Accumulation of mutant p53 proteins contributes to tumor development and metastasis, yet little is known abut how to selectively reduce the cellular load of mutant p53. Yuan and colleagues show that suppression of macroautophagy by multiple means promotes the degradation of mutant p53 through chaperone-mediated autophagy. This study provides insights into the regulation of degradation pathways mediating mutant p53 turnover in cancer cells and delineates a strategy for killing nonproliferating tumor cells that express mutant p53.
Keywords: autophagy, cancer, CMA, p53, spautin-1
Abstract
Missense mutations in the gene TP53, which encodes p53, one of the most important tumor suppressors, are common in human cancers. Accumulated mutant p53 proteins are known to actively contribute to tumor development and metastasis. Thus, promoting the removal of mutant p53 proteins in cancer cells may have therapeutic significance. Here we investigated the mechanisms that govern the turnover of mutant p53 in nonproliferating tumor cells using a combination of pharmacological and genetic approaches. We show that suppression of macroautophagy by multiple means promotes the degradation of mutant p53 through chaperone-mediated autophagy in a lysosome-dependent fashion. In addition, depletion of mutant p53 expression due to macroautophagy inhibition sensitizes the death of dormant cancer cells under nonproliferating conditions. Taken together, our results delineate a novel strategy for killing tumor cells that depend on mutant p53 expression by the activation of chaperone-mediated autophagy and potential pharmacological means to reduce the levels of accumulated mutant p53 without the restriction of mutant p53 conformation in quiescent tumor cells.
Mutations in the TP53 gene, encoding the p53 tumor suppressor, are one of the most frequent type of gene-specific alterations in human cancers (Sigal and Rotter 2000). However, unlike most other tumor suppressors, which are frequently lost in cancers due to deletions, >75% of all TP53 mutations in human cancers lead to the expression of full-length proteins with missense substitution mutations (Petitjean et al. 2007; http://www-p53.iarc.fr). While a key consequence of TP53 mutations is the loss of wild-type p53 activity, there is an increased awareness that p53 proteins with missense mutations may carry dominant phenotypes distinct from those caused by the loss of wild-type p53 function. Such phenotypes, described as mutant p53 oncogenic gain of function (GOF), include drug resistance, proproliferative anti-apoptotic functions, increased cell migration, and invasion (Jacks et al. 1994; Olive et al. 2004). In contrast to that of wild-type p53, which under normal conditions is expressed at low levels due to degradation through the ubiquitin proteasome system (UPS), a hallmark of cancer cells with TP53 mutations is the accumulation of high levels of mutant p53 protein (Kupryjanczyk et al. 1993). Accordingly, tumor-specific accumulation of mutant p53 is a critical determinant of its GOF. Thus, reducing the level of mutant p53 proteins represents an attractive anti-cancer strategy; however, pharmacological methods that are currently available are restricted to be selective toward depleting limited mutant p53 variants.
Control of wild-type p53 protein half-life is critical: p53 is rapidly degraded in normal tissue, mediated largely by the activity of the p53-targeting ubiquitin ligase MDM2, but is stabilized in response to stress (Haupt et al. 1997; Kubbutat et al. 1997). Mutant p53 expressed in normal tissues is also kept at low levels through the action of MDM2 (Terzian et al. 2008; Suh et al. 2011), although it often accumulates to high levels in tumor cells (Bartek et al. 1991). Since different mutant p53 alleles may exhibit certain unique characteristics and interact with a wide range of different proteins (Muller and Vousden 2013), multiple mechanisms might be involved to lead to the accumulation of mutant p53 proteins. One proposal is that tumor-associated stress may provoke the association of mutant p53 with molecular chaperones, such as Hsc70 and Hsp90, which leads to the stabilization of mutant p53 (Hinds et al. 1990; Whitesell et al. 1998). Consistently, inhibition of Hsp90 has been shown to promote the degradation of certain mutant p53 proteins (Li et al. 2011a,b). In addition, mutant p53 has been reported to misfold and form amyloid oligomers and fibrils (Ano Bom et al. 2012), which might be intrinsically resistant to proteasomal degradation. Indeed, interaction of mutant p53 with chaperone proteins such as Hsp70 has been shown to inhibit its ubiquitination, mediated by MDM2 and proteasomal degradation, and promote its aggregation (Wiech et al. 2012). Thus, how to effectively promote the degradation of mutant p53 in cancer cells is an important question to be investigated.
Chaperone-mediated autophagy (CMA) is a selective mechanism for degradation of protein through a lysosomal-dependent mechanism. Basal CMA activity is evident in most cells but is maximally stimulated in response to cellular stress, such as nutrient deprivation (Cuervo et al. 1995). Thus, CMA might contribute to degradation of proteins that are no longer needed under stress conditions and allow recycling to promote cell survival. Cross-talks exist among autophagy pathways and the UPS, as blockage of one pathway can lead to activation of the other (Massey et al. 2006; Kaushik et al. 2008). Cells respond to blockage of the proteasome by up-regulating macroautophagy, whereas inhibition of macroautophagy under nutritional deprivation conditions has been shown to activate CMA (Kaushik et al. 2008). However, the cellular pathway and physiological importance of CMA in cancers are currently not well defined.
In the present study, we explored the molecular mechanism of mutant p53 degradation under various growth conditions. We evaluated the significance of different cellular degradation pathways in mediating mutant p53 turnover in cancer cells cultured under confluent conditions when cells are not proliferating. We show that multiple alleles of endogenous mutant p53 cannot be effectively ubiquitinated, and, consistently, inhibition of proteasome fails to block the degradation of mutant p53 proteins in relevant cancer cells tested. Interestingly and unexpectedly, inhibition of autophagy by multiple means promotes the degradation of mutant p53. Compared with that of wild-type p53, mutant p53 degradation in confluent nonproliferating cancer cells is not mediated through the UPS. Instead, we found that mutant p53 is degraded through a lysosomal-dependent pathway involving CMA. Autophagy inhibition under cellular stress conditions when CMA is induced profoundly induces the degradation of mutant p53. Furthermore, we provide evidence that the pharmacological inhibitors of autophagy selectively induce cytotoxicity toward cancer cells with mutant p53 under nonproliferating conditions. Taken together, our study delineates a novel strategy for targeting nonproliferating cancer cells that express mutant p53.
Results
Inhibition of autophagy induces the death of select cancer cells with mutant p53 under confluent conditions
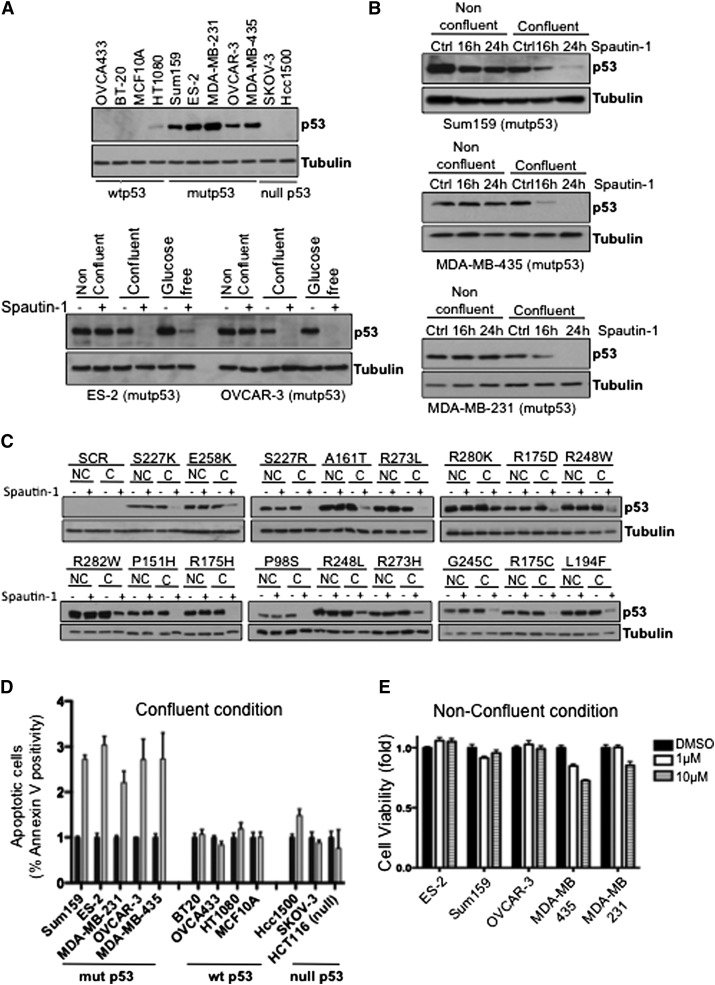
We previously developed a small molecule inhibitor of autophagy, named spautin-1 (Liu et al. 2011), which selectively inhibits the ubiquitin-specific peptidases USP10 and USP13 and blocks the deubiquitination of Vps34 complexes that leads to constitutive ubiquitination and subsequent proteasomal degradation of class III PI3 kinase (PI3K) complexes, resulting in potent inhibition of autophagy. Since USP10 is a deubiquitinating enzyme for p53 (Yuan et al. 2010) and inhibition of USP10 by spautin-1 promotes the degradation of wild-type p53 (Liu et al. 2011), we investigated whether spautin-1 also could lead to degradation of accumulated mutant p53 proteins. We compared the effect of spautin-1 on randomly selected human cancer cell lines with different mutational p53 status that either expressed wild-type or mutant p53 proteins or was null in p53 expression (Fig. 1A, top panel) under various growth conditions. A significant decrease in the mutant p53 protein levels in OVCAR-3 [p53R248Q] and ES-2 [p53S241F] cell lines was observed upon spautin-1 treatment under glucose-free and, interestingly, confluent conditions (Fig. 1A, bottom panel) when cells are not proliferating compared with that of dispersed (nonconfluent) growth conditions (Supplemental Fig. S1A). Similar results were also observed upon spautin-1 treatment in a time-dependent manner using the mutant p53-expressing breast Sum159 [p53R158InF] and MDA-MB-231 [p53R280L] and melanoma MDA-MB-435 [p53G266Q] cancer cell lines (Fig. 1B). The decrease in mutant p53 levels occurred faster in the presence of the translation inhibitor cycloheximide (CHX) (Supplemental Fig. S1B,C), while no change in the mRNA levels of p53 was found upon spautin-1 treatment regardless of growth conditions (Supplemental Fig. S1D), suggesting that the reduction in the levels of mutant p53 is due to an increase in degradation.
Figure 1.
Spautin-1 induces the death of select cancer cells with mutant p53. (A) Western blots showing the p53 expression of the untreated indicated cancer cell lines (top panel) and spautin-1-treated ovarian ES-2 and OVCAR-3 cancer cells in different growth conditions (bottom panel). Cells were treated for 6 h or 24 h during glucose-free or nonconfluent/confluent conditions, respectively. (B) Western blots of p53 levels in spautin-1-treated mutant p53 cancer cell lines under nonconfluent and confluent conditions at the indicated time points. (C) Western blots of the indicated 17 different p53 mutants expressed in HCT116 p53-null cells, followed by treatment of spautin-1 in nonconfluent (NC) and confluent (C) conditions for 24 h. (D) The apoptotic cells indicated as the percentage of Annexin V positivity of the indicated cancer cells treated with spautin-1 for 24 h in confluent conditions measured by FACS analysis. (E) Cell viability analysis of the indicated mutant p53-expressing cancer cell lines treated with different concentrations of spautin-1 for 24 h in nonconfluent conditions using ATP assay. Anti-α-tubulin antibody was used as a loading control for all Western blots. (Control) Vehicle (DMSO) alone. The error bars indicate SD.
In order to further confirm the effect of spautin-1 on mutant p53 protein expression, we tested 17 different cancer-associated mutant p53 variants (each characterized by a single amino acid substitution) by transient expression into p53-null HCT116 cells. All 17 variants of mutant p53 proteins were significantly reduced following spautin-1 treatment in confluent conditions (Fig. 1C).
We previously reported that spautin-1 had no effect on the growth and survival of Bcap-37 cells under normal culture conditions (Liu et al. 2011) but enhanced the cell death in glucose-free conditions that induced autophagy. Similarly, we found that spautin-1 induced the death of the ovarian cancer ES-2 and OVCAR-3 cells in glucose-free conditions (Supplemental Fig. S1E), whereas SKOV-3 cells (null in p53 expression) were resistant to spautin-1 during the same conditions. Thus, we next assessed the cell viability and cell death by multiple assays using actively proliferating cells or when cells reached confluency. Remarkably, spautin-1 selectively induced cell death of mutant p53-expressing cancer cell lines under confluency but not in dispersed growth conditions. No or little effect of spautin-1 on cell survival was detected in wild-type p53 or p53-null cancer cells (Fig. 1D,E; Supplemental Fig. S1F). In contrast to spautin-1, cisplatin, a conventional chemotherapeutic agent currently used in clinics, can induce the death of ES-2 cells under nonconfluent conditions but is ineffective under confluent conditions even at high concentrations (Supplemental Fig. S1G). However, when used in combination, spautin-1 can increase the efficacy of cisplatin under both confluent and nonconfluent conditions (Supplemental Fig. S1G).
To understand the long-term impact of spautin-1 treatment on cancer cell survival, we assayed the clonogenicity of select ovarian cancer cells. While spautin-1 had no effect on the clonogenic survival of SKOV-3 cells, a strong correlation between decreased clonogenic survival of mutant p53-expressing ES-2 and OVCAR-3 cells with increasing spautin-1 dosage was observed (Supplemental Fig. S1H). Taken together, these results demonstrate that spautin-1 induces cytotoxicity toward mutant p53-expressing cancer cells and concomitantly reduces the levels of mutant p53 protein expression under nonproliferative conditions.
Sensitization of missense mutant p53 expression to cell death induced by spautin-1
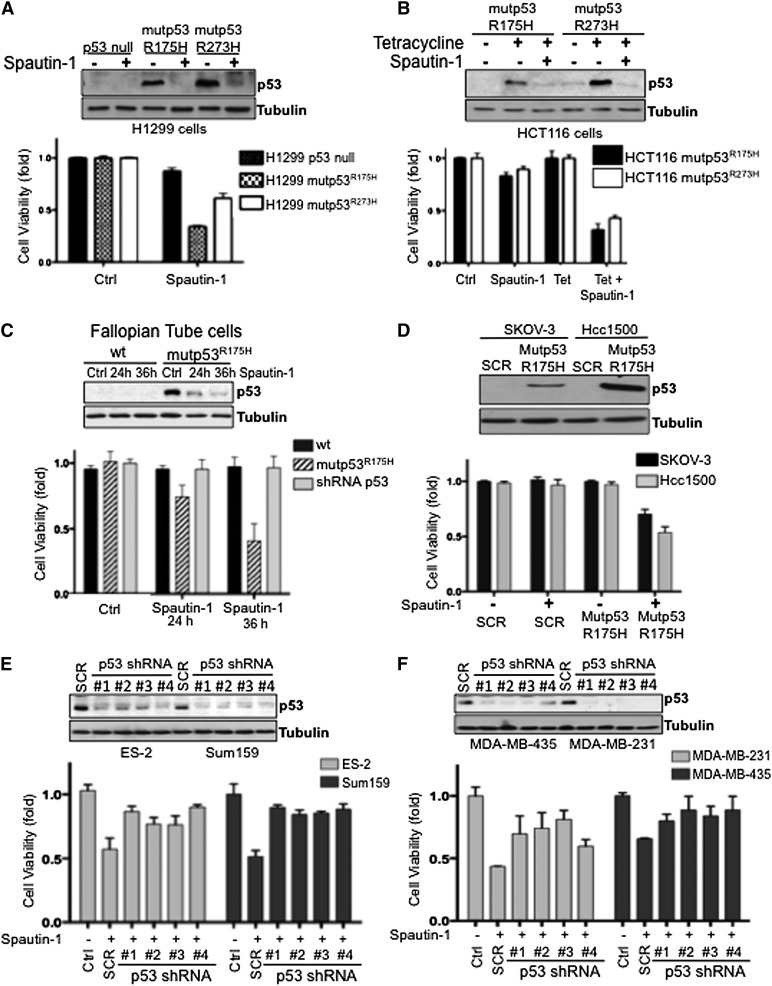
To further analyze the impact of mutant p53 expression on cell death induced by spautin-1, we next employed a set of isogenic clones of lung H1299 stably expressing R175H or R273H p53 mutants (Muller et al. 2009) and colon HCT116 cancer cell lines inducibly expressing mutant p53. Since H1299 cells are null in their p53 expression, it excludes the possibility that mutant p53 acts as a dominant-negative inhibitor on wild-type p53. Wild-type p53 was not included in this experimental set, as its expression in H1299 cells results in cell cycle arrest and apoptosis. Consistently, stable expression of mutant p53 protein sensitized nonproliferating H1299 cells to spautin-1 (Fig. 2A). In addition, isogenic clones of p53-null HCT116 cancer cells expressing the inducible R175H or R273H p53 mutant were characterized with respect to their sensitivity to spautin-1. A pronounced decrease in spautin-1-induced cell survival was detected when mutant p53 expression was induced under confluency (Fig. 2B). Again, both R175H and R273H mutant p53 proteins were degraded following spautin-1 treatment as assessed by Western blot analysis (Fig. 2A,B, top panels). To further examine whether mutant p53 expression directly conveys the sensitivity of cells toward spautin-1, we established normal human primary epithelial fallopian tube (FT) cells that either stably expressed R175H p53 mutant or in which the endogenous wild-type p53 levels were silenced by shRNA (Supplemental Fig. S2A,B). Spautin-1 specifically induced cell death of FT cells expressing mutant p53 and decreased the mutant p53 levels during confluent and glucose-free conditions (Fig. 2C; Supplemental Fig. S2C). In contrast, spautin-1 had no effect on wild-type FT cells or on FT cells in which endogenous p53 levels were depleted.
Figure 2.
The expression of mutant p53 is required for spautin-1 to sensitize cancer cells to cell death. (A) The cell viability and Western blots of p53 in p53-null and stable mutant p53-expressing (R175H or R273H) H1299 cells in the absence or presence of spautin-1. (B) The cell viability and Western blots of p53 in HCT116 cells expressing inducible mutant p53 (R175H or R273H) with or without tetracycline in the absence or presence of spautin-1. (C, top panel) Western blots of p53 of spautin-1-treated FT cells in confluent conditions. (Bottom panel) Cell viability of FT cells treated with spautin-1 in confluent conditions. (D, top panel) Western blots of p53 in SKOV-3 and Hcc1500 cells overexpressing Scrambled (SCR) or mutant p53 R175H protein. (Bottom panel;) Cell viability of Scramble (SCR) or mutant p53 R175H-expressing SKOV-3 and Hcc1500 cells treated with spautin-1. (E,F, top panels) Western blots of p53 in indicated mutant p53-expressing cells transfected with Scramble (SCR) or p53 shRNA. (Bottom panels) Cell survival of Scramble (SCR) or p53 shRNA transfected cells treated with spautin-1 under confluent conditions. Anti-α-tubulin antibody was used as a loading control for all Western blots. The cell viability was measured by ATP assay. (Control) Vehicle (DMSO) alone. The error bars indicate SD.
We also tested the effect of spautin-1 in p53-null SKOV-3 and Hcc1500 cells that were manipulated to express mutant p53 R175H. Clearly, R175H p53 expression sensitized the cells to the cytotoxic effect of spauitin-1 in confluent conditions (Fig. 2D). Conversely, silencing the expression of p53 mutant protein by shRNA prior to spautin-1 treatment markedly reduced the sensitivity of nonproliferating ES-2, Sum159, MDA-MB-231, and MDA-MB-435 cells to spautin-1 (Fig. 2E,F). These data demonstrate that spautin-1 reduces the level of mutant p53 to specifically induce cell death of a panel of nonproliferating cancer and normal cells with accumulated mutant p53 and demonstrates the sensitization effect of mutant p53 protein expression for spautin-1-mediated cytotoxicity.
Inhibition of macroautophagy and the proteasome promotes the degradation of mutant p53
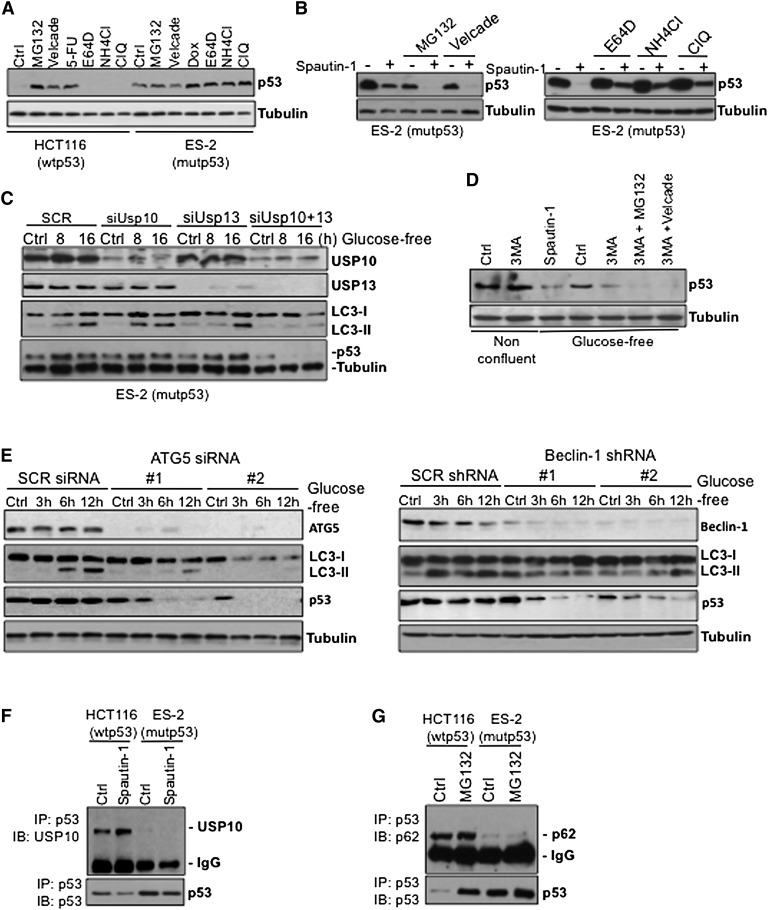
Since inhibition of USP10 promotes the degradation of wild-type p53 (Liu et al. 2011), we tested whether spautin-1-mediated degradation of mutant p53 would similarly depend on the inhibition of the deubiquitinating proteins. Wild-type p53, normally kept at low levels due to its constitutive ubiquitination and proteasomal degradation mediated by MDM2, accumulated to higher levels upon inhibition of proteasome by MG132; on the other hand, MG132 had no effect on the abundance of mutant p53 protein under nonconfluent conditions (Supplemental Fig. S3A) with or without 5-flurouracil (5-FU) or doxorubicin (Dox), two DNA-damaging agents clinically used for the treatment of colon or ovarian carcinoma (Supplemental Fig. S3A).
While the proteasome inhibitors MG132 and bortezomib (Velcade) had no effect on the levels of mutant p53 protein, cathepsin inhibitor E64d and lysosomal inhibitors ammonium chloride (NH4Cl) and chloroquine (ClQ) further stabilized mutant p53 levels in ES-2 cells, suggesting that mutant p53 might be continuously degraded through the lysosomal pathway even under nonconfluent growth conditions (Fig. 3A). Furthermore, we found that mutant p53 levels became even lower when pretreated with MG132 or Velcade prior to spautin-1 under confluent conditions (Fig. 3B, left panel). Inhibition of proteasome alone was sufficient to reduce the levels of mutant p53 compared with that of control under confluency. Conversely, pretreatment with NH4Cl, E64d, or ClQ partially attenuated spautin-1-induced mutant p53 degradation in ES-2 cells under the same conditions (Fig. 3B, right panel). Similar results were also obtained using Sum159 and MDA-MB-435 cell lines (Supplemental Fig. S3B).
Figure 3.
Inhibition of autophagy induces degradation of mutant p53. (A) Western blot of p53 in HCT116 and ES-2 cells in nonconfluent growth conditions treated with the indicated proteasomal and lysosomal inhibitors. (B) Western blots of p53 of spautin-1-treated confluent ES-2 cells in the absence or presence of proteasome inhibitors, MG132, or Velcade (left panel) or lysosome inhibitors, E64D, NH4Cl, or ClQ (right panel) for 24 h. (C) Western blots of USP10, USP13, LC3, and p53 of ES-2 cells transfected with USP10 and USP13 siRNAs alone or in combination and treated with spautin-1 under glucose-free conditions at the indicted time points 48 h post-transfection. (D) p53 levels of ES-2 cells treated with proteasome inhibitors, MG132, or Velcade in the absence or presence of 3-MA for 3 h under either normal or glucose-free conditions. (E) Western blots of ATG5, Beclin-1, LC3, and p53 of Scramble (SCR) in ATG5 siRNA (left panel) and Beclin-1 shRNA (right panel) transfected ES-2 cells treated with spautin-1 for up to 12 h in glucose-free conditions. (F) DMSO- or spautin-1-treated HCT116 and ES-2 cell lysates were subjected to immunoprecipitation using p53 antibody and immunoblotted with USP10 antibody. (G) DMSO- or MG132-treated HCT116 and ES-2 cell lysates were subjected to immunoprecipitation using p53 antibody and immunoblotted with p62 antibody. Anti-α-tubulin antibody was used as a loading control for all Western blots.
Since inhibition of USP10 and USP13 by spautin-1 promotes the degradation of p53, RNAi silencing of USP10 and USP13 is sufficient to degrade wild-type p53 under normal growth conditions (Supplemental Fig. S3C, left panel; Liu et al. 2011). However, siRNA-mediated knockdown of USP10 or USP13 was not sufficient to affect the levels of mutant p53 under the same conditions (Supplemental Fig. S3C, right panel). Thus, we studied the effect of siRNA-mediated knockdown of USP10 or USP13 on mutant p53 levels during glucose-free conditions. While the knockdown of either USP10 or USP13 alone did not affect the levels of LC3-II conversion observed in glucose-free conditions, cosuppression of USP10 and USP13 in ES-2 cells inhibited autophagy and induced degradation of mutant p53 (Fig. 3C). Prolonged starvation of these cells promoted the degradation of mutant p53 levels in a time-dependent manner. Since coknockdown of USP10 and USP13 is more effective in inhibiting autophagy than either single knockdown (Fig. 3C), our data suggest that a complete inhibition of autophagy is required to reduce the levels of mutant p53. To further assess the contribution of autophagy inhibition in mutant p53 degradation, we treated ES-2 cells with 3-methyladenine (3-MA), a class III PI3K inhibitor that blocks autophagosome formation. Mutant p53 was markedly decreased following treatment with 3-MA in a fashion similar to spautin-1 (Fig. 3D). In contrast, 3-MA had no effect on wild-type p53-expressing cells (Liu et al. 2011). In addition, in ES-2 cells, the knockdown of either ATG5 or Beclin-1, essential autophagy-related genes, led to decreased levels of mutant p53 and cell death in a time-dependent manner under glucose-free conditions (Fig. 3E; Supplemental Fig. S3D–F). The effect of ATG5 knockdown was further tested in p53-null SKOV-3 cells expressing mutant p53 R175H. Mutant p53 expression and cell viability were markedly decreased under glucose-free conditions with efficient ATG5 knockdown in a fashion similar to spautin-1 (Supplemental Fig. S3G). Combined, these data demonstrate that both pharmacological inhibition of macroautophagy and genetic removal of critical autophagy genes stimulate the degradation of mutant p53 protein in nutritient-deprived conditions and during inhibition of the proteasomes.
Next, we investigated the interaction of mutant p53 with USP10. While wild-type p53 immunoprecipitates with USP10 as reported (Yuan et al. 2010), mutant p53 failed to interact with USP10 in ES-2 and Sum159 cells (Fig. 3F; Supplemental Fig. S3H). In addition, wild-type but not mutant p53 interacted with the ubiquitin-binding protein p62 following MG132 treatment (Fig. 3G), further suggesting that spautin-1 may induce mutant p53 degradation in an ubiquitin–proteasome pathway-independent manner. These results indicate that deubiquitinating enzymes coordinately regulate the cellular levels of wild-type p53, whereas spautin-1-induced mutant p53 degradation occurs independently of the ubiquitin–proteasome pathway but may depend on regulation of autophagy. Therefore, we conclude that the mechanisms of spautin-1-induced degradation of wild-type and mutant p53 must be distinct.
CMA targets accumulated missense mutant p53 proteins to lysosomal degradation
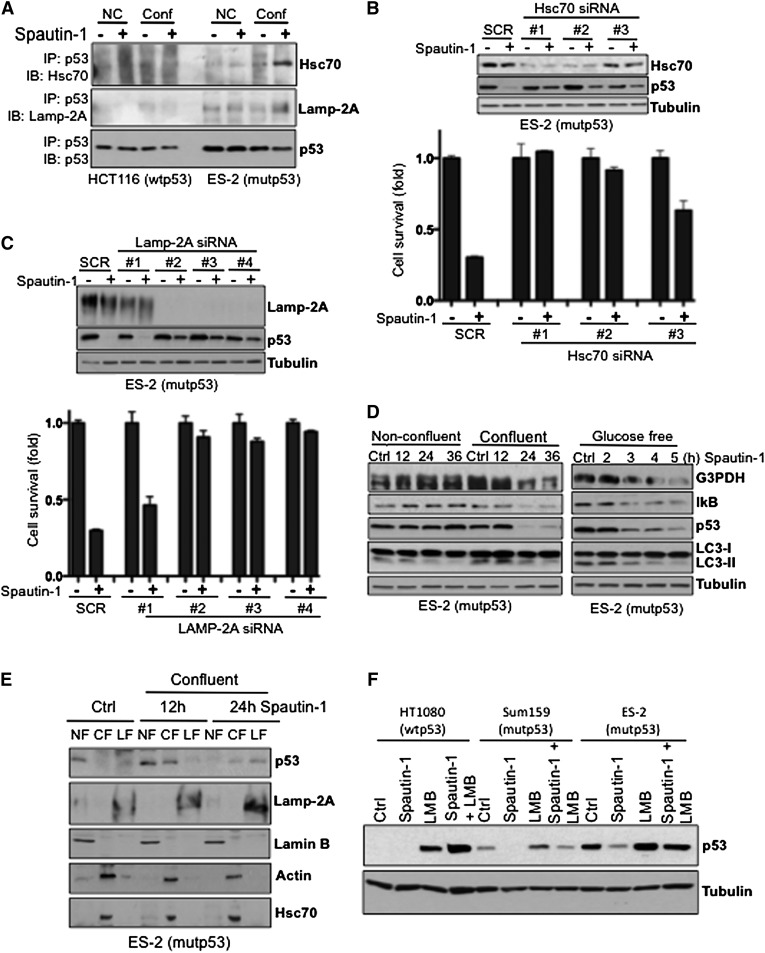
While the UPS and macroautophagy are the two critical pathways that mediate the turnover of proteins, inhibition of these protein degradation pathways can lead to the activation of CMA (Massey et al. 2006; Kaushik et al. 2008). CMA is activated under nutrient-deprived conditions, during which individual cytosolic proteins are rerouted for lysosomal degradation (Cuervo 2011). Thus, we hypothesized that mutant p53 degradation might occur via the CMA pathway when macroautophagy is inhibited by spautin-1. A cytosolic heat-shock cognate protein of 70 kDa (Hsc70) plays an essential role in CMA by recognizing KFERQ-like sequence motifs in substrate proteins. Indeed, p53 harbors two pentapeptide sequences (200NLRVE204 and 341FRELN345) that are consistent with an Hsc70 recognition motif. Our structural analysis reveals that the 200NLRVE204 motif is located on the surface of the p53 protein and is exposed, while the FRELN motif is present on the linker region and is not shown to the p53 structure (Supplemental Fig. S4A). The lysosome-associated membrane protein type 2a (Lamp-2A) is an integral membrane protein that serves as a receptor for the selective uptake and degradation of proteins by lysosomes during CMA. Mutant but not wild-type p53 bound both Hsc70 and LAMP-2A, as revealed by coimmunoprecipitation experiments (Fig. 4A). This interaction was especially observed in spautin-1-treated samples under confluency. Consistent with a possible role of CMA in mediating the degradation of mutant p53, siRNA silencing of either Hsc70 or LAMP-2A in ES-2 cells attenuated spautin-1-induced mutant p53 depletion and cell death. siRNA efficiency correlated with the amount of rescued mutant p53 levels, which also correlated with cell survival (Fig. 4B,C).
Figure 4.
CMA targets mutant p53 to lysosomal degradation. (A) HCT116 and ES-2 cells treated with spautin-1 for 16 h in either confluent or nonconfluent conditions were subjected to immunoprecipitation using antibody against p53, followed by Hsc70 and LAMP-2A Western blot. (B, top panel) Western blots of Hsc70 and p53 of Scramble (SCR) or Hsc70 siRNA transfected ES-2 cells for 48 h and treated with spautin-1 for an additional 24 h in confluent conditions. (Bottom panel) Cell viability of Scramble (SCR) or Hsc70 siRNA transfected ES-2 cells treated with spautin-1 for 24 h in confluent conditions. (C, top panel) Western blots of LAMP-2A and p53 of Scramble (SCR) or LAMP-2A siRNA transfected ES-2 cells for 48 h and treated with spautin-1 for an additional 24 h in confluent conditions. (Bottom panel) Cell viability of Scramble (SCR) or LAMP-2A siRNA transfected ES-2 cells treated with spautin-1 for 24 h in confluent conditions. (D) Western blots of G3PDH, IκB, p53, and LC3 of ES-2 cells treated with spautin-1 for the indicated time points under nonconfluent and confluent (left panel) or glucose-free (right panel) conditions. (E) Western blots of p53, Lamp-2A, Lamin B, actin, and Hsc70 showing the localization in different subcellular fractions of ES-2 cells treated with spautin-1 for the indicated time points under confluent conditions. (F) Western blot of p53 of the indicated cell lines treated with LMB or spautin-1 alone or in combination for 24 h under confluent conditions. Anti-α-tubulin antibody was used as a loading control for all Western blots. The cell viability was measured by ATP assay. (Control) Vehicle (DMSO) alone. The error bars indicate SD.
Consistent with the activation of CMA, the depletion of mutant p53 upon spautin-1 treatment under glucose-free and confluent conditions was accompanied with the degradation profile of G3PDH and IκB, the two known substrates specifically degraded by CMA (Fig. 4D; Aniento et al. 1993; Cuervo et al. 1998), supporting the hypothesis regarding the activation of the CMA pathway and degradation of mutant p53 via this pathway upon spautin-1 treatment. To further evaluate the degradation of mutant p53 by its potential uptake into the lysosomes during CMA, we assessed the accumulation of mutant p53 protein in the lysosomes by immunoblotting (Kaushik and Cuervo 2009). We pretreated cells with E64d, a cathepsin (lysosomal protease) inhibitor, prior to spautin-1, since any lysosome internalized substrate is rapidly degraded by the luminal proteases. While mutant p53 proteins are primarily nuclear, our subcellular fraction experiments revealed that under glucose deprivation or confluency, spautin-1 treatment decreased mutant p53 protein in the nuclear fraction (NF) and increased the cytosolic fraction (CF) levels of mutant p53 proteins prior to its uptake into the lysosome-enriched fraction (LF) (Fig. 4E; Supplemental Fig. S4B). As shown in Figure 4E and Supplemental Figure S4B, the amount of mutant p53 was higher in the LF of spautin-1-treated cells compared with the LF from control cells. In addition, the accumulation of the well-known CMA substrates IκB and G3PDH (Cuervo et al. 1998) from the homogenate fraction into the LF was observed upon spautin-1 treatment (Supplemental Fig. S4B) This translocation occurred concomitant with mutant p53 translocation into the LF following spautin-1 treatment. While no increase in the Hsc70 and Lamp-2A levels was detected upon spautin-1 treatment in the whole-cell lysates (Supplemental Fig. S4C), an increase of Lamp-2A levels in the LF was detected following spautin-1 treatment (Fig. 4E; Supplemental Fig. S4B). Taken together, these results confirm the higher proteolysis rates of CMA substrates observed in lysosomes of spautin-1-treated cells.
Culture density has been shown to regulate Hsc70 nuclear transport, with cells in high-density culture being unable to accumulate Hsc70 proteins in nuclei (Chu et al. 2001). Our results demonstrate that regardless of spautin-1 treatment, Hsc70 maintains its cytosolic localization during confluent conditions in ES-2 and Sum159 cancer cells (Fig. 4E; Supplemental Fig. S4D,E). Accordingly, mutant p53 is able to interact with Hsc70 solely under confluent conditions when a nuclear to cytosolic shift in the mutant p53 levels is observed upon spautin-1 treatment (Supplemental Fig. S4D,E). In order to further test the role of the nuclear export of mutant p53, we treated cells with leptomycin B (LMB), which blocks the formation of nuclear export complexes consisting of CRM1, RanGTP, and NES-containing proteins. The addition of LMB, a nuclear export inhibitor, to both wild-type p53 HCT116 and mutant p53-expressing cell lines increases the cellular levels of the p53 proteins, which localize in the cell nucleus (Fig. 4F; Supplemental Fig. S4F). Furthermore, the effect of spautin-1 on mutant p53 was diminished upon cotreatment with LMB (Fig. 4F), suggesting that nuclear export is required for the efficient degradation of mutant p53 upon spautin-1 treatment.
Taken together, we conclude that inhibition of autophagy in nonproliferating confluent cancer cells leads to increased cytosolic levels of mutant p53 proteins that might permit Hsc70 to recognize their KFERQ-like sequence motifs, which in turn promotes the degradation of mutant p53 in a lysosome-dependent manner.
Susceptibility to ubiquitination determines the degradation pathways of wild-type and mutant p53 proteins
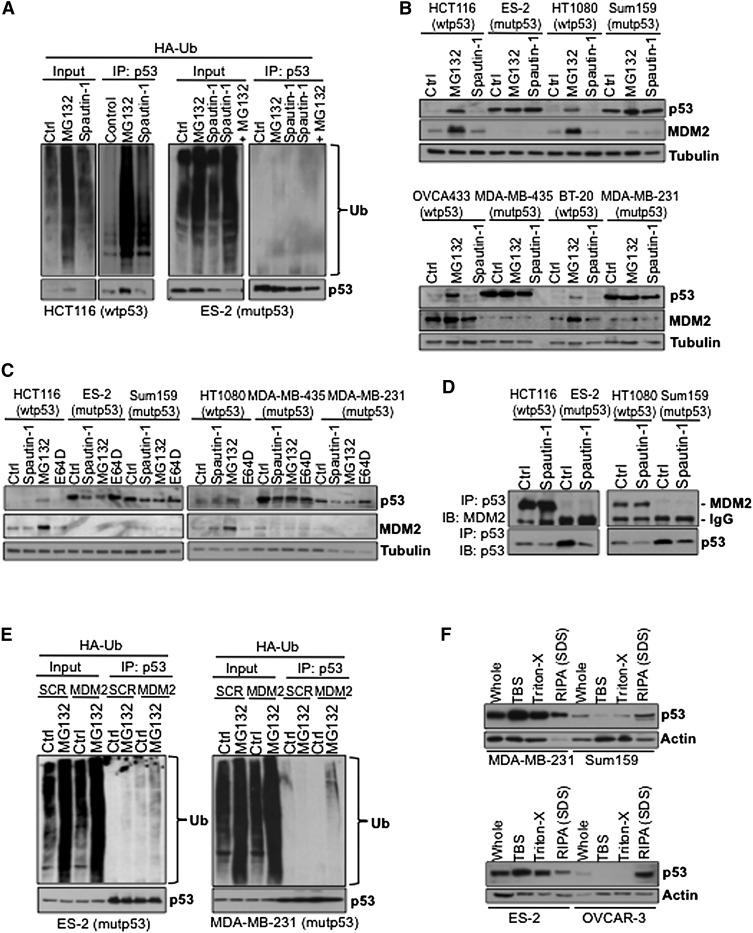
In order to understand why mutant p53 proteins are degraded through the lysosomal pathway under confluent conditions while wild-type p53 proteins are subjected to proteasomal degradation upon spautin-1 treatment, we investigated the susceptibility of mutant p53 proteins to ubiquitination. Consistent with its mode of inhibiting USP10, a deubiquitinase regulating the ubiquitination of p53, spautin-1 treatment led to the accumulation of ubiquitinated wild-type p53 in HCT116, HT1080, and OVCA433 cells, similar to treatment with MG132 (Fig. 5A; Supplemental Fig. S5A). In ES-2, Sum159, MDA-MB-231, and OVCAR-3 cells, however, mutant p53 protein was not ubiquitinated regardless of the presence or absence of MG132 or spautin-1 (Fig. 5A; Supplemental Fig. S5A), suggesting that certain alleles of mutant p53 might display resistance to UPS-dependent protein degradation.
Figure 5.
Spautin-1 induces mutant p53 degraded independently of the ubiquitin–proteasome pathway. (A) HA-ubiquitin transfected HCT116 and ES-2 cells treated with MG132 or spautin-1 cells for 24 h in confluent conditions. Cell lysates were subjected to immunoprecipitation using antibody against p53, followed by ubiquitin Western blot. (B) Western blots of p53 and Mdm2 of the indicated MG132- or spautin-1-treated cell lines under nonconfluent conditions. (C) Western blots of p53 and Mdm2 of the indicated MG132-, E64D-, or spautin-1-treated cell lines under confluent conditions. (D) The indicated DMSO- or spautin-1-treated cell lysates were subjected to immunoprecipitation using p53 antibody and immunoblotted with Mdm2 antibody. (E) The indicated HA-ubiquitin transfected mutant p53 cells overexpressing Mdm2 were treated with MG132 for 24 h in confluent conditions. Cell lysates were subjected to immunoprecipitation using antibody against p53, followed by ubiquitin Western blot. (F) Western blots showing the solubility of mutant p53 and actin in the indicated mutant p53-expressing cell lines treated with increasing detergent concentrations. Anti-α-tubulin antibody was used as a loading control for all Western blots.
As seen in Figure 5B and Supplemental Figure S5B, while the protein levels of MDM2 were increased in all wild-type p53 cells upon MG132 treatment, no change can be seen in any of the mutant p53-expressing cell lines in nonconfluent conditions. Spautin-1 treatment had no effect on the expression levels of MDM2 in any of the wild-type p53 cancer cell lines in nonconfluent conditions. Similarly, the expression levels of MDM2 remained unaffected in all wild-type and mutant p53 cancer cell lines upon spautin-1 or E64D treatments also during confluency (Fig. 5C; Supplemental Fig. S5C), which makes it unlikely that the effect of spautin-1 in promoting the degradation mutant p53 levels is mediated through MDM2. MDM2 knockdown prevents spautin-1-induced degradation of wild-type p53 in H4-LC3-GFP cells (Liu et al. 2011) yet failed to affect spautin-1-induced mutant p53 degradation in ES-2 and Sum159 cells (Supplemental Fig. S5D). The ability of MDM2 siRNA to knock down MDM2 was shown in a wild-type p53- and MDM2-expressing HCT116 cell line (Supplemental Fig. S5E).
As previously reported, MDM2 associated with wild-type p53-expressing HCT116 and HT1080 cells, while no interaction of MDM2 with mutant p53 could be detected in ES-2 and Sum159 cells (Fig. 5D). The mutant TP53 alleles expressed in ES-2, OVCAR-3, and MDA-MB-231 harbor point mutations that directly disturb the DNA-binding ability of p53 protein, categorized as so-called contact p53 mutants (Supplemental Fig. S5E), whereas MDA-MB-435 and Sum159 cells express mutant TP53 alleles that disturb the wild-type confirmation of the p53 protein, categorized as structural p53 mutants (Supplemental Fig. S5F). Regardless of their mutational status, in these cell lines, the mutant p53 proteins have lost their function to transcriptionally activate the MDM2 gene (Supplemental Fig. S5G), which explains why no or little expression of MDM2 was observed. Furthermore, we compared the MDM2 levels in HCT116 cell lines that either express wild-type, inducible R175H, or R275H mutant proteins or are null in their p53 expression. Again, the expression of both the mRNA and protein levels of MDM2 was only induced in wild-type HCT116 cell lines in the presence of MG132 or 5-FU, while no increase in the expression levels of MDM2 was detected in the p53-null or the mutant p53-expressing HCT116 cells upon tetracycline treatment (Supplemental Fig. S5H). These data indicate that an impairment of MDM2 expression might be a factor for the lack of ubiquitination of mutant p53 in cancer cells.
Next, we examined the susceptibility of mutant p53 to MDM2-mediated ubiquitination by ectopic overexpression of MDM2 in mutant p53-harboring cancer cells. However, in only two mutant p53 cell lines—MDA-MB-231 and ES-2—a modest accumulation of ubiquitinated mutant p53 could be detected upon expression of MDM2 (Fig. 5E). In OVCAR-3 and Sum159 cells, mutant p53 remained unaffected, and ubiquitination was undetectable even after MG132 treatment (Supplemental Fig. S5I). This suggests that factors other than MDM2 inactivation might play a role in the inability of mutant p53 to be ubiquitinated in cancer cells.
It has recently been shown that mutant p53 proteins may form amyloid-like aggregates in cancer cells (Ano Bom et al. 2012), and the ubiquitination and proteasomal degradation of mutant p53 is inhibited by chaperone proteins that promote their aggregation (Wiech et al. 2012). Since different alleles of mutant p53 showed a different propensity to form amyloid-like structures (Ano Bom et al. 2012), we hypothesize that the aggregation of mutant p53 may show allele specificity, which may affect its ubiquitination. To test this hypothesis, we analyzed the detergent solubility of endogenous mutant p53 proteins in the mutant p53-expressing cell lines. Using a differential buffer extraction assay, we found that mutant p53 proteins in ES-2 and MDA-MD-231 cells can be more easily extracted by buffer with gentler detergent than in Sum159 and OVCAR3 cell lines (Fig. 5F). Since mutant p53 proteins in Sum159 and OVCAR3 cells are more resistant to MDM2-mediated ubiquitination, these results suggest that differential degrees of mutant p53 protein misfolding, their binding partners, and their aggregation status may affect their susceptibility to ubiquitination and determine their degradation pathway. Regardless of their mutational status, however, a portion of mutant p53 in different cancer cells can only be extracted by RIPA buffer, which contains 1% SDS, suggesting the presence of a subset of mutant p53 proteins that may be highly misfolded and therefore are resistant to ubiquitination and degradation through the proteasomal systems.
Discussion
In this study, we describe how inhibition of autophagy by a small molecule tool compound, spautin-1, as well as by genetic knockdown of autophagy-related genes can promote the degradation of accumulated missense mutant p53 proteins through the CMA pathway in nonproliferating cancer cells. We present a strategy that might be applied to reduce the load of mutant p53 proteins without the restriction of mutant p53 conformation. In addition, since cancer cells with missense TP53 mutations display high-energy demands, they appear particularly sensitive to inhibition of autophagy when cultured under nutrient-deprived and nonproliferating conditions. Our findings suggest that degradation of mutant p53 is specifically mediated by the CMA–lysosome pathway during stress conditions and denotes the involvement of CMA in a unique pathway to induce mutant p53 expression-dependent cell death.
Our data suggest that wild-type and mutant p53 proteins differ in their sensitivity to ubiquitination and deubiquitination. While the direct inhibition of USP10 by spautin-1 leads to ubiquitination and degradation of wild-type p53 through the proteasome (Liu et al. 2011), the inhibition of USP10 and USP13 by spautin-1 in mutant p53-expressing cells indirectly regulates the degradation of mutant p53 by inhibiting autophagy. Degradation of accumulated misfolded and aggregated proteins, such as mutant Tau and mutant Huntingtin (Htt) protein, has been demonstrated to occur through CMA (Wang et al. 2009; Bauer et al. 2010), while wild-type Htt is targeted to the UPS. In addition, while degradation of proteins such as the regulator of calcineurin 1 (RCAN1), a gene identified from the critical region of Down syndrome, is mediated by the UPS, in conditions where macroautophagy is reduced, RCAN1 is targeted for degradation through CMA, suggesting that a protein may be degraded through alternative pathways under different physiological conditions (Liu et al. 2009).
It has been reported that prolonged glucose deprivation can lead to the degradation of mutant p53 through macroautophagy (Rodriguez et al. 2012). However, prolonged glucose starvation has been shown to lead to reduction of autophagy (Fuertes et al. 2003). On the other hand, CMA is activated after long-term nutritient deprivation in confluent cultured cells and certain tissues of fasted animals (Dice 2007). Basal CMA activity can be detected in most types of mammalian cells, but the maximal activation of this pathway is triggered in response to cellular stress, such as nutrient deprivation (Cuervo 2011). A reduction in macroautophagy has been shown to lead to a concomitant and progressive increase in CMA activity (Kaushik et al. 2008). Our study suggests that normal levels of macroautophagy are rather ineffective in the removal of mutant p53, as evidenced by their accumulation; on the other hand, the activation of CMA due to inhibition of macroautophagy by multiple means leads to effective removal of mutant p53, suggesting the difference in substrate selectivity between macroautophagy and CMA. Our result also suggests that the interaction of Hsc70 with mutant p53 is important for mediating their degradation through CMA: Accumulated mutant p53 proteins are largely localized in the nucleus and do not interact with Hsc70 under normal growth conditions. Spautin-1 treatment under confluency or nutritient deprivation conditions appears to facilitate the nuclear–cytosolic translocation of mutant p53 proteins, which may promote its recognition by and interaction with Hsc70, which leads to its uptake by the lysosome.
Our study proposes that in addition to inactivation of MDM2 and CHIP leading to impairment of mutant p53 degradation through the UPS (Li et al. 2011a,b), the mutational status of p53 also may play a determinant role in its ability to be ubiquitinated. It has been shown that mutant R248Q p53 proteins are found in an amyloid-like aggregate state in breast cancer samples (Ano Bom et al. 2012). In addition, the R273H mutants display a higher proportion to form amyloid-like aggregates than the R175 mutants. This suggests that different mutant p53 alleles may determine its aggregation status, which in turn may affect its susceptibility to ubiquitination. Consistently, a recent study demonstrated that the expression of Hsp70 in cancer cells mediates the stabilization of mutant p53 and protects against MDM2-mediated ubiquitination by promoting mutant p53 aggregation (Wiech et al. 2012). Since inhibition of Hsp90 has been shown to promote the degradation of certain mutant p53 proteins (Li et al. 2011a,b), our results are consistent with the proposal that the association of mutant p53 with molecular chaperones under normal growth conditions renders their resistance to proteasomal degradation, and we show a new pathway that can promote the degradation of mutant p53 without inhibiting molecular chaperones upon CMA activation under confluency and starvation conditions when autophagy is inhibited.
Altogether, our findings suggest that the regulation of the stability of mutant p53 differs from that of wild-type p53 and identify the possibility of activating the CMA pathway by inhibiting autophagy to promote the degradation of mutant p53 under nutrient deprivation and confluent conditions. However, the spautin-1 class of compounds shows cardiac toxicity (data not shown), which makes it unsuitable for in vivo studies; however, since inhibition of autophagy in general under confluent conditions can promote the degradation of mutant p53, we propose that the development of pharmacological agents that can inhibit autophagy and mimic confluent conditions might be able to selectively activate the CMA pathway for the removal of mutant p53 as a novel anti-cancer therapeutic strategy.
Materials and methods
Cell lines and culture conditions
The ovarian ES-2, OVCAR3, OVCA433, and SKOV-3; breast BT20 and HCC1500; and fibrosarcoma HT1080 cancer cell lines were cultured in RPMI (10% fetal bovine serum [FBS]), 1% penicillin/streptomycin). The SUM159 breast cancer cell line was cultured in Ham's F12 (5% FBS, 1% penicillin/streptomycin, 5 mg/mL insulin, 2 mg/mL hydrocortisone). The HEK293T, the breast cancer MDA-MB-231, the melanoma MDA-MB-435, all H1299 cell lines, and the null and inducible mutant p53-expressing HCT116 cell lines [pcDNA4-Flag-HA-p53(R175H) or pcDNA4-Flag-HA-p53(R273H)] were cultured in Dulbecco's modified Eagle's medium (DMEM; 10% FBS, 1% penicillin/streptomycin). The isolation of primary normal human FT cell lines and the culture medium was performed and provided by Dr. Tan A. Ince. All cells were grown in a humidified 5% CO2 atmosphere at 37°C and maintained in a logarithmic growth phase. For experiments performed in glucose-free conditions, we used DMEM and RPMI cell culture media with no glucose with dialyzed FBS and 1% penicillin/streptomycin. For experiments performed under normal or glucose-free conditions, cells were plated and treated in nonconfluent conditions. For nonproliferative experiments, cells were plated and treated in contact-inhibited confluent conditions. Cell proliferation measurement was performed using the CellTrace CFSE cell proliferation kit by flow cytometry.
Chemical compounds and treatments
Throughout the experiments (if not stated otherwise), cells were treated with 10 μM spautin-1 for the indicated time points for cell viability/cell death analysis and Western blotting. In selected samples, 5 μM CHX , 20 nM LMB, 500 μM DNA-damaging agent 5-FU, and 1.2 μg/μL doxorubucin were used. In some samples, 10 μM MG132, 0.1–1 μM Velcade, 2 mM 3-MA, 5 μg/μL E64D, 20 mM NH4Cl, and 10 μM ClQ were added 30 min before spautin-1 treatment. To induce the expression of mutant p53 R175H or R273H proteins in HCT116 p53-null cells (which express a tetracycline repressor by pcDNA6), cells were treated with 0.2 μg/mL tetracycline.
Cell viability was measured using CellTiterGlo ATP assay (Promega), and cellular apoptosis was detected by Annexin V/PI staining using a flow cytometer using FITC AnnexinV apoptosis detection kit I (BD Pharmingen). The error bars indicate standard deviation (SD). Data are presented as fold change over control (DMSO-treated) cells and are an average of at least three independent experiments or are representative of independent experiments. For differential solubility experiments, cells were sequentially centrifuged, supernatants were removed, and pellets were resuspended in harsher buffer conditions. Initially, cells were homogenized in Tris-buffered saline (TBS) using a microhomogenizer, and the pellet was suspended in TBS containing 1% Triton. The final pellet was resuspended in modified RIPA buffer. Equal protein amounts were used for Western blot analysis.
shRNAs, siRNAs, and plasmids
siRNA targeting Mdm2, LAMP2A, Hsc70, and ATG5 were purchased from Dharmacon, whereas Scramble, USP10, and USP13 siRNA were purchased from GenePharma. Forty nanomolar siRNA was used, and the transfection was performed using Lipofectamine RNAiMAX reagents according to the manufacturer's instructions. shRNA targeting ATG5 and Beclin-1 were kind gifts from Dr. Quan Chen. shRNA against p53 were purchased from Dharmacon. Scramble control or shRNA plasmids were transfected into 293T cells to produce the virus using FuGENE HD transfection reagent (Promega) according to the manufacturer's instructions. Viral supernatant was collected, concentrated, and added to cells. The level of expression of siRNA or shRNA was monitored 48 h post-transfection by Western blotting.
Transient transfections were performed with Lipofectamine 2000 reagent (Invitrogen), as recommended by the manufacturer, and plasmid-driven protein expression for 24 h before further experimental procedures. Cells were cotransfected with a plasmid for the expression of GFP. An empty vector, pcDNA3.1 (Invitrogen), was used as a negative control. All p53 mutant vectors exhibited the following single amino acid substitutions: P98S, P151H, A161T, R175C, R175D, R175H, L194F, S227K, S227R, G245C, R248L, R248W, E258K, R273H, R273L, R280K, and R282W. The pcDNA3 MDM2 wild-type (Addgene) HA-ubiquitin was obtained from Dr. Alfred Goldberg.
Subcellular fractionation
Subcellular fractions of cultured cells were obtained using dounce homogenization and sequential centrifugation by centrifugations. Isolation of the NF and CF was carried out using Qproteome Cell Compartment kit (Qiagen). Lysosomes were isolated using the Lysosome Enrichment kit (Pierce) according to the kit protocol.
Acknowledgments
We are grateful for the kind gifts from Dr. Karen Vousden (H1299 cells), Dr. Xinbin Chen (inducible mutant p53 HCT116 cell lines), and Dr. Quan Chen (shRNA targeting ATG5 and Beclin-1). This work was supported by a collaborative grant from Roche. H.V.N. was supported in part by a fellowship from the Swedish Society for Medical Research (SSMF). G.K. is supported by the Ligue Nationale contre le Cancer (Equipe labellisée), Agence Nationale pour la Recherche (ANR), AXA Chair for Longevity Reseach, European Commission (ChemoRes), Institut National du Cancer, Cancéropôle Ile-de-France, Fondation Bettencourt-Schueller, and LabEx Onco-Immunology.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.220897.113.
References
- Aniento F, Roche E, Cuervo AM, Knecht E 1993. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J Biol Chem 268: 10463–10470 [PubMed] [Google Scholar]
- Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, Gava LM, Ramos CH, Cepeda AO, Stumbo AC, et al. 2012. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J Biol Chem 287: 28152–28162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Bartkova J, Vojtesek B, Staskova Z, Lukas J, Rejthar A, Kovarik J, Midgley CA, Gannon JV, Lane DP 1991. Aberrant expression of the p53 oncoprotein is a common feature of a wide spectrum of human malignancies. Oncogene 6: 1699–1703 [PubMed] [Google Scholar]
- Bauer PO, Goswami A, Wong HK, Okuno M, Kurosawa M, Yamada M, Miyazaki H, Matsumoto G, Kino Y, Nagai Y, et al. 2010. Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat Biotechnol 28: 256–263 [DOI] [PubMed] [Google Scholar]
- Chu A, Matusiewicz N, Stochaj U 2001. Heat-induced nuclear accumulation of hsc70s is regulated by phosphorylation and inhibited in confluent cells. FASEB J 15: 1478–1480 [DOI] [PubMed] [Google Scholar]
- Cuervo AM 2011. Chaperone-mediated autophagy: Dice's ‘wild’ idea about lysosomal selectivity. Nat Rev Mol Cell Biol 12: 535–541 [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Knecht E, Terlecky SR, Dice JF 1995. Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am J Physiol 269: C1200–C1208 [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Hu W, Lim B, Dice JF 1998. IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol Biol Cell 9: 1995–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice JF 2007. Chaperone-mediated autophagy. Autophagy 3: 295–299 [DOI] [PubMed] [Google Scholar]
- Fuertes G, Martin De Llano JJ, Villarroya A, Rivett AJ, Knecht E 2003. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J 375: 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Hinds PW, Finlay CA, Quartin RS, Baker SJ, Fearon ER, Vogelstein B, Levine AJ 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: A comparison of the ‘hot spot’ mutant phenotypes. Cell Growth Differ 1: 571–580 [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM 2009. Methods to monitor chaperone-mediated autophagy. Methods Enzymol 452: 297–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Mizushima N, Cuervo AM 2008. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell 19: 2179–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Kupryjanczyk J, Thor AD, Beauchamp R, Merritt V, Edgerton SM, Bell DA, Yandell DW 1993. p53 gene mutations and protein accumulation in human ovarian cancer. Proc Natl Acad Sci 90: 4961–4965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Moll UM 2011a. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ 18: 1904–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, Moll UM 2011b. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 9: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang P, Song W, Sun X 2009. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J 23: 3383–3392 [DOI] [PubMed] [Google Scholar]
- Liu J, Xia HG, Kim M, Xu L, Li Y, Zhang L, Yu C, Norberg-Vakifahmetoglu H, Zhang T, Furuya T, et al. 2011. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM 2006. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci 103: 5805–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH 2013. p53 mutations in cancer. Nat Cell Biol 15: 2–8 [DOI] [PubMed] [Google Scholar]
- Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, et al. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell 139: 1327–1341 [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119: 847–860 [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629 [DOI] [PubMed] [Google Scholar]
- Rodriguez OC, Choudhury S, Kolukula V, Vietsch EE, Catania J, Preet A, Reynoso K, Bargonetti J, Wellstein A, Albanese C, et al. 2012. Dietary downregulation of mutant p53 levels via glucose restriction: Mechanisms and implications for tumor therapy. Cell Cycle 11: 4436–4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Rotter V 2000. Oncogenic mutations of the p53 tumor suppressor: The demons of the guardian of the genome. Cancer Res 60: 6788–6793 [PubMed] [Google Scholar]
- Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK, Van Pelt C, Terzian T, Lozano G 2011. Multiple stress signals activate mutant p53 in vivo. Cancer Res 71: 7168–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G 2008. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 22: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E 2009. Tau fragmentation, aggregation and clearance: The dual role of lysosomal processing. Hum Mol Genet 18: 4153–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Sutphin PD, Pulcini EJ, Martinez JD, Cook PH 1998. The physical association of multiple molecular chaperone proteins with mutant p53 is altered by geldanamycin, an hsp90-binding agent. Mol Cell Biol 18: 1517–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiech M, Olszewski MB, Tracz-Gaszewska Z, Wawrzynow B, Zylicz M, Zylicz A 2012. Molecular mechanism of mutant p53 stabilization: The role of HSP70 and MDM2. PLoS ONE 7: e51426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z 2010. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140: 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]