The conversion of male germ cell chromatin to a nucleoprotamine structure is fundamental to the life cycle. Montellier et al. find that the histone H2B variant TH2B directs the final transformation of dissociating nucleosomes into protamine-packed structures. TH2B loss up-regulates H2B and induces nucleosome instability through targeted histone modifications. After fertilization, TH2B reassembles onto the male genome during protamine-to-histone exchange. Therefore, TH2B plays a key role in the histone-to-protamine packing of the male genome.
Keywords: H2AZ, BRDT, male contraception, reprogramming, male infertility, sex chromosome inactivation, histone eviction
Abstract
The conversion of male germ cell chromatin to a nucleoprotamine structure is fundamental to the life cycle, yet the underlying molecular details remain obscure. Here we show that an essential step is the genome-wide incorporation of TH2B, a histone H2B variant of hitherto unknown function. Using mouse models in which TH2B is depleted or C-terminally modified, we show that TH2B directs the final transformation of dissociating nucleosomes into protamine-packed structures. Depletion of TH2B induces compensatory mechanisms that permit histone removal by up-regulating H2B and programming nucleosome instability through targeted histone modifications, including lysine crotonylation and arginine methylation. Furthermore, after fertilization, TH2B reassembles onto the male genome during protamine-to-histone exchange. Thus, TH2B is a unique histone variant that plays a key role in the histone-to-protamine packing of the male genome and guides genome-wide chromatin transitions that both precede and follow transmission of the male genome to the egg.
An essential feature of spermatogenesis is the generation of a transportable genome placed in the nucleus of highly specialized cells, spermatozoa, capable of leaving and surviving the parent organism. After meiosis, young spermatids (known as round spermatids) inherit a chromatin-containing transcriptionally active genome that, during subsequent stages in elongating spermatids, undergoes a genome-wide histone hyperacetylation followed by histone removal and the assembly of transition proteins (TPs) and protamines in condensing spermatids. These chromatin transitions constitute a unique feature among eukaryotes, since the universal nucleosome-based organization of the genome undergoes a metamorphosis into new and unique genome-packaging structures based on nonhistone proteins. Although essential to the life cycle, the molecular basis of these dramatic changes remains one of the most obscure issues in modern biology (Boussouar et al. 2008; Gaucher et al. 2010). Following the commitment of self-renewing proliferative spermatogonia to meiotic divisions, all of the specific drivers of meiotic and post-meiotic events are expressed through successive waves of gene activation in spermatocytes (meiotic cells) and spermatids (Gaucher et al. 2012). Actually, the gradual male genome programming has, by this stage of development, already undergone large-scale exchanges of histones in spermatocytes. De Boer's group (van der Heijden et al. 2007) discovered that the transcriptional inactivation of meiotic sex chromosomes, which occurs at the time of autosomal chromosome pairing (Montellier et al. 2012), is associated with the chromosome-wide replacement of H3.1/2 by H3.3. In spermatids, histone removal is preceded by a large-scale incorporation of new H2A variants, H2AL1/2, which are synthesized almost at the same time as TPs (Govin et al. 2007). Other studies have also shown more targeted incorporations of H2A variants such as H2A.Z and H2A.Lap1 occurring at earlier stages (Greaves et al. 2006; Soboleva et al. 2012). These observations raise two fundamental issues. First, what are the molecular mechanisms underlying such large-scale chromatin dynamics? More specifically, which molecular determinants are responsible for the partial or total disassembly of nucleosomes and for the subsequent incorporation of new histones, TPs, and protamines into chromatin? Second, what are the functions of the histone variants synthesized and incorporated into chromatin at the different stages of spermatogenic cell differentiation?
The testis-specific histone variant TH2B, or TSH2B.1 in the new unified nomenclature (Talbert et al. 2012), was discovered in mammalian testes histone extracts in 1975 and is one of the earliest histone variants to have been identified (Branson et al. 1975; Shires et al. 1975). Later studies showed that its synthesis occurs in a replication-independent manner, beginning in early spermatocytes (Brock et al. 1980). The generation of TH2B-recognizing antibodies allowed a better characterization of its expression in spermatogenic cells (for review, see Govin et al. 2004), while in vitro studies revealed some of its biochemical properties (Li et al. 2005). However, the role of TH2B in the physiological context of spermatogenic cell differentiation still remains obscure. In particular, no gene invalidation approach has yet been undertaken to probe TH2B function. This is probably due to the complex structure of the gene, whose regulatory elements are shared with a neighboring Th2a gene (Supplemental Fig. S1; Huh et al. 1991; Choi and Chae 1993), making attempts to specifically abolish Th2b gene expression without deregulating Th2a challenging.
Here, we describe the genome-wide removal of H2B, which begins in early spermatocytes, and its replacement by TH2B, demonstrating that TH2B acts on a much larger scale than any of the other known histone variants. In addition, we investigate the functions of this histone variant in spermatogenic cells. Using a knock-in approach, we added three consecutive affinity tags to the C terminus of TH2B and generated a mouse strain expressing the tagged variant in spermatogenic cells. We also generated mice where the expression of TH2B was abrogated. Overall, the results obtained by using these mouse models strongly suggest that TH2B sets nucleosome stability parameters ensuring a genome-wide transition of nucleosomes into intermediate structural entities, which in turn are required for the assembly of TPs and protamines. This study also highlights a role for TH2B in coordinating somatic-type H2B gene expression and fine-tuning histone post-translational modifications (PTMs). Additionally, we show that TH2B is also maternally expressed and replaces protamines at fertilization. Taken together, these data considerably increase our understanding of the molecular basis of male genome-wide chromatin disassembly and reassembly while assigning a function to TH2B nearly four decades after its discovery.
Results
Genome-wide replacement of H2B by TH2B
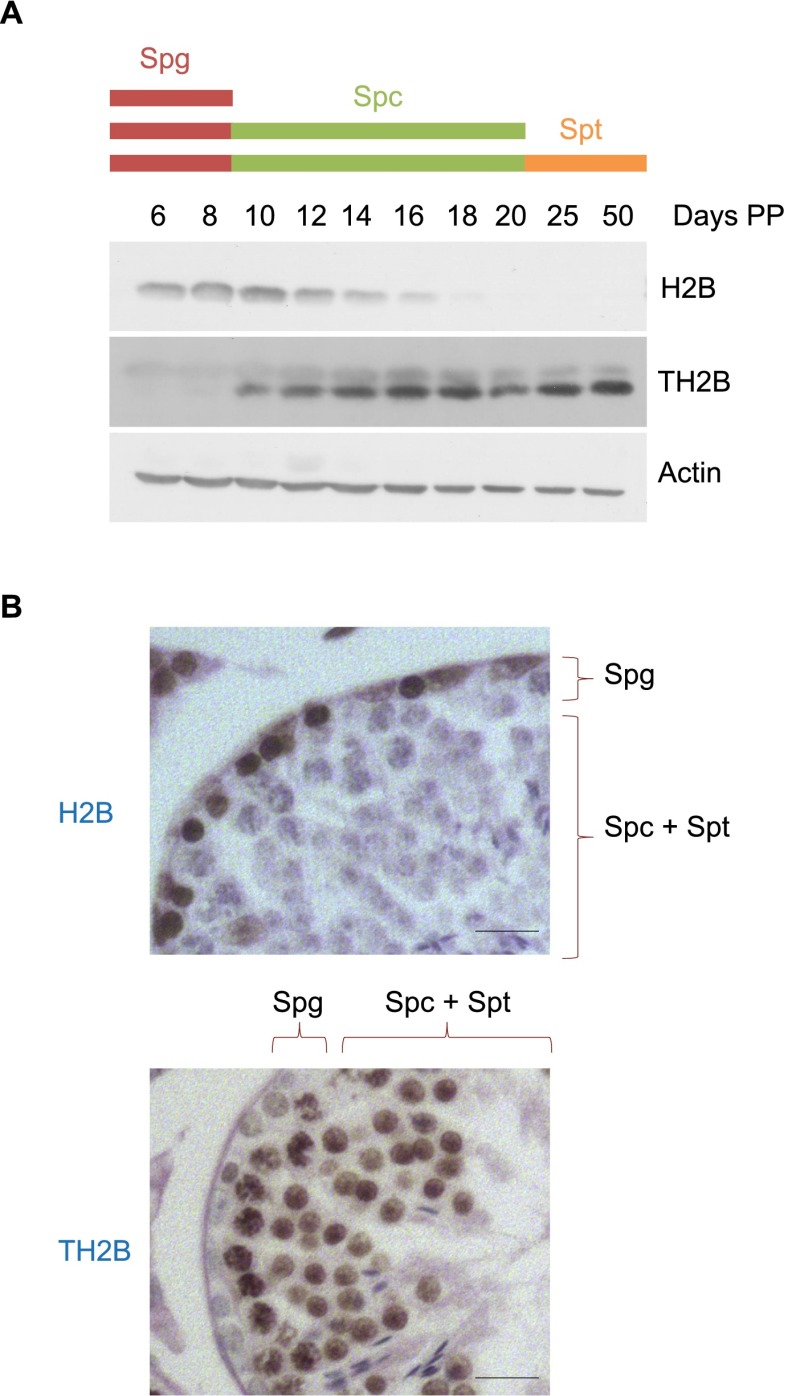
To investigate the function of TH2B, we examined the timing of TH2B protein expression in the developing testis of postnatal mice during the first wave of spermatogenesis. Samples were analyzed at time points corresponding to the appearance of successive spermatogenic cell types. Commitment to meiotic divisions occurs at 10 d post-partum (dpp) when spermatogonial divisions give rise to preleptotene spermatocytes. Spermatogenesis continues with spermatocytes undergoing meiotic divisions followed by the generation of post-meiotic haploid cells, which first appear at 20 dpp. The 50-dpp testis encompasses all spermatogenic cell types, including mature spermatozoa. Western blot analysis shows that TH2B starts to accumulate at ∼10 dpp, when pre-leptotene/leptotene spermatocytes first appear (Fig. 1A). Monitoring H2B expression in the same experiment revealed that a drastic decrease of H2B mirrors the accumulation of TH2B, with the latter largely replacing H2B by 18 dpp in spermatocytes. These findings were corroborated by immunohistochemistry of adult testis sections, confirming that H2B is expressed in spermatogonia and nearly depleted from spermatocytes and spermatids, which instead stain strongly for the presence of TH2B (Fig. 1B).
Figure 1.
A major H2B-to-TH2B transition occurs in early spermatocytes. (A) TH2B and H2B accumulation was analyzed in testes extracts at the indicated times (in days post-partum [Days PP]) by Western blots. (B) The expression of TH2B and H2B was analyzed by immunohistochemistry. Testis sections at stage II/III are represented. Spermatogonia (Spg) and spermatocytes and spermatids (Spc + Spt) are indicated. Bars, 10 μm.
A C-terminal affinity tag on TH2B causes spermatid differentiation to abort
To further explore TH2B function, the Th2b gene was tagged in embryonic stem (ES) cells following a knock-in strategy based on homologous recombination, and the corresponding mouse strain was generated. Heterozygous Th2b+/tag mice expressed the Th2b tagged allele (TH2B C-terminally fused to three consecutive affinity tags: His, Flag, and Ha) (Supplemental Fig. S1) in the same spermatogenic cells as Th2b+/+ mice and at the expected timing, showing a pattern of genome association very similar to that of wild-type TH2B (Fig. 2). In meiotic cells, both TH2B and TH2B-tag associated homogeneously with the genome, and meiotic events occurred normally. These included chromosome pairing, visualized by immunodetection of synaptonemal complexes (Sycp3) (Fig. 2B); sex body formation, visualized by the accumulation of γH2A.X on the X and Y chromosomes (Fig. 2C); and the sex chromosome-specific H3-to-H3.3 histone exchange (van der Heijden et al. 2007), as judged by the accumulation of H3.3 on the sex body (Fig. 2D). In spermatids, wild-type TH2B and the tagged protein also exhibited a highly similar appearance, with an enhanced polar localization in round spermatids (Fig. 2A).
Figure 2.
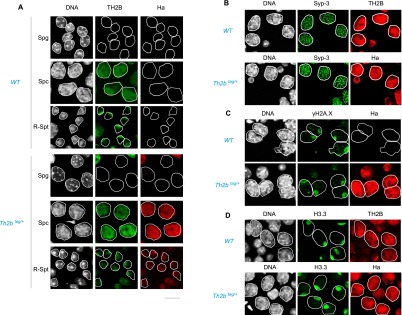
TH2B and TH2B-tag show similar intracellular distribution in wild-type and TH2B-tag-expressing spermatogenic cells. (A) Codetection of TH2B and TH2B-tag was performed by immunofluorescence on seminiferous tubule preparations from testes from the indicated genotypes using anti-TH2B or anti-Ha antibodies. The stages considered are indicated at the left of each panel: spermatogonia (Spg), pachytene spermatocytes (Spc) and round spermatids (R-Spt). (B) Sycp3 was codetected with TH2B (top panel) or TH2B-tag (anti-Ha, bottom panel) in spermatocytes of the indicated genotypes. (C) Codetection of TH2B-tag (anti-Ha panels) and γH2A.X in spermatocytes with the indicated genotypes is shown. (D) H3.3 was codetected with TH2B or TH2B-tag in spermatocytes of the indicated genotypes. Please note that the immunodetection with anti-Ha antibody is more sensitive than the detection based on the anti-TH2B antibody.
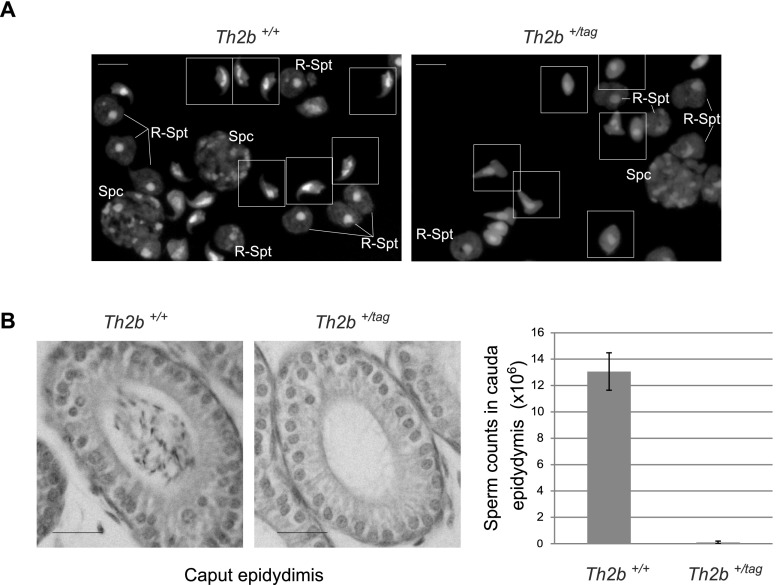
These data show that the C-terminally tagged TH2B does not interfere with any of the finely tuned events requiring dynamic chromatin, including meiotic recombination, sex chromosome inactivation and associated H2A.X phosphorylation, and the chromosome-wide H3-to-H3.3 exchange on the sex chromosome of meiotic cells. In striking contrast, at subsequent post-meiotic stages, TH2B-tag expression resulted in abnormalities occurring in elongating spermatids and a total absence of cells at later stages (Figs. 3A, 5B,C [below]; Supplemental Fig. S4). These defects lead to a total absence of epididymal spermatozoa (Fig. 3B) and complete male sterility (Supplemental Fig. S6B). Thus, C-terminally tagged TH2B causes no obvious phenotypical defects at the early stages of spermatogenesis but leads to the arrest of sperm cell development in condensing spermatids.
Figure 3.
TH2B-tag induces late arrest of spermatogenesis. (A) Different cell types stained with DAPI from the indicated genotypes are shown. Bars, 5 μm. (B) Cauda epididymis sections stained with hematoxylin are shown in the left panel, and spermatozoa counts from isolated cauda epididymis from mice with the indicated genotypes are presented as histograms in the right panel. Bars, 20 μm. Bars represent standard deviations of sperm counts from cauda epididymis of five mice of each genotype.
Figure 5.
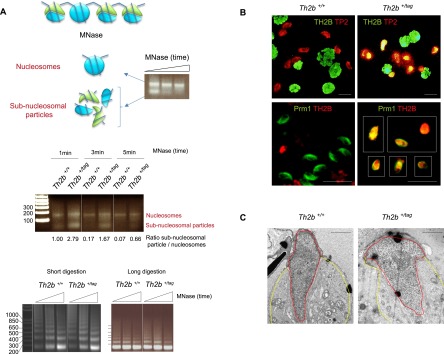
TH2B-tag affects subnucleosomal transitional states in elongating spermatids during histone replacement. (A, top panel) Nuclei isolated from elongated/condensing wild-type spermatids were digested with MNase for increasing lengths of time to release both nucleosomal MNase-resistant regions and MNase-sensitive subnucleosomal particles (histone and nonhistone proteins are represented in blue and green, respectively). (Middle panel) Nuclei from wild-type elongated/condensing spermatids or the corresponding cells isolated from TH2B-tag-expressing spermatogenic cells were extensively digested by MNase as above. The agarose gel shows nucleosomal and subnucleosomal DNA fragments released from the two cell types by MNase digestion during the indicated times. The normalized ratios of intensity of subnucleosomal particles to nucleosomes obtained with ImageJ software are indicated below each lane. This ratio was set to 1 for Th2b+/+ after 1 min of MNase digestion and was used to normalize the other values. The bottom panels show chromatin digestion by MNase of suspensions of total spermatogenic cells from wild-type and TH2B-tag mouse testes. (B) Codetection of TH2B (green) and TP2 (red) (top panel) and of Th2B (red) and protamine 1 (Prm1, green) (bottom panel) were performed on spermatogenic cell preparations from wild-type or Th2b+/tag testes by immunofluorescence using the corresponding antibodies. Bars, 10 μm. (C) Electron micrographs show representative spermatids of both genotypes. Nucleus and cytoplasm are delimited by red and yellow lines, respectively. Bars, 1 μm.
TH2B-tag is efficiently assembled into nucleosomes in meiotic and post-meiotic cells
The data described above suggest that the C-terminal tag interferes with a critical TH2B function in elongating spermatids. Since there is almost no transcription in these cells (Zhao et al. 2004), we hypothesized that the tag may perturb a distinct chromatin-related event such as the genome-wide histone replacement that occurs in elongating/condensing spermatids. To check whether the C-terminal tag could interfere with earlier events such as the assembly of TH2B or of other histones into nucleosomes, we generated mononucleosome-enriched chromatin from spermatocytes and round spermatids and used an anti-Ha antibody to immunopurify TH2B-tag-containing nucleosomes. Individual histones were then resolved by SDS-PAGE (Fig. 4A) and subjected to tryptic digestion and high-performance liquid chromatography/tandem mass spectrometry (HPLC/MS/MS) analysis. Using this approach, we confirmed that all core histones (H2A, H2B, H3, and H4) as well as linker histones were associated with TH2B-tag-containing nucleosomes. The H2B detected here probably originates from early spermatocytes during the course of H2B-to-TH2B exchange at a stage where “mixed” nucleosomes are present. Histone variants normally expressed in spermatogenic cells—including H3.3, TH3, H2A.Z, H2A.X, and macroH2A—were also identified (Fig. 4B; summarized in Supplemental Tables S1, S2; see also Supplemental Fig. S2). Thus, the C-terminal tag on TH2B does not obviously interfere with proper nucleosome assembly.
Figure 4.
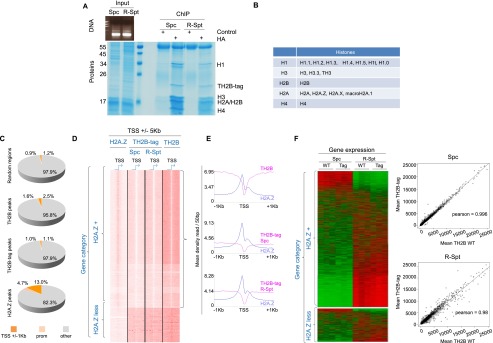
TH2B-tag is assembled into nucleosomes in spermatocytes and round spermatids and does not affect any of the fine-tuned chromatin activities. (A) Chromatin from spermatocytes (Spc) and round spermatids (R-Spt) was extensively digested with MNase (DNA gel) and subjected to immunoprecipitation using an anti-Ha antibody. The materials obtained from ChIP with anti-Ha or an irrelevant antibody were visualized after SDS-PAGE followed by Coomassie staining. The position of histones identified by MS is indicated. (B) The different variants found associated with TH2B nucleosomes are indicated (see also Supplemental Table S1). All of the unique peptides identified corresponding to each of the linker histone types are shown in Supplemental Table S2. (C) Mononucleosomes from cells isolated from Th2b+/tag (spermatocytes and round spermatids) or Th2b+/+ (total spermatogenic cell suspension) mouse testes were subjected to ChIP with anti-Ha and anti-TH2B antibodies, respectively, followed by DNA sequencing (ChIP-seq). The respective proportions of TH2B-tag and TH2B peaks (this experiment) distributed on gene regulatory regions (color-coded), including TSSs (regions covering TSSs ±1 kb) and promoter regions (from TSSs to 5 kb upstream), were compared with that of H2A.Z peaks (from spermatogenic cells, GSE29913) as well as with “random” genomic localizations (using a set of 75,000 200-bp segment regions randomly selected from the input raw read files). (D) SeqMiner software (Ye et al. 2011) was used to compare TH2B-tag's, TH2B's, and H2A.Z's respective distribution around the TSSs (±1 kb) of the same genes in the indicated spermatogenic cell populations ([Spc] spermatocytes; [R-Spt] spermatids) for the TH2B-tag and total spermatogenic cell population for TH2B. The genes were classified as either H2A.Z-positive (H2A.Z+) or H2A.Z-negative (H2A.Z-less). (E) Metagene analysis of TH2B-tag (in spermatocytes [Spc] and spermatids [R-Spt]) and TH2B (in total wild-type spermatogenic cells) together with H2A.Z distribution centered around the TSSs of the H2A.Z+ gene group. (F, left panel) Whole-genome gene expression pattern of Th2b+/+ (WT) and Th2b+/tag (Tag) spermatocytes (Spc) and round spermatids (R-Spt) are shown as heat maps for the H2A.Z+ and H2A.Z-less gene classes. (Right panel) Pearson correlation plots between individual gene expression in spermatocytes (Spc) and round spermatids (R-Spc), wild-type or expressing TH2B-tag, are shown.
TH2B-tag is assembled genome-wide
Next, we asked whether a biased TH2B-tag chromatin assembly and a subsequent defective gene expression could account for the defective histone-to-protamine replacement. To address this, we isolated mononucleosomes from TH2B-tag-expressing spermatocytes and round spermatids as described above and performed chromatin immunoprecipitation sequencing (ChIP-seq) analyses. In parallel, whole-genome transcriptome profiling was obtained from the same cell fractions comparing wild-type or TH2B-tag-expressing cells. The recent mapping of H2A.Z from spermatogenic cells (Soboleva et al. 2012) provided a useful reference for this analysis. In contrast to H2A.Z, TH2B showed no bias toward gene regulatory elements (Fig. 4C), which is consistent with a genome-wide action of TH2B in replacing H2B (Fig. 1), as opposed to gene regulatory functions. Moreover, TH2B-tag was depleted from the transcriptional start sites (TSSs) of a large group of genes. Specifically, this depletion was observed from the TSSs of genes showing H2A.Z enrichment (Fig. 4D). This anti-correlation between TH2B and H2A.Z at gene TSSs was observed in independent experiments performed in two different laboratories (in France and Australia), confirming the robustness of our mapping approach. Interestingly, Tremethick and colleagues (Soboleva et al. 2012) recently found that H2A histone variants H2A.Z and H2A.Lap1 form two adjacent nucleosomes located 200 base pairs (bp) upstream of and at the TSSs, respectively. We observed that these same regions were depleted in TH2B (Fig. 4E). Remarkably, TH2B was not depleted from TSS regions not enriched in H2A.Z (hereafter named “H2A.Z-less”) (Fig. 4D). Finally, we also performed ChIP-seq using wild-type spermatogenic cells and an anti-TH2B antibody. Comparison of the mapping of TH2B-tag with that of TH2B in wild-type cells showed a large overlap as well as a similar depletion of TH2B from H2A.Z-enriched TSSs (Fig. 4C,D). These data therefore confirm that the presence of the tag does not affect either the assembly of TH2B or its functional dynamics in chromatin. It is noteworthy that, although TH2B is excluded from H2A.Z-containing TSSs, the two variants should be able to coexist in other genomic regions, since our proteomic approach revealed the presence of H2A.Z in TH2B-containing nucleosomes (Fig. 4B).
Whole-transcriptome analysis of spermatocytes and round spermatids from wild-type or TH2B-tag-expressing mice demonstrated that the presence of TH2B-tag had no significant effect on gene expression (Fig. 4F). We also observed that, overall, genes bearing H2A.Z at their TSSs had higher transcriptional activity than H2A.Z-less genes in both spermatocytes and round spermatids (Supplemental Fig. S3A). Taken together, these results show that first, TH2B is a histone variant with genome-wide rather than locus-specific activity, and second, the C-terminal tag has no obvious effect on chromatin-dependent regulatory events in spermatocytes or round spermatids. The effect of the tag on TH2B turnover was also measured, re-enforcing this conclusion. Somatic cells stably expressing GFP-TH2B or GFP-TH2B-tag were generated and used in a FRAP (fluorescence recovery after photobleaching) experiment to compare the dynamics of both fluorescent histones. This approach did not reveal any significant effect of the tag on the turnover of TH2B (Supplemental Fig. S3B).
TH2B directs a stepwise nucleosome-to-nucleoprotamine transition
The above data suggest that the C-terminal TH2B tag does not affect nucleosome dynamics per se, since events involving nucleosome disassembly and reassembly, such as the H3-to-H3.3 transition in meiotic cells, occur normally. In contrast, the presence of the tag might disrupt certain chromatin states specific to elongated/condensing spermatids; i.e., transitional states linking the dissociating nucleosomes to the assembly of TPs and protamines. Accordingly, we investigated the DNA-packaging transitional states known to occur in condensing spermatids (Govin et al. 2007). In agreement with previous findings, extensive micrococcal nuclease (MNase) treatment of nuclei purified from elongated/condensing spermatids resulted in the isolation of MNase-resistant nucleosomal DNA as well as more sensitive subnucleosomal DNA fragments (Fig. 5A). The subnucleosome particles, previously shown to be devoid of H3 and H4 and to contain TH2B and the late-expressing H2A variant H2AL2, are thought to be transitional states preceding the complete replacement of histones by TPs (Govin et al. 2007). We monitored the time course of MNase-induced release of nucleosomal and subnucleosomal fragments in wild-type and TH2B-tag-expressing elongated/condensing spermatids. While in both wild-type and TH2B-tag-expressing cells extensive MNase digestion released the two expected nucleosomal and subnucleosomal fragments, we observed that first, slightly higher amounts of mononucleosomes were consistently obtained in TH2B-tag-expressing cells compared with wild-type cells, and second, the subnucleosomal particles from these cells were more resistant to MNase digestion than those from wild-type cells, where these particles were rapidly digested (Fig. 5A). In contrast, when the MNase sensitivity of chromatin-containing total spermatogenic cell nuclei was compared between wild-type and TH2B-tag-expressing cells, no significant difference was found (Fig. 5A, bottom panel). Altogether, these experiments suggest that the tag-dependent defects specifically occur at a time when TP and protamines should fully displace TH2B.
In support of this hypothesis, TPs and protamines are coimmunodetected with TH2B in elongating/condensing spermatids, showing that TH2B is not replaced by either TP or protamines in TH2B-tag-expressing mice. Indeed, although both nonhistone proteins are synthesized and imported into the nucleus of TH2B-tag-expressing spermatids, TH2B remains present in the same nucleus (Fig. 5B; Supplemental Fig. S4). In striking contrast, in wild-type spermatids, histones disappear when TPs and protamines accumulate (Fig. 5B; Supplemental Fig. S4, Th2b+/+ cells). Furthermore, electron microscopy shows that chromatin compaction is impaired in TH2B-tag-expressing spermatids, suggesting that TPs and protamines are unable to condense chromatin in these cells (Fig. 5C). These data suggest that the presence of the tag affects the properties of TH2B that are specifically required at the time of histone-to-TP transition.
The complete depletion of TH2B induces enhanced H2B expression and restores normal sperm cell development
The presence of the neo cassette downstream from the Th2b-tag gene (Supplemental Fig. S1) severely interferes with Th2b gene expression and generates a phenotypically null Th2b allele. Triton acid urea (TAU) gel separation of H2B and TH2B followed by immunodetection (Fig. 6A) and highly sensitive LC-MS analysis of histones extracted from testes (Fig. 6B; Contrepois et al. 2010) demonstrated a total absence of TH2B. Furthermore, both approaches showed that H2B expression is enhanced in the absence of TH2B (Fig. 6A,B). The profiling of histones from wild-type and TH2B-less testes by ultrahigh-performance LC-MS (UHPLC-MS) allowed us to define which isoforms of H2B were up-regulated (Supplemental Fig. S5A). Six H2B isoforms were readily identified (Supplemental Fig. S5A; Supplemental Table S3), whereas others, such as 1-H/1-K/2-B, could not be distinguished due to their high sequence identity. As shown in Figure 6B and Supplemental Figure S5A, the peak corresponding to TH2B is the most intense peak in the spectrum obtained from wild-type cells, while it completely disappears from TH2B-less cells. To better visualize the cross-talk between TH2B and H2B isoforms, the peak heights corresponding to these isoforms from Supplemental Figure S5A were normalized relative to the peak heights of the major H4 forms (unacetylated H4 + monoacetylated H4) whose total abundance is expected to remain constant (Supplemental Fig. S5B). As a control, we determined the ratio of histone H2A isoforms 2-A to histone H4, which remained unchanged between the two samples. Most histone H2B isoforms exhibited a twofold to fourfold increase in TH2B-less spermatogenic cells compared with wild-type cells, except for histone H2B isoforms 1-P, which showed an approximately sevenfold increase (Supplemental Fig. S5B).
Figure 6.
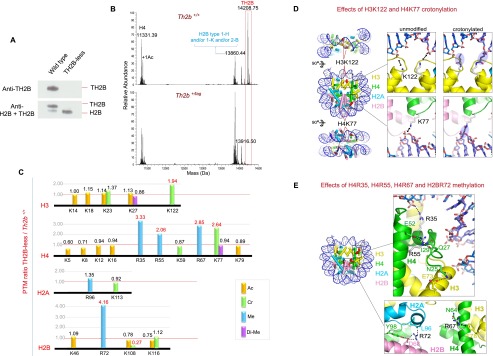
A highly specific compensation mechanism is activated in the absence of TH2B. (A) Histone extracts from wild-type and TH2B-less spermatogenic cells were subjected to electrophoresis on TAU gels to allow separation of TH2B and H2B and were subsequently used for the immunodetection of the two histones by Western blotting with a specific anti-TH2B antibody (top panel) and an antibody recognizing both TH2B and H2B (bottom panel). (B) Profiling of histones from wild-type and TH2B-less testes was performed by UHPLC-MS. The figure shows the deconvoluted electrospray mass spectra of H4, TH2B, and H2B (see Supplemental Fig. S5A,B for additional information). (C) Histones from wild-type and TH2B-less testes were subjected to in vitro isotopic labeling followed by HPLC/MS/MS analysis of histone peptides. The relative abundance of the identified histone PTMs is expressed as TH2B-less/wild-type ratio (for details see Supplemental Table S4; Supplemental Fig. S5D). Lysine acetylation (Ac), crotonylation (Cr), and dimethylation (Di-Me) and arginine monomethylation (Me) are color-coded as indicated. (D) Lysine residues exhibiting enhanced crotonylation. (Left) Nucleosome core particle (Protein Data Bank [PDB] code 3AZH). Histones H2A, H2B, H3, and H4 are in cyan, pink, yellow, and green, respectively; DNA is in blue. Modified lysines are shown as red spheres. The dyad axis is vertical in the middle view. (Insets) Close-up of H3K122 (top) and H4K77 (bottom). Hypothetical crotonyl groups are also shown (blue ovals). (E) Arginine residues displaying enhanced methylation. (Left) Nucleosome showing modified arginines as red spheres. (Top inset) Close-up of H4R35 and H4R55. H4R35 forms a direct hydrogen (H) bond to the DNA backbone by adopting an orientation that is itself stabilized by an H bond with H4 residue Tyr51 (not shown). Disruption of either H bond by methylation would weaken DNA binding. H4R55 is in an intricate H bond network with H4 residues Gln27 and Glu52 and the backbone carbonyl of Ile29. This network orients a stretch of N-terminal H4 residues (residues 25–29) such that residue Asn25 is ideally placed to H-bond with H3 residue Glu73. R55 methylation would likely destabilize this network and perturb the precise orientation of the H4 N-terminal stretch, thereby weakening the H3–H4 interface. (Bottom inset) Close-up of H4R67 and H2BR72. Residue H4R67 is stabilized by an H bond with Asn64 in a conformation allowing it to form a cation–π interaction with H3 residue Phe78. Methylation would destabilize the latter interaction by disrupting the H bond and/or delocalizing the positive charge on the R67 guanidino group due to the electron-releasing inductive effect of the added methyl group. H2BR72 is in an H bond network with H2B residue Asp68, H4 residue Tyr98, and the backbone carbonyl of H2A residue Leu96. Methylation would disrupt this network and thus destabilize the H2B–H2A and/or H2B–H4 interfaces.
Unexpectedly, the absence of TH2B led to the total rescue of the defects observed in TH2B-tag-expressing elongating spermatids (data not shown) and to the restoration of normal mature sperm production and male fertility (Supplemental Fig. S6A). Indeed, breeding experiments showed that wild-type and TH2B-less male mice were equally fertile, in striking contrast to Th2b+/tag male mice, which failed to produce any offspring (Supplemental Fig. S6A,B). Thus, whereas modifying TH2B with a C-terminal affinity tag specifically causes sperm cell development to abort, the complete ablation of TH2B appears to have no phenotypic consequences, suggesting the existence of mechanisms that compensate for the loss of TH2B function.
TH2B depletion induces the epigenetic reprogramming of nucleosomes in spermatogenic cells
The notion that TH2B depletion might be rescued by simply up-regulating H2B expression seems improbable, as this would argue against TH2B having any specialized function in spermatogenesis. We therefore conjectured that H2B up-regulation was only part of the rescue mechanism and that additional chromatin alterations were needed to recapitulate the functions ordinarily sustained by TH2B—specifically, nucleosome destabilization (Govin et al. 2007). Since certain histone PTMs are known to modulate nucleosome stability (Tropberger et al. 2013), we wondered whether altered histone PTMs, by increasing nucleosome instability, could restore an efficient histone-to-protamine exchange in the absence of TH2B. To verify this, we used in vitro isotopic labeling followed by HPLC/MS/MS analysis to quantify histone PTMs in wild-type and TH2B-less cells. Briefly, histone extracts from wild-type and TH2B-less testes were digested with trypsin and propionylated using light (12C6) and heavy (13C6) propionic anhydride, respectively. Subsequently, the light and heavy propionylated tryptic peptides were mixed in equal amounts, resolved into 12 fractions by isoelectric focusing (IEF), and analyzed by HPLC/MS/MS. Using this procedure, we identified 12 Lys acetylation sites, two Lys dimethylation sites, seven Lys crotonylation sites, and five Arg methylation sites in core histones (Fig. 6C). The MS/MS spectra of all PTM-containing propionylated peptides are presented in Supplemental Figure S5D. We then determined the relative abundance of histone marks at every PTM site by normalizing the peak intensities from TH2B-less cells to those from wild-type cells (Fig. 6C; Supplemental Table S4).
In the absence of TH2B, several enhancements of histone PTMs were observed. All of these localized to the histone fold domains as opposed to the flexible N-terminal tails, which, in contrast, showed little or no alteration (Fig. 6C). In TH2B-less cells, four sites (H4R35, H4R55, H4R67, and H2BR72) displayed enhanced arginine methylation, while two (H3K122 and H4K77) displayed enhanced crotonylation. Crotonylation is a newly discovered lysine PTM that adds a bulky four-carbon moiety to the lysine amino group and (like acetylation) neutralizes the positive charge (Tan et al. 2011). The two crotonylated sites, located near (H4K77) or next to (H3K122) the dyad axis on opposite sides of the nucleosome, both mediate electrostatic interactions with the DNA backbone (Fig. 6D; Iwasaki et al. 2011). Crotonylation would abolish these interactions and hence considerably destabilize the nucleosome core. Indeed, the acetylation of H3K122 is known to induce significant nucleosome instability (Manohar et al. 2009; Tropberger et al. 2013). Similarly, the enhanced methylation observed for the four Arg sites is also expected to have a destabilizing effect. These (unmodified) residues either participate in the histone–DNA interface (H4R35) or directly (H2BR72 and H4R67) or indirectly (H4R55) stabilize histone–histone interfaces (Fig. 6E). The specific geometry of all of these residues is such that their methylation is predicted to weaken the corresponding interface (see the legend for Fig. 6E for details). Thus, the absence of TH2B in spermatogenic cells seems to induce the epigenetic reprogramming of nucleosomes whereby strategically positioned histone residues are modified so as to enhance nucleosome instability. We also monitored nucleosome disassembly and the occurrence of transitional states in wild-type and TH2B-less elongating/condensing spermatids following extensive MNase digestion, as described above. In contrast to what was observed in TH2B-tag-expressing cells, the genome of TH2B-less cells displayed exactly the same sensitivity to MNase as the genome of wild-type cells (Supplemental Fig. S5C).
We next examined the genome-scale gene expression of our TH2B-less mouse model to test for changes in gene expression consistent with the up-regulated H2B expression and histone PTM enhancements observed upon TH2B depletion. Spermatocytes and round spermatids were purified from wild-type and TH2B-less testes, and the corresponding whole-genome gene expression measurements were compared. Only a small number (25) of genes were significantly up-regulated in the absence of TH2B (data not shown). None of these were obvious candidates that might explain the changes in histone PTMs induced by TH2B depletion. In contrast, five were H2B-encoding genes (h2bm, h2bf, h2bj, h2bk, and h2bc), accounting for the observed increase in H2B expression upon TH2B depletion.
TH2B is assembled into the male pronucleus at fertilization
Taking advantage of our ability to detect TH2B with enhanced sensitivity using high-affinity anti-tag antibodies, we also investigated TH2B expression in TH2Btag/+ female mice. We observed that TH2B-tag is present in metaphase oocytes and the female pronucleus at fertilization and is also rapidly incorporated into the male pronucleus (Fig. 7A, top panel). This was confirmed in wild-type cells using an anti-TH2B antibody (Fig. 7B). TH2B remains associated with embryonic cell chromatin (Fig. 7A, bottom panel) until later developmental stages, when it completely disappears in favor of H2B (Supplemental Fig. S7). As in spermatocytes, we once again observed that TH2B is the major H2B species in the mature oocyte, as the metaphase oocyte genome is only stained with an anti-TH2B antibody and not with an anti-H2B antibody, although the latter efficiently detects contaminating somatic nuclei in the field (Fig. 7C). This observation also confirmed the absence of any defective activity of TH2B-tag, further highlighting the highly stage-specific and restricted effects of the tag seen in elongated/condensing spermatids. Also of note, embryos obtained from TH2B-less females and males developed normally and did not show any noticeable defects, as reflected by litter sizes comparable with those of wild-type mice (Fig. 7D; Supplemental Fig. S6A). In summary, TH2B not only replaces H2B during spermatogenesis, but may also play a role following fertilization.
Figure 7.
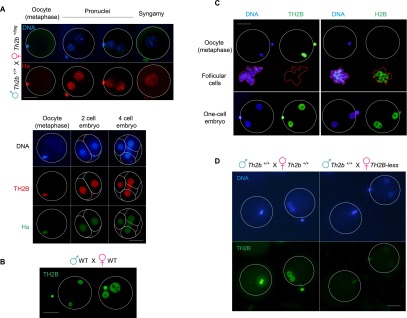
TH2B is assembled on the male genome at fertilization. (A, top panel) Th2b+/tag females were crossed with wild-type males, and TH2B-tag was detected with an anti-Ha antibody. Metaphase oocytes (indicated) and embryos at different stages are shown in the top and bottom panels. In the bottom panels, a codetection of TH2B-tag (anti-Ha) and TH2B (anti-TH2B) was performed. (B) TH2B was detected in wild-type embryos. (C) Specific antibodies against TH2B and H2B were used to detect the corresponding histones in metaphase oocytes or male and female pronuclei. The contaminating somatic follicular cells are highlighted (red line). (D, right panel) TH2B was detected in embryos from wild-type males and TH2B-less females. (Left panel) As a control, immunodetection was carried out in parallel on wild-type embryos. A specific anti-TH2B antibody was used in both cases. Bars, 40 μm.
Discussion
This study not only presents the first thorough characterization of an H2B variant in its physiological context, but also highly increases our understanding of the nucleosome-to-protamine transition that characterizes spermatogenesis. We show that, prior to this transition, the histone variant TH2B almost entirely replaces H2B in male germ cells. This activity differentiates TH2B from other known histone variants, which instead constitute only a fraction of the corresponding histones and display “region-specific” action potentials (Talbert and Henikoff 2010; Bonisch and Hake 2012). TH2B-tag/TH2B ChIP-seq analyses and a comparison of the genomic elements occupied by H2A.Z and TH2B show that H2B replacement by TH2B affects the whole genome. Since the histone-to-protamine transition also affects almost the entire male genome, we hypothesized a role for TH2B in the genome-scale nucleosome disassembly that precedes TP and protamine assembly. Our findings not only confirmed this prediction, but also highlighted the stepwise nature of histone replacement. In a previous study, we observed the occurrence of H3/H4-less subnucleosomal particles in elongated/condensing spermatids containing TH2B and H2AL2 (Govin et al. 2007). The present study shows that C-terminally tagged TH2B disrupts the processing of these particles, indicating that these transitional states are specific intermediary structures generated after nucleosome disassembly and are required for full TP/protamine loading and final histone displacement. The disruptive effect of TH2B-tag is highly specific to this particular step among the changes in genome organization that accompany histone removal, since all other chromatin-directed events occur normally. Specifically, profiling the meiotic and post-meiotic transcriptome of TH2B-tag-expressing spermatogenic cells showed no remarkable effect of the tag on gene expression during these stages. Furthermore, genome-wide ChIP-seq analysis demonstrated that TH2B-tag and wild-type TH2B behave similarly and are both excluded from H2A.Z-containing gene TSSs, suggesting normal chromatin dynamics. The chromosome-wide eviction of H3 and incorporation of H3.3 on the sex chromosomes also occur normally in TH2B-tag-expressing pachytene cells. Finally, the presence of TH2B-tag in male and female pronuclei and early embryonic cell chromatin causes no noticeable defects in embryonic development. These findings suggest that the tag interferes uniquely and very precisely with TH2B function during the processing of subnucleosomal transitional states. However, since the nature and structure of these transitional states are unknown at the present time, one can only speculate on how the TH2B-tag might hamper the final histone displacement. Nevertheless, these data demonstrate a stepwise displacement of histones and the occurrence of a critical TH2B-dependent transitional state required for the full assembly of TP and protamines.
The observation that TH2B depletion induces the activation of several h2b genes suggests that H2B replacement by TH2B primarily leads to a shutdown of gene expression in the histone gene cluster domain, with specific effects on h2b genes. Remarkably, TH2B depletion also induces dramatic changes in the PTM pattern of core histones, as revealed by the HPLC/MS/MS analysis of histones from TH2B-less mice. Specific sites within the globular domains of histones H3, H4, and H2B exhibit an enhanced degree of modification, while no particular changes were observed in the accessible N-terminal tails. Two of these sites, H3K122 and H4K77, are lysine residues that directly contact DNA and exhibit enhanced crotonylation. The acetylation of H3K122 was previously shown to induce transcriptional derepression and significant nucleosome instability (Hyland et al. 2005; Manohar et al. 2009; Hainer and Martens 2011; Tropberger et al. 2013). The presence of the bulkier, charge-neutralizing crotonyl group is expected to destabilize nucleosome structure in a manner similar to, if not more severe than, acetylation (Manohar et al. 2009; Tropberger et al. 2013). The fact that TH2B depletion can be compensated by enhanced levels of destabilizing PTMs suggests that the normal function of TH2B is to mediate nucleosome instability. Such a role for TH2B is corroborated by previous biochemical studies, which showed that, upon expression of TH2B and H2AL2 in somatic cells, the nucleosomes containing both variants are dissociated at salt concentrations that do not affect regular somatic chromatin (Govin et al. 2007). Furthermore, in vitro reconstituted octamers containing human TH2B have also been found to be particularly unstable (Li et al. 2005). These observations agree with earlier studies of pachytene cell chromatin, which revealed that nucleosomes in these cells were less stable than in somatic chromatin, as shown by their enhanced accessibility to DNase I (Rao et al. 1983). A more detailed mapping revealed that pachytene-specific DNase I-hypersensitive regions mostly correspond to H2B-contacting nucleosomal DNA fragments (Rao and Rao 1987), consistent with a specific role for TH2B in the alteration of chromatin structure. Altogether, these data strongly support the idea that the primary function of TH2B is to create a more dynamic chromatin, thereby facilitating the large-scale exchange of histones, which occurs in meiotic and post-meiotic cells. Our mouse models fully support this hypothesis. Indeed, in TH2B-tag-expressing spermatogenic cells, a decrease in the ability of nucleosomes to dissociate leads to a specific blockade of histone replacement, whereas in the total absence of TH2B, an increase in nucleosome-destabilizing PTMs allows H2B to functionally replace TH2B. Additionally, we found that TH2B is involved in the generation of transitional states, which precede genome-wide nucleosome disassembly in elongating spermatids. Other factors, such as the late-expressing H2AL2 variant (Govin et al. 2007) as well as histone hyperacetylation, which induces the action of the bromodomain-containing factor Brdt (Gaucher et al. 2012), should also be required for full nucleosome disassembly in these cells.
Finally, the assembly of TH2B on the male genome at fertilization while this variant is already present in the female pronucleus indicates a need to cover the whole embryonic cell genome with TH2B during early development. Our models did not allow us to identify a specific role for TH2B in oogenesis or during early development. Neither the TH2B-tag nor TH2B depletion revealed any major or minor defects in oogenesis or embryonic development. However, based on our spermatogenesis data, a chromatin-destabilizing role for TH2B is also conceivable during early embryonic development, when there is a strong requirement for genome plasticity.
Materials and methods
Histone preparation
Histone extraction was performed on germ cell nuclei using H2SO4 at 4°C for 1 h followed by centrifugation at 16,000g for 10 min at 4°C to isolate solubilized histones (for germ cell nuclei preparation, see “Chromatin Preparation and ChIP”). Histone precipitation was performed with TCA 33% final concentration for 30 min at 4°C, followed by centrifugation at 16,000g for 10 min at 4°C. The histone pellet was washed three times with ice-cold acetone and resuspended in H2O.
Proteomic-based approaches
All of the experimental procedures regarding the proteomic approaches are detailed in the Supplemental Material.
Chromatin preparation and ChIP
Mononucleosomes were prepared from fractionated spermatocytes and round spermatids and used for ChIP-seq analyses following the protocols described in Gaucher et al. (2012) and detailed in the Supplemental Material.
Step 12–16 condensed spermatid isolation and chromatin preparation
Step 12–16 spermatid fractionation and chromatin preparation are detailed in the Supplemental Material.
Embryo and oocyte collection and immunofluorescence
Superovulation and collection of oocytes and one-cell embryos were performed as described in Wu et al. (2008). Briefly, collected cells were treated by 0.1% hyaluronidase in M2 medium for 2 min to remove follicular cells and washed in M2 medium. Removal of the zona pellucida was performed by a short incubation in Tyrode solution. After washes in M2 medium, cells were deposited on concavaline A-pretreated slides. Cells were fixed with 4% PFA in PBS overnight at 4°C, permeabilized with 0.2% Triton X-100 for 10 min at room temperature, and blocked with 1% BSA in M2 for 30 min at room temperature. Incubation with primary antibody diluted in blocking buffer was performed for 1 h at 37°C for rabbit anti-TH2B antibody (1/700; Abcam, ab23913), for rat anti-HA antibody (1/700; Roche, 3F10), and for rabbit anti-H2B antibody (1/200; Abcam, 61245). The cells were washed with blocking buffer and subjected to secondary antibody incubation for 30 min at 37°C. After the final wash, cells were mounted with VectaShield mounting medium with DAPI.
The two-cell to four-cell embryos were collected and subjected to immunofluorescence analysis as described in Torres-Padilla et al. (2006).
Mouse models
Th2b tap-tagging was performed following the recombineering technique as previously described (Liu et al. 2003) to construct the Th2b tap tag targeting vector for homologous recombination in ES cells (AT1 line). Animal experiments were approved by ad hoc committees, and all of the investigators directly involved have an official animal-handling authorization obtained after 2 wk of intensive training and a final formal evaluation. Knock-in through homologous recombination in ES cells is described in detail in Gaucher et al. (2012). The crossing of Th2bneo mice with a CMV-driven Cre-expressing mouse line lead to the deletion of the neo cassette and the rescue of TH2B-tag expression (see Supplemental Fig. S1 for details).
Testis and epididymis histology and immunohistochemistry
Testis and epididymides were AFA-fixed overnight and paraffin-embedded. Slides were either counterstained with eosin hematoxyline or subjected to immunohistochemistry as described in Hazzouri et al. (2000). Polyclonal anti-TH2B antibody (1/500; Millipore, 07-680) and polyclonal anti-H2B antibody (1/500; Abcam, ab61245) were used.
Sperm count and purification of germ cells
Cauda epididymides were dilacerated in DMEM medium and incubated for 10 min at room temperature. Spermatozoa were counted on Malassez slides under a microscope.
Fractions enriched in spermatogenic cells at different stages of maturation (spermatocytes and round spermatids) were obtained by sedimentation on a BSA gradient as previously described in Pivot-Pajot et al. (2003). Pure fractions of condensed spermatids at steps 12–16 were obtained by sonication of total mouse spermatogenic cell preparation as described by Marushige and Marushige (1983).
Spermatogenic cell preparation, immunofluorescence, and FRAP experiments
Staged seminiferous tubules and testis imprints were prepared as described in Gaucher et al. (2012) and detailed in the Supplemental Material. FRAP experiments were performed on H1299 cell lines stably expressing GFP-TH2B and GFP-TH2B-tag and photobleaching, and the measurement of fluorescence recovery and the calculation of different parameters were carried out essentially as previously described (Moriniere et al. 2009).
RNA preparation and transcriptomic and statistical analyses
RNA from fractionated cells (spermatocytes and round spermatids) was extracted using the Qiagen RNAeasy minikit and analyzed on the Illumina whole-genome chip as performed in Gaucher et al. (2012). The global gene expression level in spermatocytes and round spermatids was compared between TH2B-tag-expressing and wild-type cells, and Pearson correlation coefficients were calculated.
Protein sample preparation and Western blotting
For total protein extracts, whole testes were homogenized in 8 M urea and sonicated at 200 J. Protein dosage was assessed by Bradford. Western blots with SDS-PAGE or TAU gels were carried out using anti-TH2B antibody (1/5000; Millipore, 07-680), anti-actin antibody (1/5000; Sigma, A5441), and anti-H2B C-term antibody (1/2500; Millipore, 07-371).
Acknowledgments
We specially thank Dr. Shunsuke Ishii (Laboratory of Molecular Genetics, RIKEN Tsukuba Institute) for insightful discussions on TH2B and for sharing unpublished information on the identification of TH2B in the egg and in early embryonic cell chromatin. We are also grateful to Dr. Karolin Luger (Colorado State University) for helpful discussions. The Th2B tap tag project was initiated in the frame of the ANR-Regulome Consortium in the S.K. and M.G. laboratories. Mice were bred in the High Technology Animal Facility (PHTA) of Grenoble University with the precious help of Rachel Balouzat and Maryline Cossin. Paraffin embedding was performed on the histology platform of Grenoble Medical School. This project is supported by ANR EpiSperm2 grant, and its cancer-oriented developments are supported by INCa and “ARC libre” funds. E.M. was a recipient of a PhD fellowship by “MTR bourse fléchée” for three years and an ARC fellowship for her fourth year. H.S. is a recipient of a Marie Curie Initial Training Network funded by European Commission (FP7-PEOPLE-2011-ITN and PITN-GA-289880). F.B. was a recipient of “contrat jeune chercheur” INSERM from 2006 to 2011. High-throughput sequencing was performed at the TGML Platform, supported by grants from IBiSA, Aix-Marseille Université, and ANR-10-INBS-0009-10.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.220095.113.
References
- Bonisch C, Hake SB 2012. Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res 40: 10719–10741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussouar F, Rousseaux S, Khochbin S 2008. A new insight into male genome reprogramming by histone variants and histone code. Cell Cycle 7: 3499–3502 [DOI] [PubMed] [Google Scholar]
- Branson RE, Grimes SR Jr, Yonuschot G, Irvin JL 1975. The histones of rat testis. Arch Biochem Biophys 168: 403–412 [DOI] [PubMed] [Google Scholar]
- Brock WA, Trostle PK, Meistrich ML 1980. Meiotic synthesis of testis histones in the rat. Proc Natl Acad Sci 77: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YC, Chae CB 1993. Demethylation of somatic and testis-specific histone H2A and H2B genes in F9 embryonal carcinoma cells. Mol Cell Biol 13: 5538–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contrepois K, Ezan E, Mann C, Fenaille F 2010. Ultra-high performance liquid chromatography-mass spectrometry for the fast profiling of histone post-translational modifications. J Proteome Res 9: 5501–5509 [DOI] [PubMed] [Google Scholar]
- Gaucher J, Reynoird N, Montellier E, Boussouar F, Rousseaux S, Khochbin S 2010. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J 277: 599–604 [DOI] [PubMed] [Google Scholar]
- Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte AL, et al. 2012. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J 31: 3809–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S 2004. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem 271: 3459–3469 [DOI] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. 2007. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol 176: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ 2006. The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis. Mol Cell Biol 26: 5394–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Martens JA 2011. Identification of histone mutants that are defective for transcription-coupled nucleosome occupancy. Mol Cell Biol 31: 3557–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sele B, Khochbin S, Rousseaux S 2000. Regulated hyperacetylation of core histones during mouse spermatogenesis: Involvement of histone deacetylases. Eur J Cell Biol 79: 950–960 [DOI] [PubMed] [Google Scholar]
- Huh NE, Hwang IW, Lim K, You KH, Chae CB 1991. Presence of a bi-directional S phase-specific transcription regulatory element in the promoter shared by testis-specific TH2A and TH2B histone genes. Nucleic Acids Res 19: 93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol 25: 10060–10070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki W, Tachiwana H, Kawaguchi K, Shibata T, Kagawa W, Kurumizaka H 2011. Comprehensive structural analysis of mutant nucleosomes containing lysine to glutamine (KQ) substitutions in the H3 and H4 histone-fold domains. Biochemistry 50: 7822–7832 [DOI] [PubMed] [Google Scholar]
- Li A, Maffey AH, Abbott WD, Conde e Silva N, Prunell A, Siino J, Churikov D, Zalensky AO, Ausio J 2005. Characterization of nucleosomes consisting of the human testis/sperm-specific histone H2B variant (hTSH2B). Biochemistry 44: 2529–2535 [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG 2003. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ 2009. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J Biol Chem 284: 23312–23321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marushige Y, Marushige K 1983. Proteolysis of somatic type histones in transforming rat spermatid chromatin. Biochim Biophys Acta 761: 48–57 [DOI] [PubMed] [Google Scholar]
- Montellier E, Rousseaux S, Zhao Y, Khochbin S 2012. Histone crotonylation specifically marks the haploid male germ cell gene expression program: Post-meiotic male-specific gene expression. Bioessays 34: 187–193 [DOI] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461: 664–668 [DOI] [PubMed] [Google Scholar]
- Pivot-Pajot C, Caron C, Govin J, Vion A, Rousseaux S, Khochbin S 2003. Acetylation-dependent chromatin reorganization by BRDT, a testis-specific bromodomain-containing protein. Mol Cell Biol 23: 5354–5365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao BJ, Rao MR 1987. DNase I site mapping and micrococcal nuclease digestion of pachytene chromatin reveal novel structural features. J Biol Chem 262: 4472–4476 [PubMed] [Google Scholar]
- Rao BJ, Brahmachari SK, Rao MR 1983. Structural organization of the meiotic prophase chromatin in the rat testis. J Biol Chem 258: 13478–13485 [PubMed] [Google Scholar]
- Shires A, Carpenter MP, Chalkley R 1975. New histones found in mature mammalian testes. Proc Natl Acad Sci 72: 2714–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboleva TA, Nekrasov M, Pahwa A, Williams R, Huttley GA, Tremethick DJ 2012. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol 19: 25–30 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S 2010. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 11: 264–275 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Ahmad K, Almouzni G, Ausio J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, et al. 2012. A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. 2011. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M 2006. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol 50: 455–461 [DOI] [PubMed] [Google Scholar]
- Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Buhler M, et al. 2013. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152: 859–872 [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Posfai E, Giele M, Pelczar P, Ramos L, Wansink DG, van der Vlag J, Peters AH, de Boer P 2007. Chromosome-wide nucleosome replacement and H3.3 incorporation during mammalian meiotic sex chromosome inactivation. Nat Genet 39: 251–258 [DOI] [PubMed] [Google Scholar]
- Wu F, Caron C, De Robertis C, Khochbin S, Rousseaux S 2008. Testis-specific histone variants H2AL1/2 rapidly disappear from paternal heterochromatin after fertilization. J Reprod Dev 54: 413–417 [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L 2011. seqMINER: An integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, Behringer RR, Boissonneault G, Yanagimachi R, Meistrich ML 2004. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis 38: 200–213 [DOI] [PubMed] [Google Scholar]