The PIWI-interacting RNA pathway is the major defense line against selfish genetic elements in the animal germline. Recent studies indicate that nuclear Piwi proteins trigger transcriptional target silencing, yet the composition of the silencing complex is unknown. Dönertas et al. now identify the zinc finger protein Gtsf1, which is essential for Piwi-mediated transcriptional repression in Drosophila. They show that Gtsf1 is a key factor in the nuclear Piwi silencing complex and propose that only a fraction of nuclear Piwi is actively engaged in target silencing.
Keywords: transposon control, piRNA pathway, Piwi, Gtsf1, transcriptional silencing, heterochromatin formation
Abstract
The PIWI-interacting RNA (piRNA) pathway is a small RNA silencing system that keeps selfish genetic elements such as transposons under control in animal gonads. Several lines of evidence indicate that nuclear PIWI family proteins guide transcriptional silencing of their targets, yet the composition of the underlying silencing complex is unknown. Here we demonstrate that the double CHHC zinc finger protein gametocyte-specific factor 1 (Gtsf1) is an essential factor for Piwi-mediated transcriptional repression in Drosophila. Cells lacking Gtsf1 contain nuclear Piwi loaded with piRNAs, yet Piwi's silencing capacity is ablated. Gtsf1 interacts directly with a small subpool of nuclear Piwi, and loss of Gtsf1 phenocopies loss of Piwi in terms of deregulation of transposons, loss of H3K9 trimethylation (H3K9me3) marks at euchromatic transposon insertions, and deregulation of genes in proximity to repressed transposons. We propose that only a small fraction of nuclear Piwi is actively engaged in target silencing and that Gtsf1 is an essential component of the underlying Piwi-centered silencing complex.
Selective silencing or repression of selfish genetic elements such as transposable elements (TEs) is an ancient evolutionary challenge (Kazazian 2004). TE silencing is of particular importance in reproductive cells of multicellular organisms. In plants and animals, specialized RNA silencing pathways centered on Argonaute proteins bound to small RNA guides are devoted to the silencing of TEs (Slotkin and Martienssen 2007; Ghildiyal and Zamore 2009). The central pathway acting in animal gonads is the PIWI-interacting RNA (piRNA) pathway that uses Argonaute proteins of the PIWI family (Malone and Hannon 2009; Siomi et al. 2011). In Drosophila, two cytoplasmic PIWI proteins (Aubergine [Aub] and Argonaute 3 [AGO3]) and one nuclear PIWI protein (Piwi) are loaded with a diverse population of 22- to 30-nucleotide (nt) RNA guides (piRNAs) that are mostly derived from heterochromatic transposon-rich loci (piRNA clusters) and TE mRNAs (Senti and Brennecke 2010). The three PIWI family proteins repress the expression of RNAs with piRNA complementary sequences via a combination of transcriptional silencing (Piwi) and post-transcriptional silencing (Aub and AGO3).
The nuclear Piwi–piRNA complex inhibits transcription of TEs by an unknown mechanism. Target silencing is accompanied by histone H3 Lys 9 trimethylation (H3K9me3) and appears to require the HP1 family member Su(var)205 (Shpiz et al. 2011; Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). The piRNA pathway factor Maelstrom (Mael) is required for Piwi-guided transcriptional silencing, yet loss of Mael (in contrast to loss of Piwi) does not result in significant loss of H3K9me3 marks at repressed TE insertions (Sienski et al. 2012). Whether H3K9me3, a mark for heterochromatin, is required for transcriptional silencing or accompanies it is therefore not fully understood.
Conceptually, TE silencing by Piwi might resemble the much better understood process of RNAi-induced heterochromatin formation in Schizosaccharomyces pombe (Moazed 2009; Grewal 2010; Castel and Martienssen 2013). In both systems, a nuclear Argonaute protein guides the transcriptional silencing of targets, and this is accompanied by H3K9 methylation. Fission yeast AGO1 assembles in the trimeric RNA-induced transcriptional silencing (RITS) complex and recruits the histone methyltransferase complex CLRC that mediates H3K9 methylation. In analogy, vertebrate and invertebrate nuclear PIWI proteins are likely to assemble a silencing complex that interacts with nascent RNA at the target locus and induces transcriptional silencing by recruiting factors impacting RNA polymerase II (Pol II) recruitment and/or the chromatin status. However, besides Mael and possibly Su(var)205, no factor is known to be required for this process, and no protein has been reported as bona fide component of a nuclear PIWI silencing complex. Here, we demonstrate that the evolutionarily conserved zinc finger protein gametocyte-specific factor 1 (Gtsf1) directly interacts with a subpool of nuclear Piwi and that Gtsf1 is required for Piwi-mediated transcriptional trans-silencing and H3K9me3 in Drosophila.
Results
CG3893 is required for transposon silencing in ovarian somatic and germline cells
In a transgenic RNAi screen for uncharacterized piRNA pathway factors in ovarian follicle cells (Handler et al. 2013), we identified CG3893 as one of the most strongly scoring candidates (note that CG3893 was also identified in two independent screens for piRNA pathway factors by the Hannon group [Czech et al. 2013; Muerdter et al. 2013]). Knockdown of CG3893 via tj-GAL4-driven Vienna Drosophila RNAi Stock Center (VDRC) RNAi hairpins or an shRNA in follicle cells (soma knockdown [SKD]) resulted in gypsy-lacZ reporter derepression to an extent comparable with knockdown of the essential piRNA pathway factors Piwi or Mael (Fig. 1A,B, top panels; Sarot et al. 2004; Olivieri et al. 2010). To probe for a role of CG3893 also in germline cells, we depleted CG3893 via MTD-GAL4-driven shRNA expression (Ni et al. 2011) or NGT-GAL4-driven VDRC hairpin expression (germline knockdown [GLKD]) (Handler et al. 2011; Wang and Elgin 2011). We monitored piRNA pathway integrity in the respective crosses with two germline-specific TE repression sensors (Burdock-lacZ and HeT-A-lacZ). Silencing of both reporters was strongly impaired (Fig. 1A,B, bottom panels). The HeT-A reporter is under very stringent control of the piRNA pathway, presumably as it is targeted by numerous very abundant germline piRNAs. Only loss of a subset of the known piRNA pathway members strongly impairs silencing of this reporter (D Dönertas, unpubl.), and CG3893 was one of the strongest factors, together with Piwi and Mael.
Figure 1.
CG3893 is required for transposon silencing in ovarian somatic and germline cells. (A) Cartoon of a Drosophila ovariole, with follicle cells (soma) in green and nurse cells and oocyte (germline) in beige. To the right, lacZ sensors that report derepression of soma-specific (gypsy) or germline-specific (Burdock and HeT-A) TEs. For the soma reporter, tj-Gal4, gypsy-lacZ flies carrying a restrictive flamenco background were crossed to VDRC lines of interest (SKD). For the germline reporters, target gene knockdown was performed by either MTD-Gal4>UAS-shRNA or NGT-Gal4>UAS-Dcr-2/UAS-VDRC (GLKD). Portions of the respective reporters targeted by piRNAs are depicted in red. (B) Shown are β-Gal stainings of egg chambers expressing gypsy (top), Burdock (middle), or HeT-A (bottom) reporters. w[1118] flies (control) or indicated VDRC/shRNA lines for piwi, mael, or CG3893 were used for the analysis. (C) Displayed are fold changes in steady-state RNA levels of indicated TEs and genes in mael or CG3893 knockdown ovaries in log10 scale (soma, left panel; germline, right panel). Values are averages of three biological replicates (error bars indicate SD) and were normalized to control knockdowns via levels of the rp49 housekeeping gene. (D) Displayed are fold changes in steady-state RNA levels of indicated TEs ([green] soma-dominant; [beige] germline dominant) and genes in CG3893[GS12962] homozygous versus heterozygous ovaries in log10 scale.
We confirmed that tissue-specific knockdowns of CG3893 also led to derepression of endogenous marker TEs at the steady-state RNA level in the respective cell types (mdg1 and gypsy in follicle cells; HeT-A and blood in germline cells) (Fig. 1C).
Independent genetic support for the validity of CG3893 as a TE silencing factor stemmed from the analysis of females homozygous for the GS12962 P-element (inserted into the CG3893 coding sequence) (Supplemental Fig. S1A). These females are viable but have rudimentary ovaries that resemble piwi mutant ovaries (Supplemental Fig. S1B; Cox et al. 1998). Soma- and germline-specific TEs are severely derepressed in CG3893[GS12962] ovaries (Fig. 1D), and mutant females are fully sterile. Precise excision of the GS12962 P-element reverted the sterility phenotype, supporting the claim that the sterility is linked to the CG3893 locus.
CG3893 is an essential piRNA pathway factor
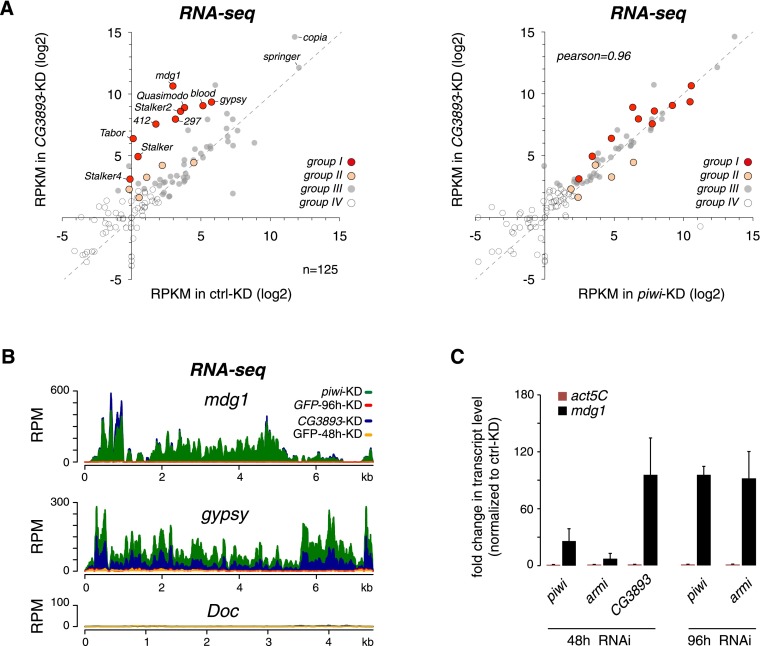
The central TE silencing system in Drosophila gonads is the piRNA pathway. We compared TE expression upon CG3893 depletion or Piwi depletion in cultured ovarian somatic cells (OSCs), which harbor a fully functional somatic piRNA pathway centered on Piwi (Niki et al. 2006; Saito et al. 2009). The set of TEs that is under piRNA pathway control in OSCs depends on the expressed piRNAs and the promoter activity of each individual TE. Based on steady-state RNA levels, depletion of Piwi leads to highly reproducible derepression of ∼15 TEs, ranging from mild to very strong (Sienski et al. 2012). CG3893 depletion resulted in very similar TE silencing defects, and TE RNA levels were highly correlated between CG3893- and Piwi-depleted cells (Fig. 2A). For example, the extent and pattern of derepression of the top elements mdg1 and gypsy were nearly identical (Fig. 2B). These observations classify CG3893 as a core factor of the somatic piRNA pathway. While we did not systematically analyze TE RNA levels in GLKD ovaries, the quantitative PCR (qPCR) analysis of selected TEs (Fig. 1C) indicates that, also in germline cells, loss of CG3893 highly resembles loss of Piwi-mediated silencing.
Figure 2.
CG3893 is a core member of the somatic piRNA pathway. (A) Scatter plot of RNA-seq reads per kilobase per million mapped reads (RPKM) values (log2; ribozero OSC RNA) for annotated Drosophila melanogaster TEs (n = 125) in control knockdown (control-KD) versus CG3893 knockdown (CG3893-KD) (left) and piwi knockdown (piwi-KD) versus CG3893 knockdown (CG3893-KD) (right). Four TE groups are color-coded according to the classification in Sienski et al. (2012). (Red and orange) Piwi-repressed; (gray) nonrepressed; (open circles) nonexpressed. (B) Density profiles of normalized RNA-seq reads mapping to mdg1, gypsy (group I), or Doc (group III). The orange profile indicates the control knockdown (GFP-48h-KD) for the CG3893 knockdown (CG3893-KD; blue); the red profile indicates the control knockdown (GFP-96h-KD) for the piwi-knockdown (piwi-KD; green). (C) Time-course experiment showing fold changes in steady-state RNA levels of mdg1 (black bars) and act5C (red bars) after 48 h or 96 h of RNAi for the indicated genes in OSCs.
Efficient depletion of Piwi in OSCs requires double transfections of siRNAs for a total of 4 d (96 h) (Saito et al. 2009). After 96 h, Piwi-depleted cells exhibit mdg1 derepression in the range of >100-fold, cease proliferation, and show signs of cell death. Similar knockdown procedures for CG3893 resulted in a loss of cells due to widespread cell death. Remarkably, knockdown of CG3893 for just 2 d (48 h) resulted in very strong mdg1 derepression (Fig. 2C). In fact, after 48 h of CG3893 depletion, derepression of mdg1 was much stronger compared with parallel Piwi or Armi depletions (Fig. 2C). Only after 96 h did Piwi- or Armi-depleted cells exhibit a comparable mdg1 derepression. As the piwi siRNA is highly effective and allows very robust Piwi depletion, we propose that the stability and abundance of Piwi–piRNA complexes require continuous knockdown for 96 h to result in full TE desilencing. Since CG3893 depletion led to a much faster derepression of TEs, we interpreted this as a first indication that CG3893 acts downstream from piRNA biogenesis.
To exclude that CG3893 acts independently of the piRNA pathway in a parallel silencing system, we performed an epistasis experiment and compared Piwi or CG3893 single knockdowns with the Piwi/CG3893 double knockdown (experimental strategy shown in Supplemental Fig. S2A). The double knockdown resembled fully the Piwi-only knockdown in terms of derepressed TEs (mdg1 and gypsy) and nonregulated TEs (F-element). qPCR analysis of the piwi and CG3893 transcripts indicated consistent levels of target knockdowns (Supplemental Fig. S2B). This strongly suggests that CG3893 acts specifically in the piRNA pathway.
CG3893 is not required for piRNA biogenesis
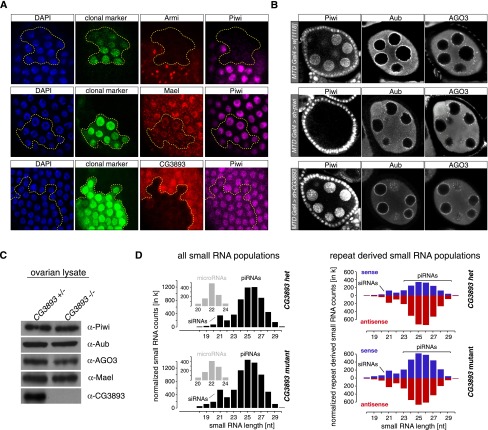
Three major classes of piRNA pathway factors can be distinguished: (1) factors required for piRNA biogenesis, (2) factors required for the specification of piRNA precursor RNAs, and (3) factors required for PIWI-mediated silencing. Loss of group 1 or 2 factors typically results in defective or impaired loading of PIWI family proteins. As unloaded PIWI proteins are reported to be unstable, this results in a measurable reduction in their levels (mostly Piwi and Aub) or in their delocalization from perinuclear nuage (AGO3 and Aub) (Klattenhoff et al. 2009; Haase et al. 2010; Olivieri et al. 2010, 2012; Saito et al. 2010; Zamparini et al. 2011). We therefore analyzed levels and localization of PIWI proteins in cells lacking CG3893. In contrast to Armi knockdown, knockdown of CG3893 in clones of follicular cells had no measurable impact on Piwi levels and in this resembled the silencing factor Mael (Fig. 3A; Sienski et al. 2012). Similarly, knockdown of CG3893 in germline cells (despite severe derepression of TEs) (Fig. 1C) did not impact Piwi/Aub/AGO3 levels or their subcellular localization (Fig. 3B).
Figure 3.
CG3893 is not required for piRNA biogenesis. (A) Confocal sections of follicular epithelia stained for Piwi (magenta), DNA (DAPI; blue), and the RNAi target gene (red) in which armi-RNAi, mael-RNAi, or CG3893-RNAi has been clonally activated (marked by GFP; boundary indicated by dashed yellow line). (B) Confocal sections of egg chambers with GLKDs of Piwi or CG3893 compared with a w[1118] cross (control), stained for Piwi, Aub, or AGO3. (C) Western blots showing protein levels of Piwi, Aub, AGO3, Mael, or CG3893 in CG3893[GS12962] heterozygous or homozygous ovaries. (D) To the left, length profiles of all small RNAs (normalized to 1 million miRNAs; small insets) isolated from ovaries of CG3893[GS12962] heterozygous or homozygous flies are shown (small RNAs mapping to rRNA, tRNAs, or snoRNAs have been computationally removed; siRNA and piRNA populations are indicated). Panels to the right show length profiles of repeat-derived ovarian small RNAs (normalized to 1 million miRNAs) from CG3893[GS12962] heterozygous or homozygous flies. (Red) Antisense; (blue) sense.
Also, in CG3893[GS12962] homozygous animals, subcellular localization of PIWI proteins was unaffected, supporting the GLKD data (shown for Piwi in Supplemental Fig. S3A). Western blot analysis of ovarian lysate from CG3893[GS12962] heterozygous and homozygous flies confirmed that Piwi, Aub, AGO3, and Mael levels are independent of the level of CG3893 (Fig. 3C). CG3893 was undetectable in lysate from CG3893[GS12962] ovaries, indicating that this is a very strong mutant allele if not a null.
To probe for piRNA levels, we first evaluated steady-state piRNA levels in OSCs after depletion of Armi, Piwi, Mael (96 h), or CG3893 (48 h) compared with respective GFP knockdowns. This indicated that loss of CG3893—similar to loss of Mael—does not impact piRNA levels in OSCs (Supplemental Fig. S3B,C).
We extended the piRNA analysis by sequencing total ovarian small RNAs in the 18- to 29-nt range from CG3893[GS12962] mutant and heterozygous animals. This indicated no apparent loss in the population of piRNAs if total small RNA populations were normalized using microRNA (miRNA) reads as normalization (Fig. 3D, left). When only repeat-derived small RNA populations were analyzed, only a very moderate reduction in antisense piRNAs was observed, while the population of sense piRNAs increased considerably (Fig. 3D, right). This resembled the small RNA profiles of mael mutants (Sienski et al. 2012) and might indicate that the strong accumulation of sense TE transcripts results in increased sense piRNA levels. When piRNA populations mapping to individual TEs or to individual piRNA clusters (only genome-unique) were analyzed, we observed very little impact of CG3893 loss on the somatic piRNA pathway and a more pleiotropic impact on the germline pathway. For example, levels of flamenco-derived piRNAs (soma-specific/dominant) (Supplemental Fig. S3D) and those mapping to soma-dominant elements (Supplemental Figs. S3E [green dots], S4A; Li et al. 2009; Malone et al. 2009) were essentially unchanged, while piRNAs mapping to the germline-specific 42AB cluster were moderately reduced (Supplemental Fig. S3D). In addition, antisense populations of piRNAs mapping to a subset of germline-dominant TEs (such as HeT-A, TART, and mdg3) were strongly reduced, while the corresponding sense piRNAs were unchanged or even increased (Supplemental Figs. S3E, S4B). Other TEs, such as the R1-element, exhibited strong increases in sense and antisense piRNA levels (Supplemental Fig. S4C). These patterns are in stark contrast to those observed upon loss of piRNA biogenesis factors and resemble those observed upon loss of the silencing factor Mael (Sienski et al. 2012). We therefore conclude that CG3893 is not integrally involved in the process of piRNA biogenesis.
CG3893 physically interacts with Piwi, and its subcellular localization depends on Piwi
While biogenesis and loading of piRNAs into Piwi occurs in the cytoplasm, Piwi-mediated transcriptional repression is a nuclear process (Siomi et al. 2011; Ge and Zamore 2013). Consistent with a role of CG3893 in the silencing process, CG3893 localizes to the nucleus of germline and somatic support cells of ovaries expressing N-terminally GFP-tagged CG3893 under its endogenous regulatory regions (Fig. 4A). Similarly, N-terminally or C-terminally GFP-tagged CG3893 ectopically expressed in OSCs is enriched in the nucleus (data not shown). We also raised a monoclonal antibody against CG3893 and analyzed the subcellular localization of the endogenous protein. In wild-type ovaries, CG3893 was enriched in germline and somatic nuclei (Fig. 4A). Expression of a CG3893 shRNA in germline cells ablated CG3893 protein, confirming the knockdown efficiency as well as the specificity of the monoclonal antibody (Fig. 4A).
Figure 4.
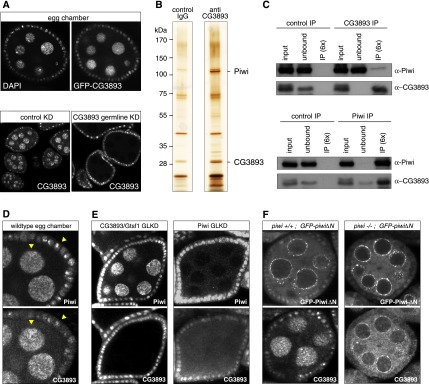
CG3893 physically interacts with Piwi, and its nuclear localization depends on Piwi. (A, top panels) Confocal sections of an egg chamber expressing GFP-CG3893 from a genomic rescue construct stained for DNA (DAPI) and analyzed for GFP fluorescence. (Bottom panels) Confocal sections of egg chambers stained for CG3893 in control and CG3893 GLKDs. (B) SDS-PAGE of a CG3893 and IgG control immunoprecipitation (IP) from nuclear OSC lysate stained with silver. A molecular weight marker is indicated to the left, and CG3893 and Piwi bands are indicated. (C) Western blots showing co-IP of endogenous CG3893 with endogenous Piwi from nuclear OSC lysates. Rabbit IgG and mouse IgG were used as control immunoprecipitations for Piwi and CG3893, respectively. (D) Confocal images of a wild-type egg chamber stained for Piwi (top) and CG3893 (bottom). Arrowheads indicate areas with pronounced colocalization. (E) Confocal images of egg chambers stained for Piwi (top panels) and CG3893 (bottom panels). The target of the respective GLKD is indicated at the top. (F) Confocal images of egg chambers expressing GFP-tagged N-terminally truncated Piwi (top panels) stained for CG3893 (bottom panels). The left panels show ovaries with a wild-type piwi background, and the right panels show ovaries with a piwi[1]/[2] background.
The monoclonal CG3893 antibody recognized a band of expected size in Western blot analysis from OSC lysate. This band was strongly reduced upon CG3893 knockdown (Supplemental Fig. S3B) and was absent in ovarian extract from CG3893[GS12962] mutants (for example, see Fig. 3C). We took advantage of this highly specific reagent and performed immunoprecipitation experiments from OSC nuclear lysate to identify copurifying factors via mass spectrometry. Two major bands differing between the CG3893 immunoprecipitation and a control IgG immunoprecipitation were apparent after silver staining (Fig. 4B). Mass spectrometry identified these unambiguously as CG3893 and Piwi, respectively. Manual inspection of the three most prominent peptide peaks for each protein in two independent mass spectrometry runs indicated an exclusive identification of CG3893 in the lower band compared with the corresponding control lane and an ∼600-fold to1000-fold enrichment of Piwi in the CG3893 immunoprecipitation compared with the control (Supplemental Fig. S5A).
We confirmed the specific physical interaction between CG3893 and Piwi by immunoprecipitation-Western experiments. Immunodepletion of CG3893 under these conditions led to a significant copurification of Piwi (Fig. 4C, top panels). Importantly, a reciprocal immunoprecipitation experiment in which we immunodepleted Piwi resulted in a very pronounced copurification of CG3893 (Fig. 4C, bottom panels). Under these conditions, we repeatedly observed (Fig. 4C) that depletion of Piwi also led to significant depletion of CG3893, indicating that most of the nuclear CG3893 pool physically interacts with Piwi, potentially even in a stoichiometric complex. In agreement with this are the observed intensities of the CG3893 and Piwi bands in the silver gel (Fig. 4B). These experiments hinted strongly at the possibility that only a very small fraction of the nuclear Piwi pool is complexed with CG3893 at a given time point. Indeed, CG3893 steady-state RNA levels were ∼20-fold lower in OSCs compared with Piwi RNA levels (Supplemental Fig. S5B). In addition, we compared fluorescence intensities of GFP-CG3893 and GFP-Piwi, both expressed under their endogenous regulatory control elements, side by side (Supplemental Fig. S5C). Although not a quantitative assay, this indicated that nuclear Piwi protein levels by far exceed CG3893 levels in vivo.
Consistent with a strong physical tie between CG3893 and Piwi, we observed a clear colocalization of the two endogenous proteins in somatic and germline nuclei of wild-type ovaries (Fig. 4D, arrowheads). More significantly, depletion of Piwi in germline cells by efficient transgenic RNAi had a pronounced effect on CG3893 subcellular localization, as we observed many egg chambers with greatly reduced nuclear CG3893 accumulation (Fig. 4E; see Supplemental Fig. S5D for the degree of variability in this phenotype). Similar results were obtained upon Piwi depletion in germline clones for the strong piwi[1] mutation as well as in somatic cells (Supplemental Fig. S5E,F). Of note, overall CG3893 levels appeared to be unaffected in Piwi-depleted cells (cf. CG3893 knockdowns in Fig. 4E).
Piwi that lacks its N terminus is loaded with piRNAs but does not translocate to the nucleus (Saito et al. 2009; Klenov et al. 2011; Sienski et al. 2012). In germline cells, it accumulates in perinuclear nuage instead. We took advantage of flies expressing a GFP-tagged PiwiΔN transgene (Sienski et al. 2012) and analyzed CG3893 localization in these ovaries. Strikingly, while ovaries expressing GFP-Piwi-ΔN in a wild-type piwi background exhibited unchanged CG3893 localization, expression of the same construct in flies mutant for endogenous piwi resulted in a strong relocalization of CG3893 to nuage, where it almost entirely overlapped with GFP-Piwi-ΔN (Fig. 4F). Piwi's localization therefore seems to dictate the localization of CG3893 in vivo.
CG3893/Gtsf1 is a zinc finger protein that interacts with Piwi via its C-terminal tail
Protein domain search algorithms identified two CHHC zinc fingers in the first 60 amino acids of CG3893, followed by an ∼100--amino-acid C-terminal tail that appears mostly unstructured and lacks any discernable domains (Fig. 5A). The very few CHHC zinc finger-containing proteins belong to U11-48K proteins (one CHHC domain), certain tRNA methyltransferases (one CHHC domain), and a conserved protein family defined by mouse GTSF1 (two CHHC domains) (Andreeva and Tidow 2008; Yoshimura et al. 2009). Reciprocal BLAST searches indicated that CG3893 belongs to the GTSF1 family that is conserved in invertebrates and vertebrates, and we therefore named CG3893 Drosophila Gtsf1. Of note, mouse GTSF1 has been reported to be essential for spermatogenesis and retrotransposon control (Yoshimura et al. 2009).
Figure 5.
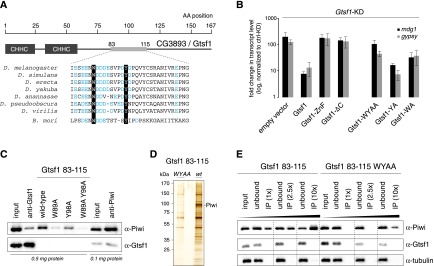
A C-terminal peptide in CG3893/Gtsf1 interacts with Piwi. (A) Cartoon of the CG3893/Gtsf1 protein showing the N-terminal zinc fingers and the unstructured C-terminal tail. Shown is the highly conserved amino acid stretch (residues 83–115) with negatively charged amino acids (blue) and the highly conserved aromatic W89 and Y98 residues. (B) Shown are derepression levels for the mdg1 and gypsy TEs upon CG3893 knockdown with simultaneous transfection of plasmids for the expression of siRNA-resistant CG3893 versions with indicated mutations. (ZnF) Zinc finger mutations; (ΔC) deletion of C-tail. (C) Shown are immunoprecipitation experiments (anti-Gtsf1 and anti-Piwi) from OSC nuclear lysate (amounts of total protein indicated) and pull-down experiments with indicated Gtsf1 peptides (residues 83–115) probed for Piwi (top blot) and Gtsf1 (bottom blot). (D) SDS-PAGE of Gtsf1 peptide (83–115) pull-downs (WY mutant, left; wild type, right) from nuclear OSC lysate stained with silver. A molecular weight marker is indicated to the left, and the Piwi band confirmed by mass spectrometry is marked. (E) Shown are peptide pull-down experiments with indicated Gtsf1 peptides where peptide amounts (1×–10×) have been varied. Input was OSC nuclear lysate, and eluates were analyzed by Western blot against Piwi (top), Gtsf1 (middle), and tubulin (bottom).
Given the domain organization of Gtsf1, we investigated the importance of the zinc fingers and the C-tail for Gtsf1 function. We performed rescue experiments in OSCs in which we depleted endogenous Gtsf1 and transfected siRNA-resistant Gtsf1 expression constructs (Fig. 5B; Supplemental Fig. S6A; Saito et al. 2009). While expression of wild-type Myc-tagged Gtsf1 rescued TE derepression >10-fold, neither the zinc finger mutant nor the Gtsf1-ΔC construct exhibited measurable activity (expression of rescue constructs was confirmed by Western analysis or immunofluorescence; expression of the zinc finger mutant was consistently lower) (Fig. 5B; Supplemental Fig. S6B,C). The CHHC zinc fingers are predicted RNA-binding domains (Andreeva and Tidow 2008), and we therefore turned our attention to the C-tail in order to identify the portion that is responsible for the Piwi interaction. The C-tail contains a highly conserved stretch of negatively charged amino acids with two embedded aromatic residues (W89 and Y98) (Fig. 5A; Supplemental Fig. S6A). We synthesized four biotinylated peptides encompassing amino acids 83–115 (wild type, W89A, Y98A, and W89A/Y98A) and tested these for Piwi interaction. Strikingly, the wild-type peptide efficiently pulled Piwi from nuclear OSC lysate, and the retrieved Piwi amount was even higher compared with a coimmunoprecipitation (co-IP) experiment in which Gtsf1 was immunodepleted (Fig. 5C). The single point mutant peptides were less efficient in this experiment (W>Y), and the double-mutant peptide showed very little Piwi retrieval. Interestingly, while Piwi immunoprecipitations readily show co-IP of endogenous Gtsf1, the peptide immunoprecipitation showed no detectable Gtsf1, strongly arguing that the peptide encompasses that part of Gtsf1 that is required for the Piwi interaction (Fig. 5C). Rescue experiments strongly confirmed the importance of the W89 and Y98 residues, as Gtsf1 constructs with single mutations were weakly impaired in activity (W>Y), while the double-mutant construct was essentially inert in this assay (Fig. 5B; Supplemental Fig. S6B,C).
While we cannot exclude that the Gtsf1 83–115 peptide interacts with Piwi via a third factor, silver staining of the peptide pull-downs coupled to mass spectrometric analysis reveals that the major protein whose retrieval is sensitive to the W/Y residues is Piwi (Fig. 5D). We therefore conclude that Gtsf1 interacts with Piwi most likely directly via a peptide that harbors two conserved aromatic residues embedded in a stretch of negatively charged amino acids.
Gtsf1 levels are considerably lower than Piwi levels, and only a subpool of Piwi is complexed with Gtsf1. We therefore asked whether increasing amounts of the Gtsf1 peptide would enrich increasing Piwi amounts or whether the amount of Piwi that can interact with the Gtsf1 peptide is limited by an unknown factor/process. As increasing peptide amounts recovered increasing amounts of Piwi, we conclude that Gtsf1 in vivo is the limiting factor in the assembly of a Piwi–Gtsf1 complex (Fig. 5E).
Gtsf1 is required for Piwi-guided transcriptional silencing and H3K9me3
Recent evidence indicates that the Piwi-RISC guides transcriptional silencing of piRNA complementary targets in ovarian germline cells and OSCs (Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). Transcriptional silencing is accompanied by H3K9me3 at target transposon insertion sites and their genomic surroundings. Loss of the Piwi-RISC leads to a significant increase in RNA Pol II occupancy on piRNA-repressed TEs and substantial decreases in H3K9me3 levels. We took advantage of the OSC system in which the Piwi-mediated silencing process can be studied at high resolution (Sienski et al. 2012). We measured RNA Pol II occupancy patterns genome-wide using chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) in OSCs knocked down for Gtsf1 and compared this with previously analyzed data sets for GFP control knockdowns and Piwi knockdowns that were prepared in parallel (Sienski et al. 2012). Similar to the RNA sequencing (RNA-seq) analysis (Fig. 2A), the very same set of strongly and intermediately derepressed elements exhibited increased Pol II occupancy in Gtsf1 knockdowns, highly comparable with Piwi knockdowns (Supplemental Fig. S7A). Supplemental Figure S7B depicts the RNA Pol II ChIP-seq patterns of the various knockdowns on the two most strongly derepressed TEs (mdg1 and gypsy) in comparison with a nontargeted TE (Doc).
Also at the level of H3K9me3 marks, Gtsf1 loss or Piwi loss triggered almost identical reductions in this heterochromatic mark on piRNA targeted TEs but not on nontargeted TEs, indicating that Gtsf1 is tightly linked to the piRNA pathway and not a general factor of heterochromatin formation (Supplemental Fig. S7C). We note that also for these experiments, Gtsf1 knockdowns were for only 48 h, while Piwi knockdowns were for 96 h. Loss of Gtsf1 therefore results in a very rapid recruitment of Pol II to piRNA-repressed TE copies and a rapid loss of the H3K9me3 mark.
To extend these findings to an in vivo setting, we depleted Gtsf1 in ovarian somatic or germline cells by tissue-specific RNAi and performed RNA Pol II ChIP-qPCR on chromatin from whole ovaries (Supplemental Fig. S7D). We determined Pol II occupancies at an active control gene (act-5C), a heterochromatic gene (light), “soma-dominant” TEs (ZAM and gypsy), the “germline-dominant” TE HeT-A, and the “intermediate” TE blood (TE classification from Malone et al. [2009]). As predicted and similar to loss of Piwi-RISC (Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013), increases in RNA Pol II occupancy were highly correlated with the TE type and the knockdown tissue, while no impact on the euchromatic or heterochromatic control genes was observed (Supplemental Fig. S7D). Our results strongly indicate that loss of Gtsf1 renders the nuclear Piwi-RISC inactive.
Gtsf1 is required for Piwi-mediated trans-silencing of individual TE insertions
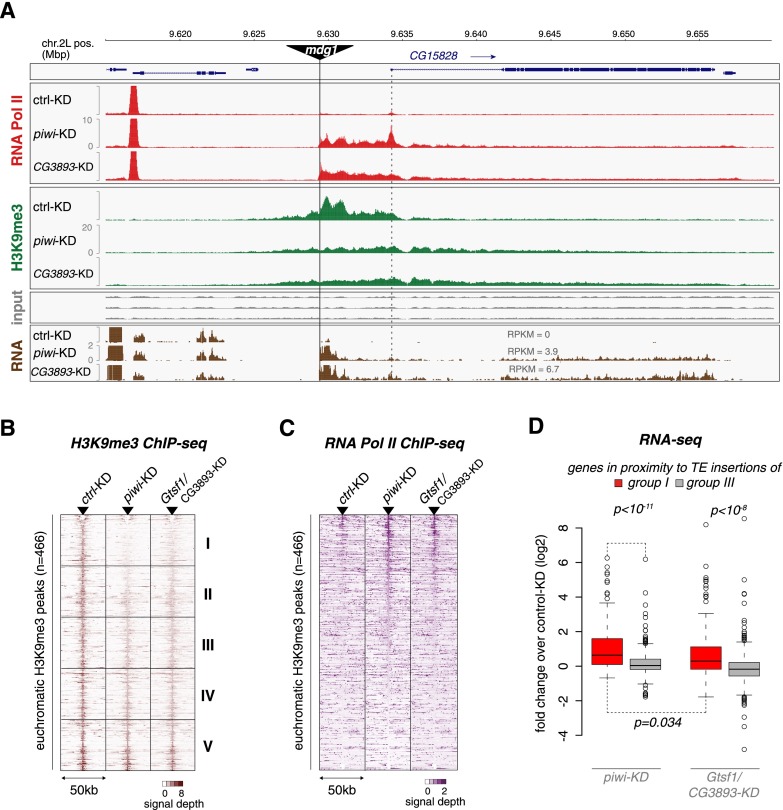
The OSC system permits a very detailed analysis of the Piwi-guided targeting events, as the uniform cell population and knowledge of the precise position of euchromatic TE insertions allow analysis of individual silencing loci at the transcriptional and chromatin level (Sienski et al. 2012). Also at this resolution, knockdown of Gtsf1 accurately mirrored knockdown of Piwi. For example, in both knockdowns, euchromatic mdg1 insertions exhibited pronounced and directional RNA Pol II “bleeding” into flanking genomic regions (example shown in Fig. 6A). In both cases, this was accompanied by a marked decrease in H3K9me3 marks, preferentially downstream from the mdg1 insertion.
Figure 6.
CG3893/Gtsf1 is required for Piwi-dependent H3K9me3 domains and trans-silencing of individual TE insertions. (A) Transcriptional activity (Pol II ChIP-seq), H3K9me3 signal (H3K9me3 ChIP-seq), and steady-state RNA levels (RNA-seq) at the CG15828 locus are shown for OSCs with the indicated knockdowns (input tracks for the respective ChIP experiments are in gray). An OSC-specific mdg1 insertion is indicated at the top; the transcriptional start site (TSS) of CG15828 is indicated with a dashed line. (B,C) Heat maps showing normalized signal of H3K9me3 (B) or Pol II occupancy (C) in windows of 50 kb centered on summits of euchromatic H3K9me3 peaks (n = 466) in control knockdown, piwi knockdown (piwi-KD), or CG3893/Gtsf1 knockdown (CG3893/Gtsf1-KD). For the H3K9me3 heat map, peaks were sorted according to the loss of signal in the 1-kb window around the peak summit in piwi-KD cells; for the Pol II heat map, peaks were sorted according to gain of signal in the 1-kb window around the peak summit in piwi knockdown cells. The H3K9me3 heat map was split into five equally sized bins as in Sienski et al. (2012), and their average signals are shown in Supplemental Fig. S8A. (D) Box plot showing the fold changes (log2) in RNA-seq RPKM values for the set of expressed genes (RPKM > 5 in any of the libraries) with a nearby insertion of a group I TE in sense orientation (red) or a nearby insertion of a group III TE in sense orientation (gray) in piwi knockdown (piwi-KD; left) or CG3893/Gtsf1 knockdown (CG3893/Gtsf1-KD; right) OSCs. P-value was calculated according to Wilcoxon rank-sum test.
At a global scale, we determined levels of H3K9me3 marks as well as RNA Pol II occupancy at euchromatic H3K9me3 domains (defined in wild-type cells) (Sienski et al. 2012) in Piwi knockdown, Gtsf1 knockdown, or control knockdown cells (Fig. 6B,C). Loss of Piwi or Gtsf1 resulted in nearly identical decreases in H3K9me3 marks and increases in RNA Pol II occupancy at these loci, whose summits are highly enriched in piRNA-repressed TE insertions (Fig. 6B,C; Supplemental Fig. S8A depicts density plots for the five bins indicated in Fig. 6B; Sienski et al. 2012).
Finally, while overall mRNA levels were highly correlated between Gtsf1 knockdown cells and control cells (Supplemental Fig. S8B), loss of Gtsf1 led to significantly increased expression of the several dozen genes that are in proximity to Piwi-repressed TE insertions and that have been shown to be derepressed upon Piwi loss (Fig. 6D; Sienski et al. 2012). Two striking examples of the previously defined classes of “repressive chromatin influence” and “promoter addition” are shown in Figure 6A and Supplemental Figure S8C, respectively.
Taken together, our genetic and genome-wide profiling studies indicate that Gtsf1 is required for all known aspects of Piwi-mediated trans-silencing of targets.
Discussion
With the recent demonstration that Piwi-mediated silencing occurs at the transcriptional level, the analogies of this piRNA pathway process to the nuclear fission yeast RNAi pathway where AGO1 guides heterochromatin formation and transcriptional silencing are apparent. In yeast, AGO1 forms a trimeric silencing complex (RITS) (Verdel et al. 2004) with the chromodomain protein Chp1 and the GW protein Tas3 (Moazed 2009; Grewal 2010). RITS interacts with nascent target RNAs and is thought to recruit via Stc1 the Clr4 complex that catalyzes H3K9 methylation to induce heterochromatin formation (Bayne et al. 2010; He et al. 2013).
However, none of the yeast AGO1-interacting factors is conserved in flies or vertebrates, and not a single protein has been described as a core component of a PIWI-guided nuclear silencing complex [but note the proposed interaction of Piwi with Su(var)205] (Brower-Toland et al. 2007; Wang and Elgin 2011). Moreover, unequivocal demonstration that Piwi is recruited to chromatin—likely via interactions with nascent target RNA—is missing (but note Ge and Zamore 2013; Huang et al. 2013). We believe that the identification of Gtsf1 as a Piwi-interacting factor provides key entry points into understanding the composition and molecular function of the transcriptional Piwi silencing complex.
The most significant finding of our study is that the majority of nuclear Gtsf1 is complexed to Piwi, but this corresponds to only a very small fraction of the total nuclear Piwi pool. Given that Gtsf1 is absolutely required for Piwi's silencing function, we envision two major scenarios. Either Gtsf1 interacts transiently with Piwi and licenses it for a downstream silencing process or the Gtsf1–Piwi interaction is regulated in vivo and occurs only upon efficient target interaction by Piwi. In a similar scenario, Gtsf1 might lock Piwi into a target-bound state, potentially via interacting with the target RNA via its zinc finger domains. We note that Piwi cross-linking immunoprecipitation (CLIP) experiments did not result in clear enrichments of piRNA complementary target RNAs (G Sienski, unpubl.). This might indicate that the portion of nuclear Piwi that is engaged in active target silencing is exceedingly small, potentially explaining also why ChIP experiments with Piwi or Gtsf1 have been uninformative (G Sienski, unpubl.). It is also possible that target binding induces a conformational shift in Piwi triggered by dislodging the piRNA 3′ end from the PAZ domain (in analogy to structural insight on bacterial Argonaute proteins) (Wang et al. 2009). This might in turn lead to the buildup of a functional silencing complex. In agreement with this, artificial tethering of Piwi to DNA (Le Thomas et al. 2013) or RNA (D Dönertas, unpubl.) fails to induce target silencing, as tethered Piwi would not be bound to a piRNA complementary target RNA.
Outlook
Central questions for the further dissection of the Piwi silencing complex are the characterization of putative additional factors and their precise molecular functions. We note that in our co-IP experiment, no other prominent band besides Piwi was apparent. Maybe factors actively involved in chromatin modification or transcriptional repression are only weakly bound to the Piwi–Gtsf1 complex and are difficult to purify. In this regard, it will be important to dissect the precise role that Gtsf1 plays in the silencing complex. Gtsf1 either bridges target-bound Piwi to active silencing activities such as histone-modifying enzymes or is an essential factor to lock Piwi into a stable target-bound state. In this context, it will be important to dissect the molecular function of the N-terminal zinc finger domains.
Materials and methods
X-Gal staining
Ovaries from 4- to 5-d-old flies were dissected into ice-cold PBS, fixed in 0.5% glutaraldehyde/PBS (15 min at room temperature), and washed with PBS. Ovaries were incubated with staining solution (10 mM PBS, 1 mM MgCl2, 150 mM NaCl, 3 mM potassium ferricyanide, 3 mM potassium ferrocyanide, 0.1% Triton, 0.1% X-Gal) at room temperature. Stainings were performed overnight for gypsy sensor and HeT-A sensor and 2 h for Burdock sensor.
Transposon qPCR analysis
cDNA was prepared via random priming of 1 μg of total RNA isolated from ovaries of 4- to 5-d-old flies. qPCR was performed using Bio-Rad IQ SYBR Green Super Mix. Each experiment was performed in biological triplicates with technical duplicates. Relative RNA levels were calculated by the 2−ΔΔC(T) method (Livak and Schmittgen 2001) and normalized to rp49 levels. Fold enrichments were calculated in comparison with respective RNA levels obtained from heterozygous flies or from flies not harboring a knockdown hairpin.
Northern blot
Total RNA was isolated from respective knockdowns and separated on a 15% polyacrylamide urea gel. RNA was transferred to Amersham Hybond-NX (RPN303T) membrane and cross-linked by EDC [1-ethyl-3-(3- dimethylaminoprophy) carbodiimide] for 1 h (Pall and Hamilton 2008). The membrane was prehybridized in Church buffer and hybridized to probes overnight at 37°C. The membrane was washed three times for 10 min with 2× SSC and 0.1% SDS and exposed.
Small RNA cloning
Small RNA cloning and sequencing was performed as described in Jayaprakash et al. (2011). In brief, 20 μg of total RNA was isolated from GS12962/GS12962 or GS12962/TM3SbSer ovaries by TRIzol and phenol/chloroform extraction. RNA was resolved on a denaturing polyacrylamide gel, and RNAs corresponding to 18–29 nt were isolated and subjected to ligations of 3′ and 5′ adaptors followed by RT–PCR amplification; libraries were sequenced on the GAII platform (Illumina).
Cell culture
OSCs were cultured as described (Niki et al. 2006) and transfected with Cell Line Nucleofector kit V (Amaxa Biosystems; program T-029). For rescue experiments, siRNA-resistant Gtsf1 was cloned into pAcM vector and cotransfected to OSCs with siRNA targeting endogenous Gtsf1 transcript as described (Klattenhoff et al. 2009; Haase et al. 2010; Olivieri et al. 2010, 2012; Saito et al. 2010; Zamparini et al. 2011).
Antibodies
α-Piwi and α-Armi (rabbit) were described in Olivieri et al. (2010), and mouse α-Piwi was described in Saito et al. (2006). α-Mael was described in Sienski et al. (2012). Mouse monoclonal α-CG3893 was raised against 6xHis-tagged full-length protein. α-Rpb3 was described in Adelman et al. (2005), and α-H3K9me3 was from Abcam (ab8898). Control IgGs were from Abcam: rabbit (ab46540) and mouse (ab18413). α-Myc antibody was from Sigma (M4439).
Immunoprecipitation
OSCs were harvested by trypsinization, and nuclear extract was prepared with Triton lysis buffer (50 mM Tris at pH 7.5, 0.5% Triton, 137.5 mM NaCl, 5 mM EDTA, 10% glycerol). The soluble nuclear fraction was incubated with α-Piwi (rabbit), α-CG3893 (mouse), control IgG (rabbit), or control IgG (mouse) coupled to Protein G Dynabeads (Invitrogen) or biotynylated peptide coupled to Streptavidin T1 Dynabeads (Invitrogen) for 2 h at 4°C and washed with wash buffer (30 mM Hepes at pH 7.5, 500 mM NaCl, 2 mM MgCl2, 10% glycerol, 0.25% Triton, 0.5% NP40). Purified complexes were eluted with SDS sample buffer and analyzed by gel electrophoresis and silver staining or Western blotting.
ChIP and RNA-seq
The ChIP and RNA-seq experiments were done as described in Sienski et al. (2012).
Computational analysis
Everything was done as described in Sienski et al. (2012).
Data access
All Illumina data sets were deposited at Gene Expression Omnibus (data set GSE48317).
Acknowledgments
We thank all Brennecke laboratory members for support and discussions. We thank the Max F. Perutz Laboratories Monoclonal Antibody Facility, particularly Stefan Schuechner and Franziska Mueller for her outstanding work on generating the Gtsf1 antibody; the VDRC and the Drosophila Genomics Resource Center for fly lines; the Campus Science Support Facilities Next-Generation Sequencing unit for Illumina sequencing; Karl Mechtler, Ines Steinmacher, and Richard Imre for mass spectrometry; Mathias Madalinski for synthesis of peptides; the Institute of Molecular Pathology/IMBA bio-optics unit for support; Sarah Lopez for fly injections; Ravi Sachidanandam, Alex Schleiffer, and Daniel Jurczak for bioinformatics support; and Mikiko Siomi for OSCs, α-Piwi, α-Aub, and pAcM plasmid. The Brennecke laboratory is supported by the Austrian Academy of Sciences, the European Community's 7th Framework Programme (FP7/2007-2013; ERC grant no. 260711EU), and the Austrian Science Fund (FWF; Y 510-B12). G.S. is supported by the Boehringer Ingelheim Fonds. All authors planned the experiments. D.D. and G.S. performed all experiments, and G.S. performed the computational analyses. All authors analyzed the data and wrote the paper.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.221150.113.
References
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17: 103–112 [DOI] [PubMed] [Google Scholar]
- Andreeva A, Tidow H 2008. A novel CHHC Zn-finger domain found in spliceosomal proteins and tRNA modifying enzymes. Bioinformatics 24: 2277–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, et al. 2010. Stc1: A critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA 2013. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14: 100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Preall JB, McGinn J, Hannon GJ 2013. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell 50: 749–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge DT, Zamore PD 2013. Small RNA-directed silencing: The fly finds its inner fission yeast? Curr Biol 23: R318–R320 [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Olivieri D, Novatchkova M, Gruber FS, Meixner K, Mechtler K, Stark A, Sachidanandam R, Brennecke J 2011. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J 30: 3977–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J 2013. The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50: 762–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Pillai SS, Taglini F, Li F, Ruan K, Zhang J, Wu J, Shi Y, Bayne EH 2013. Structural analysis of Stc1 provides insights into the coupling of RNAi and chromatin modification. Proc Natl Acad Sci 110: E1879–E1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24: 502–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaprakash AD, Jabado O, Brown BD, Sachidanandam R 2011. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res 39: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, Gvozdev VA 2011. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci 108: 18760–18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Toth KF 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−Δ Δ C(T) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D 2009. Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Guzzardo PM, Gillis J, Luo Y, Yu Y, Chen C, Fekete R, Hannon GJ 2013. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol Cell 50: 736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. 2011. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8: 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y, Yamaguchi T, Mahowald AP 2006. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc Natl Acad Sci 103: 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J 2010. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J 29: 3301–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D, Senti KA, Subramanian S, Sachidanandam R, Brennecke J 2012. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell 47: 954–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ 2008. Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J 2010. The piRNA pathway: A fly's perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpiz S, Olovnikov I, Sergeeva A, Lavrov S, Abramov Y, Savitsky M, Kalmykova A 2011. Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons. Nucleic Acids Res 39: 8703–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R 2007. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SC 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ 2009. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461: 754–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura T, Toyoda S, Kuramochi-Miyagawa S, Miyazaki T, Miyazaki S, Tashiro F, Yamato E, Nakano T, Miyazaki J 2009. Gtsf1/Cue110, a gene encoding a protein with two copies of a CHHC Zn-finger motif, is involved in spermatogenesis and retrotransposon suppression in murine testes. Dev Biol 335: 216–227 [DOI] [PubMed] [Google Scholar]
- Zamparini AL, Davis MY, Malone CD, Vieira E, Zavadil J, Sachidanandam R, Hannon GJ, Lehmann R 2011. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 138: 4039–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]