During the later stages of male germ cell differentiation, the genome undergoes dramatic post-meiotic reorganization and global compaction. In this issue of Genes & Development, Montellier et al. report that the testis-specific histone variant TH2B regulates post-meiotic genome-wide histone-to-protamine replacement in male germ cells. Here, Bošković and Torres-Padilla discuss this novel insight into the physiological function for one of the oldest known histone variants.
Keywords: H2AZ, BRDT, male contraception, reprogramming, male infertility, sex chromosome inactivation, histone eviction
Abstract
Producing competent gametes is essential for transmitting genetic information throughout generations. Spermatogenesis is a unique example of rearrangements of genome packaging to ensure fertilization. After meiosis, spermatids undergo drastic morphological changes, perhaps the most dramatic ones occurring in their nuclei, including the transition into a protamine-packaged genome. In this issue of Genes & Development, Montellier and colleagues (pp. 1680–1692) shed new light on the molecular mechanisms regulating this transition by ascribing for the first time a function to a histone variant, TH2B, in the regulation of this process.
The formation of the germline is a tightly regulated process that starts with germ cell specification and includes subsequent reprogramming, meiosis, and differentiation. Out of the 200 different cell types in an adult mammal, sperm packages its genome in the most unique way. The process of spermatogenesis entails global changes in the chromatin structure of the germ cells. During spermatid elongation and condensation, the DNA is stripped of most of its nucleosomal packaging and first becomes wrapped around so-called transition proteins (TPs), which are later replaced by protamines. Protamines are small, highly basic proteins that bind DNA with high affinity and wrap it in a toroidal structure, which is necessary for the high condensation of the genome in the mature sperm (Balhorn 2007). This transition is essential, and defects in this process can lead to infertility. It is also worth noting that although the nucleosome-to-protamine exchange is global, it is not complete. Some regions of the genome remain packaged with histones, with the extent of nucleosome retention differing between species (Wykes and Krawetz 2003; Hammoud et al. 2009; Brykczynska et al. 2010; Erkek et al. 2013). Nucleosomes are particularly enriched in the regulatory regions of genes that are important for the earliest developmental stages post-fertilization (Hammoud et al. 2009). Likewise, some heterochromatic regions, such as centromeres and telomeres, preserve their histones, while the rest of the sperm genome becomes highly compacted (Govin et al. 2007). Despite being at the center of intense research, the molecular mechanisms behind histone-to-protamine replacement remain obscure, making it one of the lasting questions in reproductive and chromatin biology.
Histone variants and subnucleosomal structures
Apart from the canonical histones, which are incorporated into chromatin during DNA synthesis, a number of histone variants have been described in recent decades, providing an additional layer for the regulation of chromatin function. Expression of many histone variants in a highly regulated manner is one of the hallmarks of spermatogenesis (Kimmins and Sassone-Corsi 2005). Many of the variants have been identified in the testis, and some are almost exclusively expressed during male gametogenesis. For instance, H2A.Lap1 (lack of acidic patch) is highly abundant in late pachytene and round spermatids during the time of intensive transcription (Soboleva et al. 2012). During this time, H2A.Lap1 sits on the transcription start site (TSS) of active genes and was suggested to facilitate transcription, probably owing to its biochemical properties, which lead to the formation of less stable nucleosomes in vitro. Together with H2A.Z, another histone variant present at the −2 nucleosome of active genes in round spermatides, H2A.Lap1 creates a unique chromatin environment that is important for activation of late spermatid genes (Soboleva et al. 2012). Furthermore, two other H2A variants—H2AL1 and H2AL2 (Govin et al. 2007)—were identified in the testis and were shown to form structures that protect shorter, subnucleosomal DNA fragments in condensing spermatids. These variants preferentially dimerize with yet another specific germline histone variant, TH2B. When ectopically expressed in somatic cells, H2AL2 and TH2B are able to form canonical nucleosomes, but these are less stable compared with H2A–H2B-containing nucleosomes. However, the role of TH2B in vivo had not been addressed before this study.
TH2B was one of the earliest histone variants identified (Kumaroo et al. 1975; Meistrich et al. 1985), but its physiological role during spermatid maturation has not been investigated. Now, in this issue of Genes & Development, Montellier et al. (2013) document that TH2B is expressed in spermatocytes and spermatids, and its presence inversely correlates with the presence of canonical H2B, suggesting that TH2B gradually replaces H2B on chromatin, making it the first histone variant to be deposited on a genome-wide scale during spermiogenesis.
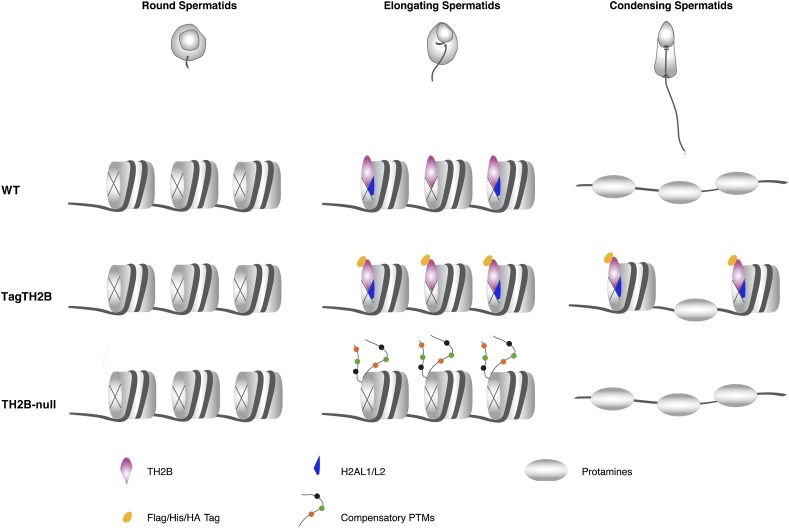
The investigators used two different approaches to address TH2B function using mouse models. In the first one, the addition of a C-terminal tag resulted in a dominant-negative phenotype whereby TH2B remained abnormally bound to the chromatin until later stages of spermatogenesis in elongating spermatids. Second, the investigators generated a phenotypically null TH2B allele. Counterintuitively, the tagged model leads to complete male sterility, whereas the TH2B-null model seems to be compensated through alterations in the post-translational histone modifications of H2B, H3, and H4 (Fig. 1). The molecular and physiological characterization of the two models allowed the investigators to pinpoint the essential role of TH2B in restructuring nucleosomal organization to allow for subsequent replacement by TPs, which are essential for male fertility.
Figure 1.
Changes in histone variant composition allow a smooth transition from a nucleosome-based toward a protamine-packed DNA during spermatogenesis. In elongating spermatids, histone variants are enriched in the sperm chromatin. Among them, TH2B becomes incorporated into chromatin on a global scale, and this precedes histone replacement in condensing spermatids. The addition of a His/Flag/HA tag in the C terminus of TH2B disturbs its replacement and results instead in defective protamine incorporation, which leads to male sterility. In the TH2B-null mouse model, the process of protamine incorporation occurs normally, and TH2B absence is suggested to be balanced through the acquisition of compensatory histone modifications on the core histones H2B, H3, and H4.
Adding a His/Flag/HA C-terminal tag on TH2B interferes with the late stages of spermiogenesis, with severe defects first observed at the time when TPs accumulate in elongating spermatids. The presence of tagged TH2B caused complete male sterility despite proper incorporation of the histone. However, while tagged TH2B incorporation was not affected, it seems that the displacement of tagged TH2B from chromatin was impaired during the transitional stages, when nucleosomes are replaced by TPs and subsequently by protamines. The same group had previously identified the presence of subnucleosomal particles in spermatids, which contain TH2B and H2AL2 but lack H3–H4 tetramers (Govin et al. 2007). The study at hand reports that the presence of a tag on TH2B specifically affects the nature of these particles, as they become more resistant to MNase treatment. Furthermore, the investigators report that TH2B and TPs/protamines, which are not usually present in chromatin at the same time, colocalize in these mutant mice. These findings, together with electron microscopy observations, demonstrate that tagged TH2B germ cells show impaired chromatin compaction. The question remains whether the tag effect is specific for TH2B or could confer similar effects if present on the canonical H2B histone or other histones in general.
On the contrary, analysis of the TH2B-null model shows that TH2B depletion does not cause any defect in producing competent sperm cells, and mice are fully fertile. However, the levels of H2B are significantly increased in the absence of TH2B, suggesting that H2B in surplus can compensate for the lack of TH2B. Strikingly, the “compensatory” H2B is accompanied by the occurrence of differentially post-translationally modified nucleosomes, with different modifications appearing not only in the canonical H2B but also on H3 and H4. This implies that the absence of TH2B leads to a cross-talk of modifications on other histones. In a very simplistic manner of speaking, it seems that in the absence of TH2B, the germline tries to mimic its functional output by modifying histones in a way that will be disruptive to nucleosomal stability. Previous studies of chromatin fibers containing the H2A variant H2A.Lap1 showed that these fibers display folding and sedimentation properties essentially identical to those containing acetylated H3 and H4 nucleosomes (Soboleva et al. 2012). The striking accordance of the H2A.Lap1 and TH2B data in spermatids in regards to destabilizing chromatin structure makes it easy to imagine that histone variant incorporation, now at a global level, directly orchestrates the transitions from nucleosome to protamine genome packaging by regulating chromatin structure.
Chromatin post-translational modifications (PTMs) in post-meiotic germ cells
Core histone acetylation is present at different stages of spermatogenesis, with high levels both preceding and following meiosis and lower levels during the reductive division itself. Histone H4 acetylation patterns in mouse spermiogenesis show a stage-specific pattern, differing between round and elongating spermatids and disappearing in condensing spermatids prior to protamine incorporation (Hazzouri et al. 2000). Interestingly, transcription does not occur during the elongation process of spermatid maturation, suggesting that core histone acetylation in these cells is not related to gene expression. Indeed, histone hyperacetylation is often observed before histone replacement in a number of species, including flies, roosters, and rats (Grimes and Smart 1985; Oliva et al. 1987; Jayaramaiah Raja and Renkawitz-Pohl 2005). These findings led to the idea that global chromatin hyperacetylation functions as a signal for the eviction of histones to allow for tight genome packaging by protamines. Furthermore, indirect evidence of the importance of hyperacetylation in the process of chromatin remodeling in elongating and condensing spermatids emerged from the observation that species that retain their histones throughout spermiogenesis also keep them in a hypoacetylated state (Kennedy and Davies 1981).
The presence of acetylated histones in late stages of spermatogenesis led to the discovery of the double bromodomain-containing protein BRDT, which subsequently proved to be an important regulatory player in both premeiotic and post-meiotic stages of germ cell development. BRDT specifically recognizes acetylated H4 and is able to induce acetylation-dependent chromatin compaction in somatic cells and in vitro (Pivot-Pajot et al. 2003). Further investigation of BRDT function in post-meiotic germ cells showed that its first bromodomain (BD1) binds double-acetylated H4 (K5ac and K8ac) and directs genome-wide histone replacement (Moriniere et al. 2009; Gaucher et al. 2012). The mechanism of BRDT action seems to entail recognition of acetylated chromatin via BD1 in elongating spermatids, followed by a process the investigators termed “chromatin squeezing” through interaction of two (or more) BRDT molecules, which would facilitate histone eviction. A similar observation was also reported in rat spermatids (Dhar et al. 2012), but an indirect effect of Brdt on histone eviction cannot be excluded.
In their present study, Montellier et al. (2013) report that in the absence of TH2B, core histones exhibit differential PTMs. Namely, histone H3K122 and H4K77 crotonylation were more abundant, and the position of these residues relative to the DNA backbone suggests a potential disruptive effect on nucleosomal structure. Crotonylation, like acetylation, confers a negative charge to the lysine; however, crotonyl groups are bulkier than acetyl groups and potentially cause even more steric hindrance between histones and DNA (Tan et al. 2011). How this modification exhibits its function during spermiogenesis and whether there are specific readers of histone crotonylation present in maturing germ cells remain to be investigated. Also, whether BRDT can read crotonylated residues in the absence of TH2B and whether this compensates for histone replacement remain to be addressed. Importantly, the present study also underlines the importance of both the PTM charge and its position on nucleosomal properties, with the attention shifting from protruding histone tails to their buried globular domains (Tropberger et al. 2013).
Although acetylation is the most widely investigated histone PTM in spermiogenesis, the importance of other regulatory pathways, such as histone ubiquitylation, is now coming to light. The RING domain-containing E3 ubiquitin ligase RNF8 was discovered to be an important regulator of spermiogenesis. RNF8-deficient mice showed problems in DNA condensation and histone eviction during spermatid maturation without defects in TP and protamine expression (Lu et al. 2010). Dissection of the molecular mechanism of RNF8 action revealed that RNF8 ubiquitylates H2A and H2B, which in turn stabilizes MOF association with chromatin. MOF is a major H4K16 acetyltransferase, and H4K16ac is one of the first triggers of global histone eviction in elongating spermatids. Likewise, male mice lacking the ubiquitin-conjugating enzyme HR6B (homolog of yeast RAD6) are infertile and show defects at later stages of spermatogenesis. However, spermiogenesis in Hr6b−/− mice is not completely blocked. Instead, defective spermatid elongation results in a high rate of apoptosis in germ cells. It is possible to imagine that in the absence of ubiquitin-conjugating enzymes, histones are not efficiently degraded upon replacement, leading to phenotypic abnormalities in subsequent steps of spermiogenesis (Roest et al. 1996). Whether the absence of TH2B promotes compensatory mechanisms through ubiquitylation remains to be addressed.
Unexpectedly maternal: testis-specific variant on maternal chromatin
The epigenetic contribution of the sperm after fertilization has been highly debated due to the rapid protamine-to-nucleosome exchange in the earliest zygotic stages and the relatively low levels of histones that remain on the mature sperm. Indeed, most of the histones that assemble into the paternal pronucleus are of maternal origin. Here, the Kochbin team (Montellier et al. 2013). demonstrates that TH2B, previously thought to be testis-specific, is also present in the maternal chromatin prior to fertilization and that it is incorporated into the male pronucleus of the zygote rapidly thereafter. It is known that the embryonic genome undergoes extensive reprogramming during the earliest developmental stages, which presumably entails a great deal of chromatin plasticity. Taken together with the putative role of TH2B in creating a dynamic and plastic chromatin, an attractive hypothesis for the analogous function of TH2B during the remodeling process in the zygote arises. Indeed, it is possible to envisage that TH2B in the embryo serves to facilitate the opening and resetting of embryonic chromatin for subsequent development. Undoubtedly, future investigations of the TH2B histone variant in early embryogenesis will shed more light into the molecular mechanisms of its function during genome-wide epigenetic reprogramming.
Concluding remarks
Fine-tuned regulation of gametogenesis seems to involve a variety of functional interactions between different histone variants. The work of Montellier et al. (2013) shows that TH2B is depleted in the two nucleosomes that surround TSSs and are known to be enriched in another testis-specific variant, H2A.Lap1 (Soboleva et al. 2012). Similarly, the sex chromosome-specific replacement of H3 for H3.3 provides another example of regulated chromatin structure in spermatogenesis (van der Heijden et al. 2007). Thus, it seems that the process of spermatogenesis uses an increasing number of histone variants to impose the functionality of the genome at each stage of gametogenesis, which perhaps could explain why there are so many testis-specific variants described to date (Boussouar et al. 2008). Mammalian spermatogenesis is an example of orchestration of myriad cellular processes all directed toward creating cells that are able to leave the host organism to create a new life. The morphological changes after the formation of spermatids and their elongation, cytoplasmic eviction, and acrosome formation are paralleled by remarkable nuclear rearrangements resulting in a genome packaging unlike any other. It is perhaps due to such exquisite timing and variety of players—from chromatin remodelers to histone modifications—governing spermiogenesis that this process still evokes many open questions. Now, the work by Montellier et al. (2013) contributes significantly to our understanding of the highly precise molecular events necessary for the formation of competent spermatozoa. Most interestingly, this study underscores the plasticity and resourcefulness of germ cells, which employ available backup mechanisms such as programmed nucleosome disruption by histone PTMs in the absence of the factor of choice. As the formation of mature and competent sperm cells is essential for species propagation, perhaps it should not come as a surprise that evolution installed fail-safe checkpoints to ensure that spermatogenesis reaches completion with the highest efficiency possible.
Acknowledgments
We thank S. Bour for graphic work, and A. Burton for critical reading of the manuscript. Work in the Torres-Padilla laboratory is funded by ANR-09-Blanc-0114, Epigenesys NoE, ERC-2011-StG 280840 “NuclearPotency,” and Marie Curie FP7 ITN “Nucleosome4D.”
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226167.113.
References
- Balhorn R 2007. The protamine family of sperm nuclear proteins. Genome Biol 8: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussouar F, Rousseaux S, Khochbin S 2008. A new insight into male genome reprogramming by histone variants and histone code. Cell Cycle 7: 3499–3502 [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17: 679–687 [DOI] [PubMed] [Google Scholar]
- Dhar S, Thota A, Rao MR 2012. Insights into role of bromodomain, testis-specific (Brdt) in acetylated histone H4-dependent chromatin remodeling in mammalian spermiogenesis. J Biol Chem 287: 6387–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schubeler D, Vlag J, Stadler MB, Peters AH 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 20: 868–875 [DOI] [PubMed] [Google Scholar]
- Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte AL, et al. 2012. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J 31: 3809–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. 2007. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol 176: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes SR Jr, Smart PG 1985. Changes in the structural organization of chromatin during spermatogenesis in the rat. Biochim Biophys Acta 824: 128–139 [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S 2000. Regulated hyperacetylation of core histones during mouse spermatogenesis: Involvement of histone deacetylases. Eur J Cell Biol 79: 950–960 [DOI] [PubMed] [Google Scholar]
- Jayaramaiah Raja S, Renkawitz-Pohl R 2005. Replacement by Drosophila melanogaster protamines and Mst77F of histones during chromatin condensation in late spermatids and role of sesame in the removal of these proteins from the male pronucleus. Mol Cell Biol 25: 6165–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BP, Davies PL 1981. Phosphorylation of a group of high molecular weight basic nuclear proteins during spermatogenesis in the winter flounder. J Biol Chem 256: 9254–9259 [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P 2005. Chromatin remodelling and epigenetic features of germ cells. Nature 434: 583–589 [DOI] [PubMed] [Google Scholar]
- Kumaroo KK, Jahnke G, Irvin JL 1975. Changes in basic chromosomal proteins during spermatogenesis in the mature rat. Arch Biochem Biophys 168: 413–424 [DOI] [PubMed] [Google Scholar]
- Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X 2010. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 18: 371–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich ML, Bucci LR, Trostle-Weige PK, Brock WA 1985. Histone variants in rat spermatogonia and primary spermatocytes. Dev Biol 112: 230–240 [DOI] [PubMed] [Google Scholar]
- Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Héry P, Curtet S, et al. 2013. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev (this issue). doi: 10.1101/gad.220095.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461: 664–668 [DOI] [PubMed] [Google Scholar]
- Oliva R, Bazett-Jones D, Mezquita C, Dixon GH 1987. Factors affecting nucleosome disassembly by protamines in vitro. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J Biol Chem 262: 17016–17025 [PubMed] [Google Scholar]
- Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S 2003. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol 23: 5354–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roest HP, van Klaveren J, de Wit J, van Gurp CG, Koken MH, Vermey M, van Roijen JH, Hoogerbrugge JW, Vreeburg JT, Baarends WM, et al. 1996. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 86: 799–810 [DOI] [PubMed] [Google Scholar]
- Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, Tremethick DJ 2012. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol 19: 25–30 [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Buhler M, et al. 2013. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152: 859–872 [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P 2007. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 39: 251–258 [DOI] [PubMed] [Google Scholar]
- Wykes SM, Krawetz SA 2003. The structural organization of sperm chromatin. J Biol Chem 278: 29471–29477 [DOI] [PubMed] [Google Scholar]