by ATP binding and hydrolysis. The crystal structures allow identification of the amino acid residues involved in binding ssDNA and proposed to be involved in the ssDNA tracking activity. They form a channel in which several bases of ssDNA bind in pockets formed predominantly by aromatic stacking interactions (Fig. 1). It is proposed that coordinated rearrangement of the relative positions of these residues in an ATP-dependent manner results in the ssDNA being passed unidirectionally along the base-stacking pockets (8). In the model, the motor is fueled by the hydrolysis of one ATP molecule for every base of ssDNA that is pumped through the protein (8). ssDNA translocation has been detected indirectly by studying Pi release from PcrA⋅DNA complexes, confirming the view that one ATP hydrolysis event is required for each base of movement along the DNA lattice (11).
Figure 1.
Structure of the ssDNA binding site in substrate and product complexes of PcrA helicase. (Left) A ribbon representation of the two complexes with the inset at higher magnification. The protein backbone is white with amino acid residues, which contact ssDNA color-coded according to the cartoon representation (Right). The partial duplex DNA substrate that is bound to PcrA in both complexes is orange. In the product complex an area of duplex DNA is disordered and has not been modeled.
The residues that form the ssDNA binding channel are predicted by the model to be essential to helicase function. Many of the residues that contact DNA are found in the conserved helicase motifs that are characteristic of the superfamily I DNA helicases. Y257, W259, and R260 all are found within helicase motif III, which has been shown to play a critical role in coupling ATP hydrolysis to helicase activity (12–14). F64 is located in motif Ia, and H587 is found in the region between the closely aligned motifs V and VI. F192 and F626 are outside of the signature motifs but are conserved within the Rep/UvrD/PcrA family of SF1 DNA helicases (15).
Here we present the results of a mutagenesis study of the residues that form the ssDNA binding site. The results of mutating the W259 and R260 residues to alanine have been reported previously and are consistent with a role in ssDNA binding and/or translocation (12). Both mutations result in helicase and DNA-binding defects, whereas kinetic parameters for ATP hydrolysis are close to wild-type values. In this study we have created alanine mutants for all of the remaining residues in the ssDNA binding site of PcrA helicase (F64A, F192A, Y257A, H587A, F626A). The mutant proteins have been studied biochemically to determine their ability to separate duplex DNA, hydrolyze ATP, bind ssDNA and double-stranded DNA (dsDNA), and translocate on ssDNA. The results confirm the essential nature of all of the residues for helicase activity. They also clarify the contribution of individual residues to the ssDNA translocation activity and illuminate the mechanism by which it is coupled to ATP hydrolysis.
Materials and Methods
Site-Directed Mutagenesis.
Site-directed mutagenesis of the pcrA gene was performed by using a PCR technique as described (16). Mutant genes were sequenced (Alice Taylor, University of Oxford) to confirm the desired mutation and the absence of spurious mutations that may have been introduced during the PCR.
Protein Preparations.
Mutant PcrA proteins were prepared in the same manner as the wild type as described (17) and were more than 99% pure as estimated from Coomassie-stained SDS/PAGE gels. Protein concentration was determined spectrophotometrically by using a calculated extinction coefficient of 0.76 OD280 mg−1⋅ml−1⋅cm−1 (or 0.75 OD280 mg−1⋅ml−1⋅cm−1 for the Y257A protein). N-[2-(1-maleimidyl)ethyl]-7-(diethyamino) coumarin-3-carboxamide (MDCC)-modified phosphate binding protein was prepared as described (18). Mutant PcrA proteins were crystallized in either space group P65 or C2 under conditions described for the wild-type enzyme (19), suggesting that global folding is unperturbed in all of the engineered proteins.
Helicase Assays.
3′-Tailed substrates were prepared by annealing a radioactively labeled 45-mer oligonucleotide to a slight excess of ssM13mp18 DNA as described (20), creating a substrate with a 22-bp duplex region and a 23-base 3′ terminated ssDNA tail. Time-course reactions were performed at 37°C in a reaction buffer containing 20 mM Tris (pH 7.5), 50 mM NaCl, 3 mM MgCl2, 2.5 mM ATP, 4 mM DTT, and 10% glycerol with 200 nM protein and 1 nM DNA substrate. After a 2-min preincubation the reactions were started with PcrA and terminated by adding stop buffer [0.4% (wt/vol) SDS, 40 mM EDTA, 8% (vol/vol) glycerol, 0.1% (wt/vol) bromophenol blue]. Displaced and annealed oligonucleotide were separated by electrophoresis through a 10% nondenaturing polyacrylamide gel at constant voltage (150 V). Gels were dried and quantitative analysis was performed by using a PhosphorImager and imagequant software (Molecular Dynamics).
ATP Hydrolysis Assays.
Steady-state ATPase activity was measured by linking ATP hydrolysis to NADH oxidation as described (21). Reactions were performed at room temperature with 24 nM protein in a buffer containing 20 mM Tris⋅Cl (pH 7.5), 50 mM NaCl, 3 mM MgCl2, and 4 mM DTT. Values for KDNA, the concentration of dT16 required for half-maximal stimulation of ATP hydrolysis, were determined at saturating (2 mM) ATP concentrations. Values for the kcat and Km were determined at saturating dT16 concentrations (10 × KDNA value). Substrate concentrations were varied at least 5-fold above and below the measured Km and KDNA values. Data were fitted directly to the Michaelis–Menten equation by using the program graphpad prism version 2.0 (GraphPad, San Diego).
Electrophoretic Mobility-Shift Assays.
Protein at various concentrations was incubated with 2.5 nM ssDNA (dT16) or dsDNA (22-bp blunt end duplex) probe in buffer (20 mM Tris, pH 7.5/50 mM NaCl/3 mM MgCl2/4 mM DTT) at room temperature for 20 min. The sequences and preparation of probes are described elsewhere (10). A small volume of loading buffer [0.25% bromophenol blue/40% (wt/vol) sucrose for ssDNA gel shifts or 25% Ficoll/0.05% bromophenol blue for duplex DNA shifts] was added to each sample immediately before they were run on a native polyacrylamide gel (9% for ssDNA gel shifts and 6% for duplex DNA gel shifts) to separate the protein–DNA complex from the free DNA. Gels were dried and exposed to x-ray film.
Phosphate Release Assay.
Assays for ssDNA-dependent Pi release were performed essentially as described (11, 22). All data presented are single trace measurements. Stopped-flow measurements were made with a HiTech SF61MX apparatus, with excitation at 436 nm through 4-nm slits. Fluorescence emission was measured after a 455-nm cutoff filter. All concentrations quoted are the final ones after mixing and were 100 or 10 nM PcrA, 16 μM (saturating) oligonucleotide, 500 μM (saturating) ATP, and 3 μM N-[2-(1-maleimidyl)ethyl]-7-(diethyamino) coumarin-3-carboxamide (MDCC)–phosphate binding protein in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, and 3 mM MgCl2. All experiments were performed at 20°C.
Results
Strand Displacement Helicase Assays.
Wild-type and mutant PcrA proteins were tested for their ability to displace a short oligonucleotide from M13mp18 ssDNA. All of the mutant proteins were defective in helicase activity (Fig. 2). The relative helicase activities of wild-type and mutant PcrA proteins, based on the initial rates of strand displacement, are shown in Table 1. The biochemical basis for the reduced helicase activity of each of the mutant proteins then was investigated by assaying for the various subactivities that together constitute a processive DNA unwinding activity.
Figure 2.
Helicase activity of wild-type and mutant PcrA proteins measured by using a strand displacement assay (see Materials and Methods). Data shown are the average of at least two independent experiments. Error bars represent SD of the mean. Relative rates of helicase activity are shown in Table 1.
Table 1.
Relative helicase activity in wild-type and mutant PcrA proteins
| Protein | Relative rate, % |
|---|---|
| Wild type | 100 |
| F64A | 4 |
| F192A | 2 |
| Y257A | <1 |
| H587A | 5 |
| F626A | 26 |
Estimates of initial rates of strand displacement from the data shown in Fig. 2.
ssDNA Binding.
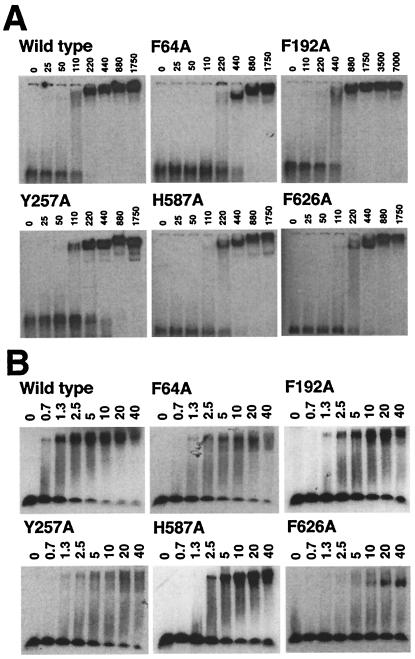
To probe the ability of wild-type and mutant proteins to bind ssDNA we used the electrophoretic mobility-shift assay technique. All of the mutants displayed a slight reduction in their affinity for ssDNA of between 2- and 4-fold difference (Fig. 3A). However, in energetic terms, these are surprisingly modest decreases in ssDNA binding affinity particularly when compared with a mutant in which residue W259 (Fig. 1) is changed to alanine, which results in a decrease of ssDNA binding affinity of around 2 orders of magnitude (12).
Figure 3.
Electrophoretic mobility-shift assays. (A) ssDNA band shifts. A total of 2.5 nM dT16 was incubated with protein at the nanomolar concentration indicated (see Materials and Methods). (B) Duplex DNA band shifts. A 22-bp blunt end duplex at 2.5 nM was incubated with protein at the micromolar concentration indicated (see Materials and Methods).
dsDNA Binding.
Affinity for a duplex DNA substrate also was studied by using the electrophoretic mobility-shift assay. Wild-type PcrA displays a considerably lower affinity for dsDNA than for ssDNA (Fig. 3). With the exception of F626A, all of the mutant proteins bound duplex DNA with an affinity comparable to that of the wild type, albeit reduced by up to ≈4-fold. For the F64A and Y257A mutants, the gel bands observed for the PcrA⋅DNA complexes were poorly defined and ran in a broad area at the top of the gel. Interestingly, in the duplex-bound “substrate” complex of PcrA, there is a direct contact between F64 and Y257 that is not present in either the apo enzyme or product complex structures of PcrA (7, 8). Therefore, it is possible that the diffuse bands seen for these mutants reflect a reduction in stability of the conformation(s) of the protein required to bind duplex DNA. The affinity of the F626A mutant for duplex DNA was reduced most significantly. Even at the highest protein concentration tested (40 μM), only a small proportion of the duplex DNA was band-shifted, suggesting a reduction in affinity of more than 50-fold relative to wild type.
Steady-State ATP Hydrolysis.
The wild-type and mutant proteins were assessed for their ability to catalyze ssDNA-dependent ATP hydrolysis by using a continuous steady-state assay in which nucleotide hydrolysis is coupled to the oxidation of NADH. All mutants displayed Michaelis–Menten kinetics (data not shown). In each case three experimental values were determined, kcat (the turnover number), Km (the concentration of ATP at which turnover is half maximal), and KDNA (the concentration of ssDNA at which ATP turnover is half maximal). The results of these assays are shown in Table 2. All of the mutant proteins have Km for ATP within 2- to 3-fold of that of the wild-type enzyme. The measured turnover rates are also within this range with the exception of Y257A, which has a kcat that is about 7-fold lower than that of the wild type. KDNA is increased in all of the mutants ranging from small changes (around 2-fold) for the F64A, H587A, and F626A mutants, to the larger changes observed for the F192A and Y257A proteins (6-fold and 5-fold, respectively). However, even these alterations in KDNA are modest when compared with the 200-fold increase observed for a W259A mutant PcrA (12), putting into perspective the relative contributions made by each of these residues to binding of ssDNA. It is worthy of note that the mutants that contact the DNA in domain 1A (F192A and Y257A) show larger effects than those in domain 2A (H567A and F626A).
Table 2.
Steady-state kinetic parameters for ssDNA-dependent ATP hydrolysis with oligonucleotide cofactor dT16
| Protein | kcat, s−1 | Km, μM | KDNA, nM |
|---|---|---|---|
| Wild type | 35 | 110 | 200 |
| F64A | 14 | 230 | 380 |
| F192A | 42 | 220 | 1,180 |
| Y257A | 5 | 310 | 930 |
| H587A | 47 | 200 | 440 |
| F626A | 18 | 70 | 450 |
Terms are defined in Materials and Methods.
Presteady-State ATP Hydrolysis and ssDNA Translocation.
We previously have developed an assay that allows us to monitor ssDNA translocation in wild-type PcrA (11). Using N-[2-(1-maleimidyl)ethyl]-7-(diethyamino) coumarin-3-carboxamide (MDCC)-phosphate binding protein, a fluorescent sensor for inorganic Pi (18), the kinetics are measured for Pi release from preformed PcrA⋅ssDNA complexes. A well defined rapid phase of Pi release is observed whose magnitude and duration are directly related to the length of ssDNA in the complex. This is followed by a slower steady state that is independent of DNA length. Typical Pi release traces for wild-type PcrA in complex with polythymidylates of 7, 12, and 17 bases in length are shown in Fig. 4. Although only three oligonucleotide lengths were used in the present study, these were representative of the full range used previously (11) and were used to estimate the qualitative effects of the mutations on the translocation properties of the protein. The rapid phase is associated with unidirectional ssDNA translocation and the data allowed the quantification of the step size (1 base/ATP) and speed (50 bases/s) of this process in PcrA. The steady state is thought to be associated with uncoupled ATP hydrolysis by PcrA molecules “stuck” at the 5′ terminus, a proposal that has been demonstrated directly using oligonucleotides labeled with the fluorescent base 2-aminopurine at the 5′ end (unpublished data). The assay has been used to demonstrate that single-strand translocation remains intact when mutations are made in the duplex DNA binding site of PcrA, because the Pi release kinetics remain essentially identical (10). By contrast, all five mutations in the ssDNA binding site lead to altered Pi release kinetics, indicating that some aspect of ssDNA translocation is defective (Fig. 4).
Figure 4.
Presteady-state Pi release from wild type and F192A, H587A, and F626A mutant PcrA⋅ssDNA complexes. All data shown are single trace measurements. Experiments were performed as described in Materials and Methods. Data for rapid-phase amplitude and duration are shown in Table 2. (Inset) An expanded view of the initial part of the trace for the dT12 oligonucleotide.
Pi release from ssDNA complexes of the H587A and F626A mutants is qualitatively similar to wild type in that a rapid phase is followed by a slower steady state (Fig. 4 and Table 3). The steady-state rates of the wild type and the mutant proteins are similar, in agreement with the coupled ATPase assay results (Table 2). In common with the wild-type enzyme, the steady-state rate of the H587A mutant is independent of DNA length, although there is a slight dependency for the F626A protein, which is discussed below. Therefore we assume that, for these mutants, the steady state is dominated by uncoupled ATP hydrolysis at the 5′ end of DNA as in the wild type. In turn, this finding implies that the mutant proteins are capable of some degree of DNA translocation, such that the majority of protein in the steady state is at the 5′ end of DNA. However, the magnitude and duration of the rapid (translocation) phase of Pi release are perturbed in both cases. For the wild-type protein, the kinetic data allowed the determination of a model describing Pi release during the ssDNA translocation process (11). The model can be adjusted to investigate the influence that each of the various ssDNA translocation parameters (e.g., speed, efficiency, processivity, and directionality) has on the fit of the traces produced by the model to those observed for the mutant proteins. The H587A protein displays a longer rapid phase with a slightly increased Pi release amplitude relative to the wild type. This can be accounted for by a slower translocation rate with some degree of uncoupling between ATP hydrolysis and unidirectional movement along the DNA, or reduced processivity of the protein. The F626A protein displays a longer rapid phase of slightly smaller amplitude than the wild-type protein. Interestingly, the steady-state rate that follows this burst shows a slight dependency on the length of oligonucleotide, unlike that of the wild-type protein. This observation is consistent with slower translocation, in which case the steady state becomes dominated less by molecules trapped at the 5′ end of the ssDNA and there is a significant contribution to the steady-state ATPase activity from translocating PcrA molecules. As discussed previously (11), in such a situation the steady-state rate becomes dependent on DNA length to an extent that depends on the proportion of the enzyme molecules in each of the two states.
Table 3.
Pi release parameters during ssDNA translocation
| Protein | Polythymidylate length, bases | Rapid-phase amplitude, P/PcrA | Rapid-phase duration, s | Rapid-phase rate, s−1 | Steady-state rate, s−1 |
|---|---|---|---|---|---|
| Wild type | 7 | 2.1 | 0.09 | 22.2 | 4.7 |
| 12 | 4.5 | 0.14 | 33.2 | 4.8 | |
| 17 | 7.0 | 0.16 | 46.5 | 5.8 | |
| F192A | 7 | 0.48* | NA | NA | 8.8 |
| 12 | 0.85* | NA | NA | 14.5 | |
| 17 | 0.80* | NA | NA | 15.2 | |
| H587A | 7 | 4.1 | 0.20 | 20.8 | 4.5 |
| 12 | 7.0 | 0.25 | 28.4 | 5.0 | |
| 17 | 6.9 | 0.32 | 21.3 | 4.6 | |
| F626A | 7 | 2.0 | 0.09 | 21.7 | 1.8 |
| 12 | 3.1 | 0.18 | 17.2 | 2.6 | |
| 17 | 4.0 | 0.23 | 17.3 | 3.1 |
Rapid-phase amplitude and duration are shown for wild type, F192A, H587A, and F626A PcrA⋅ssDNA complexes with polythymidylates of 7, 12, and 17 bases in length. NA, not available.
The rapid phase of Pi release for the F192A mutant represents a true “phosphate burst” rather than a multiple turnover linear phase as in the wild-type protein.
This relationship between enzyme molecules trapped at the 5′ end of the DNA and those actively translocating becomes even more significant for some of the other mutant proteins. For the F64A and Y257A mutants, the initial burst of rapid ATP turnover is now undetectable (data not shown), and we observe a significant relationship between the steady-state ATPase activity and the length of the ssDNA cofactor (Fig. 5). The steady-state dependence on length is not caused by an increase in the number of possible binding sites for PcrA, because the DNA concentration used is saturating and is in considerable excess over PcrA in all experiments. Unfortunately, the loss of the two phases of the Pi release data reduces the information that can be obtained regarding the translocation of the enzymes on ssDNA. However, the simplest explanation for the data is that a significant proportion of the PcrA molecules, which are hydrolyzing ATP, are actively translocating during steady state. This may be because they have not reached the 5′ end of DNA due to premature dissociation (reduced processivity), or because they no longer become stuck at the 5′ end as described for wild-type PcrA (11). However, the steady-state ATPase rates of these mutants (Table 2) show some differences. Whereas the activity of the Y257A mutant is impaired 7-fold, that of the F64A is only reduced 2- to 3-fold.
Figure 5.
Dependence of the steady-state ATPase rate on oligonucleotide length that is observed for the F64A and Y257A mutants. These data were obtained by using the phosphate release assay.
The F192A protein displays Pi release kinetics that are qualitatively different from the wild type or any of the other mutant proteins (Fig. 4). Curiously, there is an initial Pi burst equivalent to a single turnover of the enzyme that is followed by a steady state that is similar to wild type and independent of DNA length. The Pi burst indicates that the rate-limiting step in the ATP hydrolysis cycle occurs after the release of the first Pi molecule although what this step might be is unclear at the present time.
Discussion
On the basis of “substrate” and “product” complexes of PcrA, we have proposed that helicase activity results from two distinct but coupled protein functions: ssDNA translocation powered by a unidirectional tracking motor and duplex destabilization ahead of the motor that facilitates its progression (8). Subsequently, we have demonstrated and mapped the duplex destabilization activity, allowing the generation of mutant proteins with intact ssDNA translocation but poor helicase activity (10). We have demonstrated biochemically the unidirectional ssDNA translocation and confirmed that one ATP molecule is required for every base that is tracked (11). In this study, we have sought to identify the relative importance of the individual aromatic amino acid residues that form the ssDNA binding site (Fig. 1). The results also provide clues as to which residues are involved in coupling the translocation activity to the hydrolysis of ATP.
Structure and Proposed Mechanism of the ssDNA Translocation Motor.
Substrate and product structures of PcrA helicase reveal the binding site for a ssDNA tail in a partial duplex DNA substrate. It is formed in a groove across the top of the 1A and 2A subdomains and predominantly involves stacking interactions between the DNA bases and aromatic amino acids (Fig. 1). This creates a series of acceptor pockets for the DNA bases along the length of the PcrA molecule. However, the exact positioning and occupancy of the acceptor pockets is not equivalent in the two crystal structures (Fig. 1), which led to a proposal of the mechanism for unidirectional translocation along ssDNA (8). Broadly speaking, the ssDNA binding site may be thought of as consisting of two halves, each contributed by one of the RecA-like subdomains (1A and 2A). The 2A subdomain ssDNA binding site is the “front half” because it forms the leading edge of the protein relative to the DNA during translocation. It contains the H587 and F626 residues along with helicase motifs V and VI. The 1A subdomain is the “rear half” of the binding site and contains the F64 residue of motif Ia, the F192 residue, Y257, W259 and R260 of motif III, as well as motifs I and II, which contribute to the ATP hydrolysis chemistry (9). All of the residues highlighted in Fig. 1 make direct contacts with ssDNA and, consequently, were predicted to be critical for ssDNA translocation (8). The F626, Y257, and F64 residues were of particular importance because their position and/or contact with ssDNA appeared to be altered by the ATP hydrolysis cycle (ref. 8; Fig. 1).
The Roles of H587 and F626 in the Front Half of the ssDNA Binding Site.
Mutation of H587 to alanine results in a 20-fold decrease in helicase activity that cannot be attributed to defective ATP hydrolysis. Therefore, the H587A mutant, and indeed all of the mutations described herein, may be described as an “uncoupling” mutant: the basic ATP hydolysis machinery, and its mechanism of stimulation by ssDNA have essentially remained intact, whereas helicase activity is reduced disproportionately. There is no significant defect in dsDNA binding. Despite the position of H587 in the ssDNA binding site of PcrA, there is surprisingly little effect on ssDNA binding. Therefore it is reasonable to suggest that the reduction in helicase activity relates to some more dynamic aspect of the PcrA/ssDNA transactions rather than simply ssDNA binding energy. Study of the kinetics of Pi release from H587A.ssDNA complexes support this view, because the mutant protein shows an altered rapid (translocation) phase of Pi release, followed by a steady-state rate that is comparable to that of the wild-type enzyme.
Mutation of F626 to alanine results in the smallest (around 4-fold) decrease in helicase activity of any of these mutant proteins (Fig. 2). Steady-state ATP hydrolysis assays indicate that the defective helicase activity is not caused by an inability to hydrolyze ATP (Table 2), and ssDNA gel shifts and the steady-state ATPase assay KDNA measurement both indicate only a 2-fold reduced ssDNA affinity. Unexpectedly, DNA gel-shift assays indicate a large decrease in affinity of the mutant protein for duplex DNA. The F626 residue is found at the very leading edge of the ssDNA binding site, at the ssDNA/dsDNA interface (Fig. 1). Although the PcrA substrate complex clearly shows the residue involved in an aromatic stacking interaction with the first base of the ssDNA tail of the substrate, it is possible that during the reaction cycle it might interact with the final base pair of duplex DNA. Alternatively, the blunt ends of the duplex used in the gel shifts might be subject to thermal fraying, allowing access of unpaired bases to the F626 residue. In respect to binding of duplex DNA, the F626A mutant behaves in a manner that is similar to a class of mutant proteins that were made in the duplex binding site of PcrA (10). However, for those mutant proteins, ssDNA translocation, as measured by studying Pi release from DNA complexes, was essentially the same as that of the wild type (10). In contrast, the F626A mutant protein displays perturbed ssDNA translocation, which most likely relates to reduced processivity and/or translocation rate on ssDNA. In our DNA translocation model, the F626 residue is involved in binding and releasing DNA bases in response to ATP hydrolysis. The modest decrease in helicase activity observed in the mutant protein reveals this residue to be of lesser importance than many of the others that interact with the ssDNA. Our experiments indicate that the most important residues in the ssDNA motor are found toward the rear (3′ end) of the ssDNA binding site.
The Roles of F64, F192, and Y257 in the Rear Half of the ssDNA Binding Site.
The F64, F192, and Y257 residues are situated in the rear half of the ssDNA binding site. In the substrate (ATP bound) complex of PcrA, F64 and Y257 contact each other, occluding one of the DNA base binding pockets (Fig. 1). The conformational change associated with nucleotide binding results in the displacement of a base from the Y257 to the F192 binding pocket. Thus the three residues form a triad that underpins the proposed mechanism for DNA motor activity. Mutation of any of these three residues to alanine results in proteins with the most severe defects in helicase activity (25-fold for F64A, 50-fold for F192A, and at least 100-fold for Y257A). However, equilibrium binding studies and the KDNA measurement from ATP hydrolysis assays both show that the ssDNA binding affinity of the mutants is reduced by only 2- to 6-fold. Analysis of steady-state ATP hydrolysis compared with wild type reveals little change for either the F192A or F64A mutants, although the rate of ATP turnover is reduced 7-fold in Y257A protein. The presteady-state kinetics are rather different to those of the wild-type enzyme, and the proteins no longer show an initial rapid burst of ATP turnover but simply enter a steady-state rate immediately. Moreover, the ATPase rates of the F64A and Y257A proteins now show a pronounced dependence on the length of the ssDNA oligonucleotide, a property that differs from the wild-type enzyme. The simplest explanation of this observation is that the proteins are no longer getting trapped at the 5′ end of the DNA after translocation. Instead, it would seem that these mutants are now able to translocate along the ssDNA and then dissociate more rapidly either from the end or at some point before reaching the end. This means that there will be a significant contribution to the steady-state ATPase rate from molecules that are translocating along DNA and that this population will increase in proportion with increased length of the oligonucleotide. The linear dependence of kcat on the length of the ssDNA suggests that the proteins are dissociating from the DNA and then reassociating rather than translocating in a nondirectional manner because the latter would result in a nonlinear relationship. Note that ATPase activity is still stimulated in the presence of ssDNA, showing that ATP turnover is not simply uncoupled from binding to DNA.
For the F192A mutant, it seems that the enzyme is able to undergo one rapid ATP turnover when prebound to DNA, but then enters a steady-state rate of ATP turnover that is independent of the length of the ssDNA. The molecular basis for this initial single rapid turnover event remains unclear although a conformational change after ATP hydrolysis, relating to movement of a base from the Y257 binding pocket to the (now disrupted) F192 binding pocket, is a likely candidate.
In conclusion, these experiments allow a comparison of the quantitative importance of the seven residues that form the ssDNA binding channel of PcrA. The results of the helicase assays for proteins with mutations in the ssDNA binding site indicate that the major players are located in the rear half of the binding site and are contributed by domain 1A. In contrast, mutation of H587 and F626, located in domain 2A toward the front of the ssDNA binding channel, had less significant effects on helicase activity. In all cases, the mutations made relatively little difference to the various activities that together constitute a DNA translocation activity. Despite their location in the ssDNA binding channel (8), the mutations presented here have surprisingly little effect on the ability of the protein to bind ssDNA, particularly when compared with the much larger effect on ssDNA binding that is observed when the W259 residue is mutated to alanine (12). Therefore, the defective helicase activity must relate to some faulty aspect of ssDNA translocation and/or the mechanism by which it is coupled to ATP hydrolysis. In the case of the F626A and H587A mutants, defective ssDNA translocation was observable directly with the mutant⋅ssDNA complexes displaying altered rapid (translocation) phases of Pi release. The remaining mutants displayed Pi release characteristics that were qualitatively different from the wild-type protein, an observation that is also consistent with faulty ssDNA translocation, but a fuller description of the effect of those mutations on ssDNA translocation requires a more direct assay for helicase movement on DNA.
DNA Translocation Catalyzed by DNA Helicases.
The DNA helicases are a remarkably diverse group of enzymes and it will be of interest to see whether other helicases operate PcrA-like DNA translocation mechanisms. The Rep helicase of E. coli, also a member of helicase superfamily I, is proposed to translocate via DNA looping rather than by unidirectionally tracking along the contour of the nucleic acid (23, 24). The hepatitis C virus helicase, a well studied representative of helicase superfamily II, has been suggested to track ssDNA by using an inchworm-type mechanism and is thought to progress by 1–2 bases for each ATP that is hydrolyzed (25, 26). Conserved tryptophan and valine residues sandwich several bases of DNA and drive translocation by a ratchet mechanism that involves their repositioning during the ATP hydrolysis cycle. Mutagenesis has supported the proposed mechanism by confirming the importance of the tryptophan and valine residues for helicase activity (27–29). Recently, a molecular mechanism has been proposed for ssDNA translocation catalyzed by the T7 gene 4 protein, a representative of the hexameric replicative helicases, which is also a unidirectional tracking mechanism (30). Although the mechanistic step size is tentatively suggested to be 1–2 bases/ATP, this proposal awaits biochemical analysis. In the superfamily II HCV helicase (25) and possibly in the hexameric helicases (30), ssDNA binds to the protein in a fashion that is quite unlike the base-stacking arrangement seen in the superfamily I DNA helicases PcrA and Rep (8, 24). It is likely that these structural differences will relate to underlying mechanistic and/or functional differences between the helicases of various families.
Acknowledgments
We thank Jennifer Byrne and Jackie Hunter for technical assistance. This work was supported by the Wellcome Trust (to D.B.W.) and the Medical Research Council (to M.R.W.). M.S.D. is supported by a Wellcome Prize Fellowship.
Abbreviations
- ssDNA
single-stranded DNA
- dsDNA
double-stranded DNA
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Lohman T M, Bjornson K P. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 2.Soultanas P, Wigley D B. Curr Opin Struct Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 3.McGlynn P, Lloyd R G. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 4.Matson S W, Bean D W, George J W. BioEssays. 1994;16:13–22. doi: 10.1002/bies.950160103. [DOI] [PubMed] [Google Scholar]
- 5.Petit M A, Dervyn E, Rose M, Entian K-D, McGovern S, Ehrlich S D, Bruand C. Mol Microbiol. 1999;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 6.Soultanas P, Dillingham M S, Wigley D B. Nucleic Acids Res. 1999;26:2374–2379. doi: 10.1093/nar/26.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Nature (London) 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 8.Velankar S, Soultanas P, Dillingham M S, Subramanya H S, Wigley D B. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 9.Soultanas P, Dillingham M S, Velankar S S, Wigley D B. J Mol Biol. 1999;290:137–148. doi: 10.1006/jmbi.1999.2873. [DOI] [PubMed] [Google Scholar]
- 10.Soultanas P, Dillingham M S, Wiley P, Webb M R, Wigley D B. EMBO J. 2000;19:3799–3810. doi: 10.1093/emboj/19.14.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dillingham M S, Wigley D B, Webb M R. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 12.Dillingham M S, Soultanas P, Wigley D B. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brosh R M, Jr, Matson S W. J Biol Chem. 1996;271:25360–25368. doi: 10.1074/jbc.271.41.25360. [DOI] [PubMed] [Google Scholar]
- 14.Brosh R M, Jr, Matson S W. J Biol Chem. 1997;272:572–579. doi: 10.1074/jbc.272.1.572. [DOI] [PubMed] [Google Scholar]
- 15.Korolev S, Yao N, Lohman T M, Weber P C, Waksman G. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPherson M J, Quirke P, Taylor G R, editors. PCR: A Practical Approach. Oxford: IRL; 1992. pp. 207–209. [Google Scholar]
- 17.Soultanas P, Dillingham M S, Wigley D B. Nucleic Acids Res. 1998;26:2374–2379. doi: 10.1093/nar/26.10.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brune M, Hunter J L, Howell S A, Martin S R, Hazlett T L, Corrie J E T, Webb M R. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 19.Bird L E, Brannigan J A, Subramanya H S, Wigley D B. Nucleic Acids Res. 1998;26:2686–2693. doi: 10.1093/nar/26.11.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crute J J, Mocarski E S, Lehman I R. Nucleic Acids Res. 1988;16:6585–6596. doi: 10.1093/nar/16.14.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura J K, Gellert M. J Biol Chem. 1990;265:21342–21349. [PubMed] [Google Scholar]
- 22.Brune M, Hunter J L, Corrie J E T, Webb M R. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 23.Wong I, Lohman T M. Science. 1992;256:350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- 24.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 25.Porter D J T, Short S A, Hanlon M H, Preugschat F, Wilson J E, Willard D H, Consler T G. J Biol Chem. 1998;273:18906–18914. doi: 10.1074/jbc.273.30.18906. [DOI] [PubMed] [Google Scholar]
- 26.Kim J L, Morgenstern K A, Griffith J P, Dwyer M D, Thomson J A, Murcko M A, Lin C, Caron P R. Structure (London) 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 27.Preugschat F, Danger D P, Carter L H, Davis R G, Porter D J T. Biochemistry. 2000;39:5174–5183. doi: 10.1021/bi9923860. [DOI] [PubMed] [Google Scholar]
- 28.Lin C, Kim J L. J Virol. 1999;73:8798–8807. doi: 10.1128/jvi.73.10.8798-8807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paolini C, Lahm A, De Francesco R, Gallinari P. J Gen Virol. 2000;81:1649–1658. doi: 10.1099/0022-1317-81-7-1649. [DOI] [PubMed] [Google Scholar]
- 30.Singleton M R, Sawaya M R, Ellenberger T, Wigley D B. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]