Abstract
Bamboo mosaic virus (BaMV) is a single-stranded positive-sense RNA virus. One of the plant glutathione S-transferase (GST) genes, NbGSTU4, responds as an upregulated gene in Nicotiana benthamiana post BaMV infection.
In order to identify the role of NbGSTU4 in BaMV infection, the expression of NbGSTU4 was knocked down using a virus-induced gene silencing technique or was transiently expressed in N. benthamiana in BaMV inoculation.
The results show a significant decrease in BaMV RNA accumulation when the expression level of NbGSTU4 is reduced; whereas the viral RNA accumulation increases when NbGSTU4 is transiently expressed. Furthermore, this study identified that the involvement of NbGSTU4 in viral RNA accumulation occurs by its participation in the viral early replication step. The findings show that the NbGSTU4 protein expressed from Escherichia coli can interact with the 3′ untranslated region (UTR) of the BaMV RNA in vitro in the presence of glutathione (GSH). The addition of GSH in the in vitro replication assay shows an enhancement of minus-strand but not plus-strand RNA synthesis.
The results suggest that the plant GST protein plays a role in binding viral RNA and delivering GSH to the replication complex to create a reduced condition for BaMV minus-strand RNA synthesis.
Keywords: Bamboo mosaic virus (BaMV), glutathione (GSH), glutathione S-transferase (GST), in vitro RNA replication, redox, viral RNA replication, virus-induced gene silencing (VIGS)
Introduction
Glutathione S-transferases (GSTs) are grouped into a large family of both eukaryotes and prokaryotes and catalyze a variety of reactions (Sheehan et al., 2001; Dixon et al., 2002; Allocati et al., 2009). Conventionally, GSTs catalyze the transfer of a reduced tripeptide glutathione (GSH; glutamyl-cysteinyl-glycine) to various substrates containing a reactive electrophilic center, to form a polar S-glutathionylated product for reducing oxidative stress (Dixon et al., 2002). Plant GSTs were first recognized in 1970, and have been proven to play a role in the detoxification occurring in herbicide injuries (Frear & Swanson 1970). These plant GSTs are classified into eight distinct groups: phi, tau, theta, zeta, lambda, DHAR, TCHQD and microsomal (Edwards & Dixon, 2005). As more plant genomes are sequenced, the list of GSTs continues to grow. In Arabidopsis, 48 GST-like genes were grouped into 28 tau, 13 phi, 3 theta, 2 zeta and 2 lambda GSTs (Dixon et al., 2002); 42 maize GST-like genes were grouped into 12 phi, 28 tau and 2 zeta GSTs; and 25 soybean GST-like genes were grouped into 20 tau, 1 zeta and 4 phi GSTs (McGonigle et al., 2000). The members of the phi and tau family are specific to plants and are the most abundant. They are induced by various treatments that induce general oxidative stress, such as osmotic stress, temperature stress and chemical toxins (Mauch & Dudler, 1993; Marrs, 1996). Furthermore, the role of tau GSTs has been characterized in the tolerance to cold temperature and the oxidative stress that occurs when they are overexpressed in tobacco seedlings (Roxas et al., 1997). Similar responses were also observed in yeast (Kampranis et al., 2000; Kilili et al., 2004). These observations suggest that the tau GSTs can play a significant role in protecting plants against oxidative stress. Moreover, three tau GSTs (NbGSTU1, NbGSTU2 and NbGSTU3) and one phi GST (NbGSTF1) from Nicotiana benthamiana plants were examined for their roles in fungal infections (Dean et al., 2005). The expression levels of NbGSTU1 and NbGSTU3 were found to be upregulated post-infection, although those of NbGSTU2 and NbGSTF1 were unaffected. Further analysis revealed that only the NbGSTU1-knockdown plant showed more lesions compared to the others when inoculated with Colletotrichum orbiculare. These findings suggest that, although a relatively large number of different GSTs are present in plants, only a few are involved in disease development (Dean et al., 2005).
Bamboo mosaic virus (BaMV) is a single-stranded positive-sense RNA virus. An RNA genome of c. 6.4 kb with a 5′-cap and a 3′-poly(A) tail contains five open-reading frames (Lin et al., 1992, 1994). ORF1 encodes a 155 kDa replicase for viral RNA replication (Li et al., 2001; Huang et al., 2004). ORF2 to ORF4 encode movement proteins that participate in intra- and intercellular movement of the virus (Lin et al., 2004, 2006). ORF5 encodes the capsid protein for virus encapsidation. Potexvirus coat protein is also involved in virus movement (Cruz et al., 1998). The 3′ untranslated region (UTR) of BaMV has been structurally mapped (Cheng & Tsai, 1999); it contains cis-acting elements for the initiation of minus-strand RNA synthesis (Cheng & Tsai, 1999; Huang et al., 2001; Cheng et al., 2002), viral RNA long-distance movement (Chen et al., 2003), and the polyadenylation of plus-strand RNA (Chen et al., 2005). Furthermore, the host factor chloroplast phosphoglycerate kinase has been reported to interact with the 3′ UTR, and is essential for efficient BaMV accumulation in plants (Lin et al., 2007). A few differentially expressed genes from N. benthamiana plants infected with BaMV were recently identified in viral infection cycles (Cheng et al., 2010). One of these cDNA fragments had similarities to a GST from N. tabacum (Cheng et al., 2010).
This study is involved in a cloning of the full-length new tau group GTS gene NbGSTU4 from the N. benthamiana plant. NbGSTU4 not only binds to the 3′ UTR of the BaMV RNA, but also enhances the viral RNA replication in vitro. This study details the role of the NbGSTU4 in BaMV infection cycle.
Materials and Methods
Plasmid construction
An ACCA3 cDNA fragment (200 bp) screened from cDNA-AFLP (Cheng et al., 2010) showing a 94% sequence identity to C-7 mRNA sequence of N. tabacum (accession number X64399 in NCBI database) was cloned into pTRV2 using EcoRI site to generate pTRV2/ACCA3 for virus-induced gene silencing (VIGS). The primer set designed for cloning the coding region of the ortholog gene from N. benthamiana is based on N. tabacum C-7 mRNA sequence. The forward primer is Nb_C-7/BamHI/F, 5′-GGATCCATGGCTGACGAAGTTGTCC-3′ (BamHI site is underlined); and the reverse primer is Nb_C-7/XhoI/STOP/R, 5′-GTTACTCGAGGGCAATGCCCCACTTTTG-3′ (XhoI site is underlined). The PCR product was cloned into pGEM-T easy vector and sequenced. The restriction sites BamHI/SalI and BamHI/XhoI flanking the coding region were used for cloning to pBImGFP and pGEX-41 for plant and E. coli expression, respectively. For pNbGSTU4/S13A construction, the mutant fragment was amplified from pNbGSTU4 using BamHI-NbGSTU4+1, 5′-GGATCCATGGCTGACGAAGTTGTCCT-3′, and NbGSTU4-54 mutant, 5′-CCTCATCCCAAACATGGCTACATAGGTATCCAA-3′, to synthesize a 60-bp fragment which was than used as a megaprimer for a second PCR with primer mGFP-31, 5′-GTTCTTCTCCTTTACTAGTCAGATC-3′. The amplified DNA fragment was gel purified and cloned into pGEM-T easy for sequencing. A correct recombinant clone was digested with BamHI and SpeI, and inserted into the corresponding region of pNbGSTU4. For measuring the knockdown efficiency of various NbGSTs in plants, we used real-time PCR with the primer sets: NbGSTU1 (F: 5′-GATGGCAGAAGTGAAGTTG-3′ and R: 5′-GCTCCTAGCCAAAATSCCA-3′); NbGSTF1 (F: 5′-GGCTTCAAGATTAACCTGGGA-3′ and R: 5′-GCCAARATATCAGCACACC-3′); NbGSTU2 (F: 5′-GTAGGCATAAAACCAGCTGTAGT-3′ and R: 5′-GTGAGTACATTGAWGAAGTTTGG-3′); and NbGSTU3 (F: 5′-GCATAAGAATAAAACCAACTAGTAA-3′ and R: 5′-GTGAGTACATTGAWGAAGTTTGG-3′).
Virus-induced gene silencing (VIGS)
The control plasmid, pTRV2/Luc, containing a portion of Luciferase gene was constructed. Plasmids pTRV2/Luc and pTRV2/ACCA3 (the 200 bp derived from NbGSTU4) were transformed into the Agrobacterium tumefaciens C58C1 strain by electroporation. To knock down the expression level of NbGSTU4 in N. benthamiana, the A. tumefaciens C58C1 containing pTRV1, pTRV2/Luc or pTRV2/ACCA3 was cultured to OD600 = 1 at 30°C before induction by the addition of 130 μM acetosyringone in 10 mM MgCl2 for 3 h at room temperature. The pTRV2/Luc- or pTRV2/ACCA3-containing A. tumefaciens C58C1 was mixed with pTRV1-containing A. tumefaciens C58C1 at a 1 : 1 volume ratio. The 1st and 2nd leaves were infiltrated with the mixed broth at the four-leaf stage (seedlings with two cotyledons and two leaves). Then 200 ng of BaMV, PVX, CMV or TMV virion RNA was inoculated onto the 6th leaf when it was mature. Total RNAs and proteins were extracted from the leaves at 3 dpi and measured for the mRNA and viral coat protein levels, respectively. For the protoplast inoculation assay, protoplasts prepared from the 6th leaf were transfected with 1 μg of BaMV virion RNA. The levels of NbGSTU4 mRNA and viral coat proteins were measured at 24 h post-inoculation.
Protoplast inoculation and viral RNA quantification
Four grams of sliced N. benthamiana leaves were digested with 25 ml of enzyme solution (0.1% bovine serum albumin, 0.6 mg ml−1 pectinase and 12 mg ml−1 cellulase in 0.55 M Mannitol/2-(N-morpholino)ethanesulphonic acid pH 5.7) at 25°C overnight. The mesophyll cells were isolated and transfected with 2 μg of virion RNA (c. 4 × 105 cells for each sample) with the help of polyethyleneglycol. Finally, the transfected protoplasts were incubated under constant light (15 μmol m−2 s−1) at 25°C for 24 or 48 h. For Northern blotting analysis, the total RNA was extracted from protoplasts (2.5 × 105 cells), glyoxalated, electrophoresed through a 1% agarose gel and transferred to a Zeta-Probe membrane (Bio-Rad), as described previously (Tsai et al., 1999). The hybridization probe, a 0.6-kb 32P-labelled RNA transcript derived from the HindIII-linearized pBaMV-O/SB2.6 (Huang & Tsai, 1998), is complementary to the 3′-end of positive-strand BaMV RNA. The banding signals were scanned and quantified using a PhosphorImager (BAS-2500; Fujifilm, Tokyo, Japan).
The qRT-PCR technique was used to detect both BaMV plus- and minus-strand genomic RNAs. The cDNA synthesis reaction was performed according to the manufacturer's instructions using ImProm-II™ Reverse Transcriptase (Promega) with the primers Oligo dT(25T), and BaMV+1 (5′-GAAAACCACTCCAAACGAAA-3′) for the plus- and minus-strand, respectively. qPCR for quantifying the accumulation of BaMV genomic RNA and minus-strand RNA, primers BaMV+1766 (5′-CACATCCGGCACTTACCA-3′) and BaMV-2002 (5′-ATGTATCACGGAAATAAGAGTT-3′) were used in the reaction containing a 1000× dilution of SYBR green I (Cambrex Bio Science Rockland Inc., Rockland, ME, USA). qPCR was performed in 0.2-ml PCR tubes with 0.6 mM primer, 0.2 mM of each deoxynucleoside triphosphate, 10 mM Tris-HCl pH 8.8, 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 2 μl of cDNA, 3 units of Taq DNA polymerase (Promega) and RNase-free water to a final volume of 20 μl. Cycling conditions began with an initial hold at 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 30 s. Reactions were carried out in a RotorGene 3000 (Corbett Research, Sydney, Australia) with data acquisition at 72°C on the channel with excitation at 470 nm and detection at 585 nm using a high-pass filter for both plus- and minus-strand. All samples were run at least twice, and the reaction without template or reverse transcriptase was performed as a negative control, and actin gene were taken for normalization.
UV-crosslinking
The purified proteins (GST and GST-NbGSTU4) after dialysis against the buffer containing 20 mM Tris-HCl pH 7.6, 20 mM NaCl, 2 mM DTT, 5% glycerol were incubated with labeled r138/40A (the 3′ UTR of BaMV) or Ba-77 (the 3′-end of BaMV minus-strand) individually for 20 min on ice in a binding buffer (20 mM Tris-HCl pH 7.6, 2 mM MgCl2, 50 mM KCl, 2 mM DTT, 5% glycerol), and irradiated with a 254-nm-wavelength UV lamp (Stratagene, LaJolla, CA, USA, UV stratalinkerTM 1800) on ice for 10 min. After irradiation, the samples were treated with 40 μg of RNase A for 30 min at 37°C, boiled in 1X SDS sample buffer, and electrophoresed on a 12% or 14% sodium dodecylsulphate polyacrylamide gel. GSH was added at a different concentration (50, or 100 mM) in the UV-crosslinking reaction.
In vitro RdRp assay
The replication complex of BaMV used for an in vitro RdRp assay was purified from infected N. benthamiana as described previously (Cheng et al., 2001; Lin et al., 2005). To analyze the exogenous template activity, the replication complex of BaMV was 1.5% NP40-solublize- and micrococcal nuclease-treated. The RdRp assay was carried out in a 50-μl reaction containing 5 μl of 1.5% NP40-solublized RdRp fraction, 2 mM each of ATP, CTP, and GTP, 2 μM UTP, 0.066 μM [α-32P]UTP (3000 Ci mmol−1, Dupont-NEN), 4.8 mg ml−1 of bentonite, 10 mM dithiothreitol, 3 mM MgCl2, and 50 ng of RNA template and incubated at 30°C water bath for 1 h. The RNA products were extracted with phenol-chloroform and precipitated with ethanol. For test the effects of different redox agents, 50 mM of GSH, GSSG, DTT, and H2O2 were added in the reaction. Each reagent was dissolved in 50 mM Tris buffer and adjusted pH value to 7.0 before used.
Results
NbGSTU4 is required for efficient BaMV replication
The fragment of cDNA of a GST homolog upregulated in N. benthamiana following BaMV infection was identified by using the cDNA-AFLP technique in our previous study (Cheng et al., 2010). An experiment using the Tobacco rattle virus (TRV)-based virus-induced gene silencing (VIGS) technique (Lin et al., 2007) to knock down the expression of this gene (designated hereafter as NbGSTU4) in N. benthamiana showed that the accumulation levels of the BaMV coat protein in the knockdown plants was reduced to 51% of that of the control plants. A similar reduction level was also found in another member of the Potexvirus, Potato virus X (PVX) (Fig. 1a,b). However, no significant interference occurred in the accumulation of Cucumber mosaic virus (CMV) and Tobacco mosaic virus (TMV) when inoculated into the knockdown plants (Fig. 1a,b). The knockdown efficiency of this gene was also measured by quantitative real-time PCR, and was shown to be c. 34% of that of the control plants (Fig. 1c). These results suggest that this GST-homolog is involved in the infection cycle of the Potexvirus. To investigate further, the full-length coding sequence of this N. benthamiana GST homolog was amplified by RT-PCR using the primers designed according to the C-7 cDNA sequence from N. tabacum. Four clones were sequenced, and they had the same sequence of the gene that was isolated from N. benthamiana (accession number JF915552). The amino acid sequence of this cDNA has 87% identity to the GST C-7 of N. tabacum (Takahashi & Nagata, 1992) (Supporting Information Fig. S1). Because C-7 of N. tabacum has been grouped into the tau GST family (Edwards et al., 2000), and the nucleotide sequence of this cDNA clone shows an 80% identity to that of NbGSTU2 in N. benthamiana (Dean et al., 2005), this study consequently designated this gene as NbGSTU4 (the fourth tau GST gene identified from N. benthamiana). To examine whether knocking down the expression of NbGSTU4 by using VIGS would also reduce the expression of other homologous genes in N. benthamiana, an inspection, using real-time PCR with specific primers, of the mRNA levels of NbGSTU1, NbGSTU2, NbGSTU3 and NbGSTF1in the NbGSTU4-knockdown plants was required. The results indicated that the expression levels of the other homologous genes tested in the NbGSTU4-knockdown plants were insignificant unlike those of the control plants (Fig. 1c). The results suggest that the knockdown expression of NbGSTU4 is specific, and the consequence of this knockdown in the N. benthamiana plant is mainly derived from the expression reduction of NbGSTU4 (a 66% reduction, Fig. 1c). Furthermore, we could not locate any significant morphological changes in the knockdown plants in size, shape, and height compared to the control plants (inoculated with a TRV vector-carrying luciferase gene) (Fig. S2).
Fig. 1.
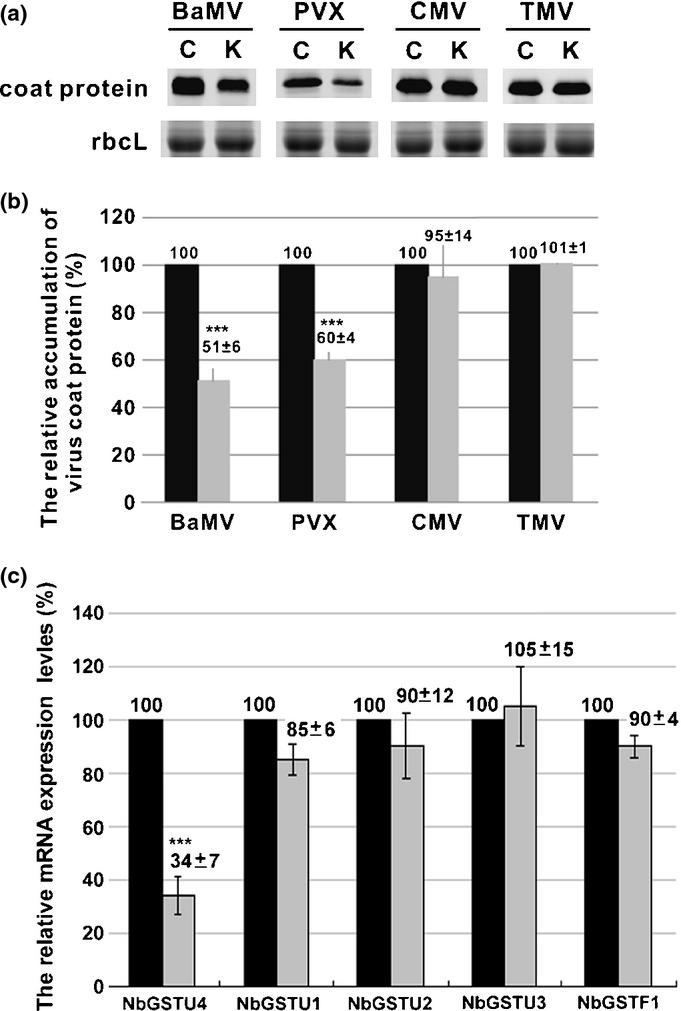
The characterization of NbGSTU4-knockdown Nicotiana benthamiana plants. (a) Western blot analysis of Bamboo mosaic virus (BaMV), Potato virus X (PVX), Cucumber mosaic virus (CMV) and Tobacco mosaic virus (TMV) coat protein in NbGSTU4-knockdown (indicated as K) and control (Luc-knockdown indicated as C) N. benthamiana plants 3 d post inoculation. Total proteins were extracted from inoculated leaf, separated on a 12% SDS-polyacrylamide gel, and probed with anti-coat protein antiserum. rbcL indicates the large subunit of RuBisCo stained with Coomassie Blue as a loading control. (b) A histogram of coat protein accumulation derived from (a). (c) The accumulation levels of different types of GST mRNA indicated in NbGSTU4-knockdown and control (Luc-knockdown) plants were measured by real-time RT-PCR. Control plants, black bars; NbGSTU4 knockdown plant, grey bars. The numbers shown above the statistic bar were the average ± SE of at least three independent experiments. Asterisks indicate statistically significant differences between the indicated group analyzed by Student's t-test (***, P < 0.001).
In order to clarify which step of the BaMV infection cycle was affected by reducing the NbGSTU4 expression, a protoplast assay was performed to exclude cell-to-cell movement. Northern blot analysis of RNA samples, derived from NbGSTU4-knockdown protoplasts transfected with the BaMV for 48 h, showed that the accumulation level of the BaMV genomic RNA was reduced compared to that of the control protoplasts (Fig. 2a). To quantify the precise accumulation levels of both the BaMV RNA and NbGSTU4 mRNA in the protoplasts, real-time PCR was performed to evaluate both the viral RNA accumulation and the mRNA knockdown efficiency, respectively. The accumulation levels of both the plus- and minus-strand BaMV RNA in the knockdown protoplasts were 64% and 58%, respectively, compared to those in the control protoplasts (Fig. 2b); the ratio of the reduction in the plus- and minus-strand BaMV RNA remained the same. The expression level of NbGSTU4 in the knockdown protoplasts was c. 32% of that of the control protoplasts (Fig. 2c). These results suggest that NbGSTU4 is involved in BaMV viral RNA replication, rather than in the virus movement.
Fig. 2.
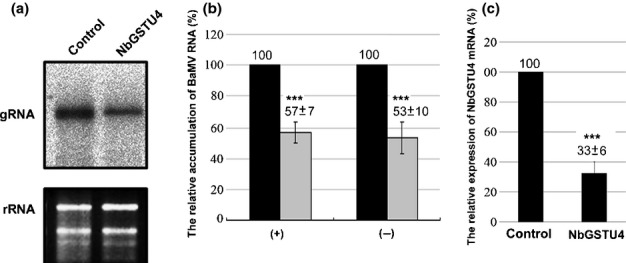
The accumulation levels of Bamboo mosaic virus (BaMV) RNA in NbGSTU4-knockdown protoplasts (a) Northern blot analysis of BaMV RNA in NbGSTU4-knockdown and control (Luc-knockdown) N. benthamiana protoplasts inoculated with BaMV RNA 48 h post-inoculation. rRNA indicates the loading control. (b) The accumulation levels of the plus- and minus-strand BaMV RNA in NbGSTU4-knockdown N. benthamiana protoplasts was measured by real-time RT-PCR. (c) The expression levels of NbGSTU4 in control and NbGSTU4-knockdown N. benthamiana protoplasts were measured by real-time RT-PCR. Control plants, black bars; NbGSTU4, grey bars. The numbers shown above the statistic bar were the average ± SE of at least three independent experiments. Asterisks indicate statistically significant differences between the indicated group analyzed by Student's t-test (***, P < 0.001).
NbGSTU4 interacts with the 3′ UTR of BaMV RNA in a glutathione dose-dependent fashion
The accumulation levels of both the plus- and minus-strands of BaMV RNA were reduced by c. 40% in the NbGSTU4-knockdown protoplasts (Fig. 2), suggesting that the synthesis of both strands was equally affected. One possible explanation for this is that an effect on minus-strand RNA synthesis could cause a similar reduction in plus-strand accumulation if the minus-strand RNA accumulated can produce only a fixed amount of plus-strand RNA (Chen et al., 2005; Wang & Nagy, 2008). We then tested whether NbGSTU4 could interact with the 3′ UTR of the BaMV genomic RNA and possibly regulate minus-strand RNA synthesis. The results derived from the UV-crosslinking assay showed that the recombinant fusion protein GST-NbGSTU4 expressed in E. coli (Fig. 3a) binds the 3′ UTR of BaMV (r138/40A), but no RNA binding with the fusion partner (GST) alone was observed (Fig. 3b, Lane 1).
Fig. 3.
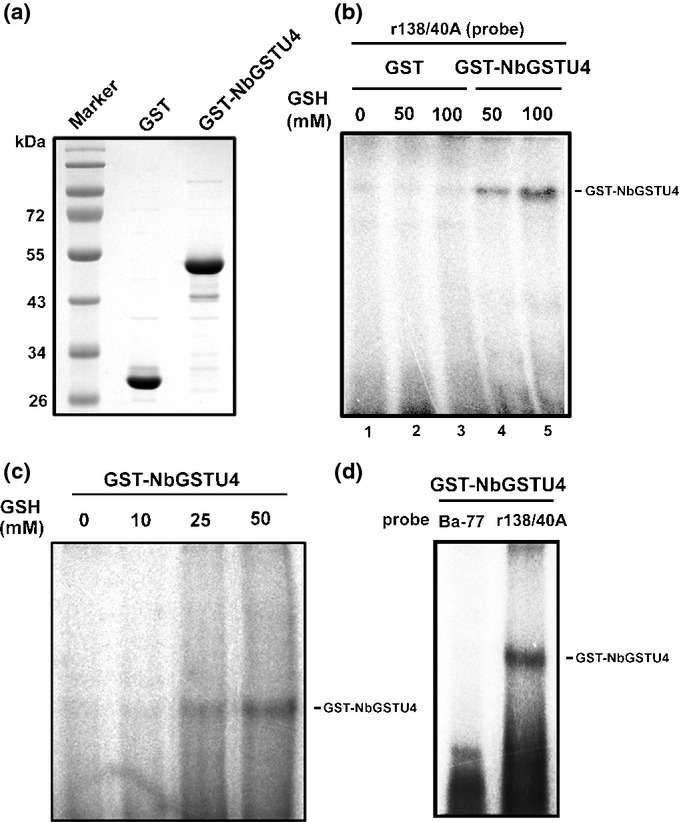
UV-crosslink of the E. coli-expressed GST and GST-NbGSTU4 with Bamboo mosaic virus (BaMV) RNAs. (a) Recombinant glutathione S-transferase (GST) and GST-NbGSTU4 proteins were expressed and purified from E. coli BL21, and resolved in a 12% SDS-polyacrylamide gel. (b) GST and GST-NbGSTU4 were UV-cross linked with r138/40A (178 nts, the 3′UTR containing the promoter for minus-strand RNA synthesis) in the presence of different concentration of glutathione (GSH) indicted on the top of each lane and resolved in a 14% SDS-polyacrylamide gel. The lane number is indicated at the bottom of each lane. (c) GST-NbGSTU4 was UV-cross linked with r138/40A in the presence of different concentrations of GSH (indicted at the top of each lane) and resolved in a 12% SDS-polyacrylamide gel. (d) GST-NbGSTU4 was incubated with BaMV r138/40A and Ba-77 (77 nts, the promoter for plus-strand RNA synthesis). The radioactive UV-crosslinked GST-NbGSTU4 protein is indicated.
Because GSTs bind GSH as a co-substrate/cofactor to form a functional homo or heterodimer (Dixon et al., 2002), GSH was added in the UV-crosslinking reactions to determine whether the interaction of GST with GSH could affect the binding. This showed that GSH could enhance the binding of NbGSTU4 with the 3′ UTR of BaMV in a higher dose concentration (50 and 100 mM; Fig. 3b, Lanes 4 and 5). Because the intracellular concentration of GSH is highly variable, ranging from c. 1 to as much as 10–20 mM (Meister & Anderson, 1983), we then tested whether the NbGSTU4 still can interact with the BaMV RNA at that concentration. The results shown in a dose-dependent manner indicated that the interaction could be observed in the presence of 10 mM GSH (Fig. 3c). We further tested whether NbGSTU4 can bind the 3′-terminal region of the minus-strand BaMV (Ba-77); the promoter for plus-strand RNA synthesis (Fig. 3d). The results indicated that the binding was specific to the 3′ UTR of BaMV.
GSH enhances replication of the BaMV at an early stage
Because GSH could enhance the binding of NbGSTU4 with the 3′ UTR of BaMV RNA, a test was conducted to determine whether a simple addition of GSH into BaMV-transfected protoplasts could enhance viral accumulation. The experiment was performed by transfecting BaMV RNA into N. benthamiana protoplasts and adding GSH into the medium at different time points post-inoculation. The accumulation levels of the BaMV plus- and minus-strand RNA of all of the treatments were examined by real-time RT-PCR at 24 h post-inoculation (hpi). The addition of GSH at 4 and 6 hpi caused a significant enhancement in the accumulation of both BaMV plus- and minus-strand RNA (Fig. 4a). Furthermore, the N. benthamiana plant infiltrated directly by GSH could also significantly enhance the accumulation of both the plus- and minus-strands of BaMV RNA at 24 hpi (Fig. 4b).
Fig. 4.
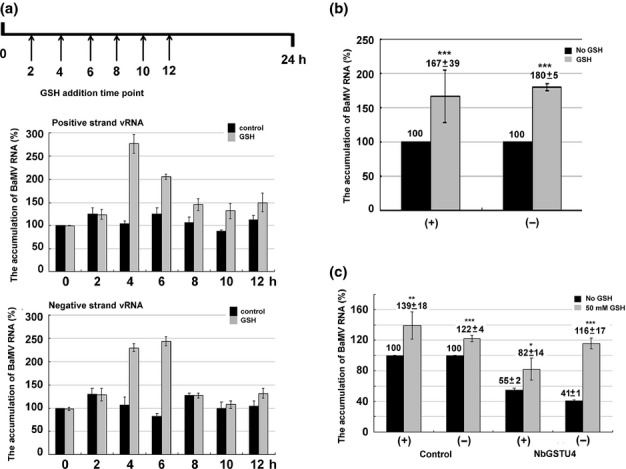
Real-time RT-PCR analysis of the accumulation of Bamboo mosaic virus (BaMV) RNA in the presence of glutathione (GSH) in protoplasts and plants. (a) Protoplasts were treated with GSH or Tris buffer (control) at different time points (h) post-BaMV RNA inoculation (hpi) indicated with arrows. Total RNAs were extracted at 24 hpi for real-time RT-PCR analysis. The accumulation levels of BaMV plus- and minus-strand RNA (Table S1) were indicated in the histogram. (b) The accumulation levels of BaMV plus-strand (+) and minus-strand (−) RNA in N. benthamiana which were co-infiltrated with the infectious BaMV cDNA clone pKB (driven by 35S promoter) and either Tris buffer as a control or with GSH (50mM). (c) The accumulation levels of BaMV plus-strand (+) and minus-strand (−) RNA in NbGSTU4-knockdown or control N. benthamiana plants which were co-infiltrated with pKB and either Tris buffer or GSH (50 mM) at 24 hpi. The numbers shown above the statistic bar were the averages ± SE of three independent experiments. Asterisks indicate statistically significant differences between the indicated group analyzed by Student's t-test (**, P < 0.01; ***, P < 0.001).
The observations that GSH could enhance the NbGSTU4 binding to the BaMV 3′ UTR and that the addition of GSH stimulated BaMV accumulation suggested that the BaMV 3′ UTR binds NbGSTU4 to recruit GSH for BaMV RNA production. One of the possible mechanisms is that GSH is required for minus-strand RNA synthesis and, thus, is achieved through its interaction with NbGSTU4. If this hypothesis is correct, the accumulation levels of the BaMV in the NbGSTU4-knockdown plants can be restored when GSH is provided. The results validated the proposed hypothesis that providing sufficient GSH can raise the accumulation of the BaMV RNA in NbGSTU4-knockdown plants more than that in the control plants, especially for minus-strand RNA (Fig. 4c).
The hypothesis that the NbGSTU4 binding the 3′ UTR of BaMV delivers GSH to the replication complex for viral RNA synthesis was tested further by creating a mutant NbGSTU4/S13A-GFP, which abolished the GSH-binding activity by substituting the conserved GSH-binding site serine to alanine (Zeng et al., 2005). We then transiently expressed NbGSTU4-GFP or NbGSTU4/S13A-GFP in BaMV-inoculated N. benthamiana leaves. The results indicated that the accumulation of the BaMV coat protein in the NbGSTU4-GFP-expressed plant is enhanced to 170% of those of the control plants (transiently expressed GFP only) (Fig. 5). However, in NbGSTU4/S13A-GFP-expressed plants the accumulation of the BaMV coat protein was similar to that of control plants. The results confirm that the GSH-binding activity of NbGSTU4 is important for BaMV accumulation.
Fig. 5.
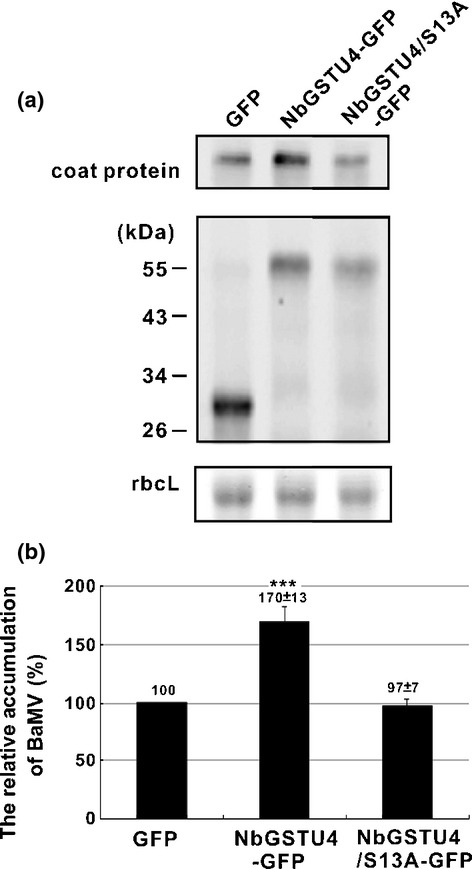
Western blotting analysis of the accumulation of Bamboo mosaic virus (BaMV) coat protein in NbGSTU4-GFP or its mutant expressed N. benthamiana plants. (a) The plants were inoculated with BaMV virion and followed by transiently expressed GFP, NbGSTU4-GFP or NbGSTU4/S13A-GFP at 2 d post-inoculation (dpi). The total proteins were extracted at 5 dpi. The protein accumulation levels were measured by western blotting with the antibody against BaMV coat protein or GFP. (b) The accumulation levels of BaMV coat protein were shown as the histogram indicated. The numbers shown above the statistic bar were the average ± SE of at least three independent experiments. Asterisks indicate statistically significant differences between the indicated group analyzed by Student's t-test (***, P < 0.001).
GSH stimulates minus-strand RNA synthesis in an in vitro RdRp assay
This study showed that NbGSTU4 and GSH can facilitate the replication of the BaMV in N. benthamiana. The effect of the GSH on the replication of the BaMV is suggested to be associated with minus-strand RNA synthesis. Further validation of this hypothesis required performing an in vitro RdRp assay with exogenous RNA templates in different concentrations of GSH (Fig. 6). The results indicate that GSH can only facilitate minus-strand RNA synthesis (using 3′ UTR of BaMV RNA, r138/40A) in a dose-dependent manner. In 100 mM GSH, r138/40A template activity could reach 2.7-fold of that without the addition of GSH in the RdRp preparation. By contrast, we could not observe the enhancement when GSH was added to plus-strand RNA synthesis (using the 3′-terminal promoter sequence of minus-strand RNA, Ba-77). We then tested whether the BaMV minus-strand RNA synthesis requires GSH specifically, or simply requires reduced conditions in an in vitro RdRp assay. The results indicated that the RNA synthesis rate could be enhanced significantly when reducing agents, such as GSH and DTT, are added to the reaction, whereas oxidizing agents, such as GSSG and H2O2, would reduce RNA synthesis (Fig. 7). We also tested whether the synthetic tripeptide, α-glutamate-cystein-glycine, mimicking the GSH structure, but without the γ-glutamate linkage, can have the same effect in the RdRp assay. The results indicated that the synthesized tripeptide reduced the RdRp activity (Fig. 7, Lane 6) and suggested the specificity of the GSH in BaMV RNA synthesis.
Fig. 6.
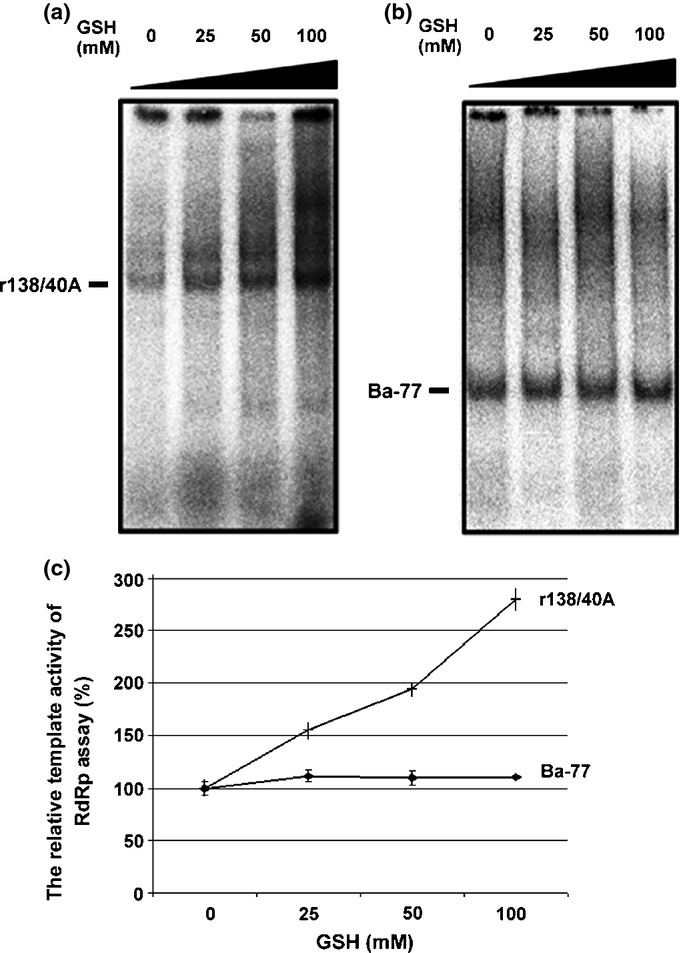
In vitro RdRp assay with exogenous RNA templates. Approximately 50 ng of RNA template r138/40A (the 3′ UTR of Bamboo mosaic virus (BaMV)) in (a) and Ba-77 (the 3′-end 77 nts of BaMV minus-strand) in (b) were incubated with BaMV RdRp complex for the in vitro RNA synthesis in the presence of different concentrations (0–100 mM) of glutathione (GSH) as indicated. The RdRp products labeled with [α-32P] UTP as indicated bands were separated on a 5% acrylamide gel and quantified by a phosphorimager. (c) The relative RdRp template activities were plotted according to the data derived from (a) and (b). The banding density of the in vitro RdRp assay with either r138/40A or Ba-77 was set as 100% in the absence of GSH (0 mM). Each spot on the plot was the average ± SE of at least three independent experiments.
Fig. 7.
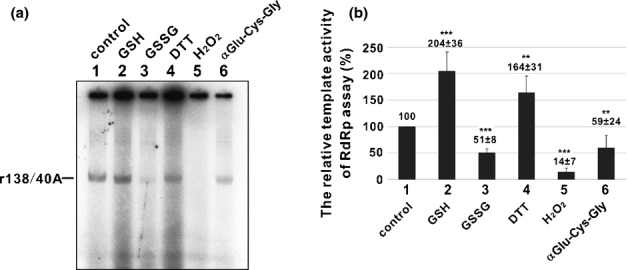
The effects of different redox agents in the in vitro RdRp assay. (a) Approximately 50 ng of RNA template r138/40A was incubated with Bamboo mosaic virus (BaMV) RdRp complex for in vitro RNA synthesis in the presence of 50 mM of different redox agents as indicated. (b) The relative RdRp template activities were plotted according to the data derived from (a). The banding density of the in vitro RdRp assay in the presence of 50 mM Tris buffer (pH 7.0) was set as 100%. The numbers shown above the statistic bar were the average ± SE of three independent experiments. Asterisks indicate statistically significant differences between the indicated group analyzed by Student's t-test (**, P < 0.01; ***, P < 0.001).
Overall, the results suggest that minus-strand RNA synthesis of the BaMV required a more reduced environment for efficient RNA synthesis. To create this condition for RNA synthesis, the viral RNA interacts with a GST, NbGSTU4, to draw the GSH into the replication complex.
Discussion
Host factors associated with the 3′ UTR of positive-sense RNA viruses may have contradictory roles in regulating viral infection cycles. Overexpression of the 3′ UTR binding protein Nsr1p/nucleolin in Nicotiana benthamiana reduced the accumulation of tombusvirus RNA, and the addition of purified Nsr1p in the replicase complex inhibited the in vitro activity (Jiang et al., 2010). Knocking down the expression level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in N. benthamiana could enhance accumulation of the BaMV, and the addition of recombinant GAPDH in the RdRp assay could inhibit minus-strand RNA synthesis, suggesting a role in negatively regulating BaMV infection (Prasanth et al., 2011). NF90 interacting with the stable 3′-terminal stem-loop structure of the dengue RNA could positively regulate its replication. A depletion of NF90 in cells could significantly diminish the production of infectious dengue viruses (Gomila et al., 2011). The translation elongation factor, eEF1A, not only binds to the 3′-end of the Tomato bushy stunt virus (TBSV) RNA, but also binds to the TBSV p33 replication protein and acts as a co-factor assisting in viral RNA replication (Li et al., 2009). In this study, we also found that a host factor, a GST, could interact with the 3′ UTR of BaMV RNA and positively regulate minus-strand RNA synthesis.
Plant GSTs are a group of enzymes playing a major role in protecting plants from various stresses, such as pathogen attacks (Mauch & Dudler, 1993; Li et al., 1997). The three known tau-class GSTs identified in N. benthamiana (NbGSTU1, -U2 and -U3) have been reported to be involved in fungal infections (Dean et al., 2005). The new member of the 4th tau GST gene, NbGSTU4, identified in this study is required for BaMV efficient replication. This study provides evidence that, by using the TRV-based VIGS technique to knock down the expression of NbGSTU4, no effect on the expression levels of other homologous genes (the other known tau GSTs), was specific in N. benthamiana (Fig. 1). A survey in the rice genome identified 79 GSTs including 52 tau GSTs (Jain et al., 2010), among which OsGSTU4 was demonstrated to play a role in herbicide resistance (Jo et al., 2011). Although none of the tau GST gene in bamboo (the natural host of BaMV) has been identified yet, we believe that bamboo belonging to Poaceae family should contain as many as tau GSTs as those in rice.
Three lines of evidence help link NbGSTU4 to minus-strand RNA synthesis: (1) the E. coli-expressed NbGSTU4 binds to the 3′ UTR, although not to the 3′-end of the minus-strand BaMV RNA (Fig. 3); (2) the reduction or enhancement of the plus- and minus-strand RNA accumulation in knockdown or transiently expressed plants, respectively, remains the same (Fig. 2); and (3) the effect of the GSH on BaMV accumulation in protoplasts occurs only at an early time point (Fig. 4). These results suggest that minus-strand RNA synthesis requires GSH. This requirement is evident from the in vitro RdRp assay, which shows that the template activity of the 3′ UTR exogenous RNA can be enhanced by the addition of GSH in a dose-dependent manner, whereas the promoter for plus-strand RNA synthesis cannot be enhanced in the presence of GSH (Fig. 6).
GSTs and GSH are involved in antioxidation processes in cells. The oxidative stress (a plant anti-pathogen infection strategy) induced by virus infections (Fodor et al., 2001; Mathew et al., 2010) can be a hindrance for viral replication. Therefore, reducing the oxidative stress by providing GSH can be a crucial step for viral RNA replication in cells after infection. A possible role of NbGSTU4 upregulated by virus infection is to provide an antioxidation environment for virus replication. The results derived from the in vitro RdRp assay indicate that providing GSH or DTT can enhance minus-strand RNA synthesis, but not plus-strand RNA synthesis. The likely role of NbGSTU4 is to bring GSH to the replication site and provide anti-oxidative conditions for minus-strand RNA synthesis. Because the concentration of GSH in cells is c. 1–20 mM, it might not provide a sufficiently strong reduced environment for viral RNA replication. The interaction of viral RNA with GST (BaMV RNA with NbGSTU4, Fig. 3) to recruit GSH and to increase the reductive condition at the viral replication site might be a good strategy.
Conclusion
This is the first study to examine the involvement of a new plant tau-class GST protein, NbGSTU4, in BaMV viral RNA minus-strand RNA synthesis. Through the binding of NbGSTU4 to the 3′ UTR of the genomic RNA, either by providing an antioxidative condition or by changing the redox status of the replicase complex, the minus-strand RNA can be synthesized efficiently. The discovery of a plant GST protein involving the replication of a plant virus could help us to gain insights into the relationship between viral RNA replication and the GST metabolic pathway.
Acknowledgments
This work was supported by grants from the National Science Council through research grants NSC 97-2752-B-005-004-PAE and NSC 99-2321-B-005-003-MY3.
Supporting Information
Additional supporting information may be found in the online version of this article.
Fig. S1 Amino acid sequence alignment of Nicotiana tabacum C-7 (accession number Q40480), and N. benthamiana NbGSTU4 (accession number JF915552).
Fig. S2 The phenotype of NbGSTU4-knockdown Nicotiana benthamiana plants.
Table S1 The relative accumulation levels of BaMV plus- and minus-strand RNA in the control (Tris buffer) and GSH (GSH treated) protoplasts
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Allocati N, Federici L, Masulli M, Di Ilio C. Glutathione transferases in bacteria. FEBS Journal. 2009;276:58–75. doi: 10.1111/j.1742-4658.2008.06743.x. [DOI] [PubMed] [Google Scholar]
- Chen IH, Chou WJ, Lee PY, Hsu YH, Tsai CH. The AAUAAA motif of bamboo mosaic virus RNA is involved in minus-strand RNA synthesis and plus-strand RNA polyadenylation. Journal of Virology. 2005;79:14 555–14 561. doi: 10.1128/JVI.79.23.14555-14561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IH, Meng M, Hsu YH, Tsai CH. Functional analysis of the cloverleaf-like structure in the 3' untranslated region of bamboo mosaic potexvirus RNA revealed dual roles in viral RNA replication and long distance movement. Virology. 2003;315:415–424. doi: 10.1016/s0042-6822(03)00560-9. [DOI] [PubMed] [Google Scholar]
- Cheng CP, Tsai CH. Structural and functional analysis of the 3′ untranslated region of bamboo mosaic potexvirus genomic RNA. Journal of Molecular Biology. 1999;288:555–565. doi: 10.1006/jmbi.1999.2716. [DOI] [PubMed] [Google Scholar]
- Cheng JH, Ding MP, Hsu YH, Tsai CH. The partial purified RNA-dependent RNA polymerases from bamboo mosaic potexvirus and potato virus x infected plants containing the template-dependent activities. Virus Research. 2001;80:41–52. doi: 10.1016/s0168-1702(01)00348-3. [DOI] [PubMed] [Google Scholar]
- Cheng JH, Peng CW, Hsu YH, Tsai CH. The synthesis of minus-strand RNA of bamboo mosaic potexvirus initiates from multiple sites within the poly(A) tail. Journal of Virology. 2002;76:6114–6120. doi: 10.1128/JVI.76.12.6114-6120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SF, Huang YP, Wu ZR, Hu CC, Hsu YH, Tsai CH. Identification of differentially expressed genes induced by bamboo mosaic virus infection in Nicotiana benthamiana by cDNA-amplified fragment length polymorphism. BMC Plant Biology. 2010;10:286. doi: 10.1186/1471-2229-10-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz SS, Roberts AG, Prior DA, Chapman S, Oparka KJ. Cell-to-cell and phloem-mediated transport of potato virus x. The role of virions. The Plant Cell. 1998;10:495–510. doi: 10.1105/tpc.10.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JD, Goodwin PH, Hsiang T. Induction of glutathione S-transferase genes of Nicotiana benthamiana following infection by Colletotrichum destructivum and C. orbiculare and involvement of one in resistance. Journal of Experimental Botany. 2005;56:1525–1533. doi: 10.1093/jxb/eri145. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. Plant glutathione transferases. Genome Biology. 2002;3:REVIEWS3004. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon DP. Plant glutathione transferases. Methods in Enzymology. 2005;401:169–186. doi: 10.1016/S0076-6879(05)01011-6. [DOI] [PubMed] [Google Scholar]
- Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends in Plant Science. 2000;5:193–198. doi: 10.1016/s1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- Fodor J, Hideg E, Kecskes A, Kiraly Z. In vivo detection of tobacco mosaic virus-induced local and systemic oxidative burst by electron paramagnetic resonance spectroscopy. Plant Cell Physiology. 2001;42:775–779. doi: 10.1093/pcp/pce096. [DOI] [PubMed] [Google Scholar]
- Frear DS, Swanson HR. Biosynthesis of s-(4-ethylamino-6-isopropylamino-2-s-triazino) glutathione: partial purification and properties of a glutathione S-transferase from corn. Phytochemistry. 1970;9:2123–2132. [Google Scholar]
- Gomila RC, Martin GW, Gehrke L. NF90 binds the dengue virus RNA 3′ terminus and is a positive regulator of dengue virus replication. PLoS ONE. 2011;6:e16687. doi: 10.1371/journal.pone.0016687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Huang YL, Meng M, Hsu YH, Tsai CH. Sequences at the 3′ untranslated region of bamboo mosaic potexvirus RNA interact with the viral RNA-dependent RNA polymerase. Journal of Virology. 2001;75:2818–2824. doi: 10.1128/JVI.75.6.2818-2824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Tsai CH. Evolution of bamboo mosaic virus in a nonsystemic host results in mutations in the helicase-like domain that cause reduced RNA accumulation. Virus Research. 1998;58:127–136. doi: 10.1016/s0168-1702(98)00109-9. [DOI] [PubMed] [Google Scholar]
- Huang YL, Han YT, Chang YT, Hsu YH, Meng M. Critical residues for GTP methylation and formation of the covalent m7GMP-enzyme intermediate in the capping enzyme domain of bamboo mosaic virus. Journal of Virology. 2004;78:1271–1280. doi: 10.1128/JVI.78.3.1271-1280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Ghanashyam C, Bhattacharjee A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics. 2010;11:73. doi: 10.1186/1471-2164-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Li Z, Nagy PD. Nucleolin/Nsr1p binds to the 3′ noncoding region of the tombusvirus RNA and inhibits replication. Virology. 2010;396:10–20. doi: 10.1016/j.virol.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo HJ, Lee JJ, Kong KH. A plant-specific tau class glutathione S-transferase from Oryza sativa with very high activity against 1-chloro-2,4-dinitrobenzene and chloroacetanilide herbicides. Pesticide Biochemistry and Physiology. 2011;101:265–269. [Google Scholar]
- Kampranis SC, Damianova R, Atallah M, Toby G, Kondi G, Tsichlis PN, Makris AM. A novel plant glutathione S-transferase/peroxidase suppresses Bax lethality in yeast. Journal of Biological Chemistry. 2000;275:29 207–29 216. doi: 10.1074/jbc.M002359200. [DOI] [PubMed] [Google Scholar]
- Kilili KG, Atanassova N, Vardanyan A, Clatot N, Al-Sabarna K, Kanellopoulos PN, Makris AM, Kampranis SC. Differential roles of tau class glutathione S-transferases in oxidative stress. Journal of Biological Chemistry. 2004;279:24 540–24 551. doi: 10.1074/jbc.M309882200. [DOI] [PubMed] [Google Scholar]
- Li YI, Chen YJ, Hsu YH, Meng M. Characterization of the AdoMet-dependent guanylyltransferase activity that is associated with the N terminus of bamboo mosaic virus replicase. Journal of Virology. 2001;75:782–788. doi: 10.1128/JVI.75.2.782-788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pogany J, Panavas T, Xu K, Esposito AM, Kinzy TG, Nagy PD. Translation elongation factor 1A is a component of the tombusvirus replicase complex and affects the stability of the p33 replication co-factor. Virology. 2009;385:245–260. doi: 10.1016/j.virol.2008.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZS, Alfenito M, Rea PA, Walbot V, Dixon RA. Vacuolar uptake of the phytoalexin medicarpin by the glutathione conjugate pump. Phytochemistry. 1997;45:689–693. doi: 10.1016/s0031-9422(97)00031-9. [DOI] [PubMed] [Google Scholar]
- Lin JW, Chiu HN, Chen IH, Chen TC, Hsu YH, Tsai CH. Structural and functional analysis of the cis-acting elements required for plus-strand RNA synthesis of Bamboo mosaic virus. Journal of Virology. 2005;79:9046–9053. doi: 10.1128/JVI.79.14.9046-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Ding MP, Hsu YH, Tsai CH. Chloroplast phosphoglycerate kinase, a gluconeogenetic enzyme, is required for efficient accumulation of Bamboo mosaic virus. Nucleic Acids Research. 2007;35:424–432. doi: 10.1093/nar/gkl1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Chang BY, Liao JT, Lin NS, Hsu YH. Arg-16 and Arg-21 in the n-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. Journal of General Virology. 2004;85:251–259. doi: 10.1099/vir.0.19442-0. [DOI] [PubMed] [Google Scholar]
- Lin MK, Hu CC, Lin NS, Chang BY, Hsu YH. Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the bamboo mosaic virus satellite RNA-mediated expression system. Journal of General Virology. 2006;87:1357–1367. doi: 10.1099/vir.0.81625-0. [DOI] [PubMed] [Google Scholar]
- Lin NS, Lin FZ, Huang TY, Hsu YH. Genome properties of Bamboo mosaic-virus. Phytopathology. 1992;82:731–734. [Google Scholar]
- Lin NS, Lin BY, Lo NW, Hu CC, Chow TY, Hsu YH. Nucleotide sequence of the genomic RNA of bamboo mosaic potexvirus. Journal of General Virology. 1994;75:2513–2518. doi: 10.1099/0022-1317-75-9-2513. [DOI] [PubMed] [Google Scholar]
- Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1996;47:127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- Mathew SS, Bryant PW, Burch AD. Accumulation of oxidized proteins in herpesvirus infected cells. Free Radical Biology and Medicine. 2010;49:383–391. doi: 10.1016/j.freeradbiomed.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Dudler R. Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiology. 1993;102:1193–1201. doi: 10.1104/pp.102.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle B, Keeler SJ, Lau SM, Koeppe MK, O'Keefe DP. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiology. 2000;124:1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A, Anderson ME. Glutathione. Annual Review of Biochemistry. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- Prasanth KR, Huang YW, Liou MR, Wang RY, Hu CC, Tsai CH, Meng M, Lin NS, Hsu YH. Glyceraldehyde 3-phosphate dehydrogenase negatively regulates the replication of bamboo mosaic virus and its associated satellite RNA. Journal of Virology. 2011;85:8829–8840. doi: 10.1128/JVI.00556-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxas VP, Smith RK, Jr, Allen ER, Allen RD. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nature Biotechnology. 1997;15:988–991. doi: 10.1038/nbt1097-988. [DOI] [PubMed] [Google Scholar]
- Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochemical Journal. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Nagata T. Differential expression of an auxin-regulated gene, parC, and a novel related gene, C-7 from tobacco mesophyll protoplasts in response to external stimuli and plant tissues. Plant Cell Physiology. 1992;33:779–787. [Google Scholar]
- Tsai CH, Cheng CP, Peng CW, Lin BY, Lin NS, Hsu YH. Sufficient length of a poly(A) tail for the formation of a potential pseudoknot is required for efficient replication of bamboo mosaic potexvirus RNA. Journal of Virology. 1999;73:2703–2709. doi: 10.1128/jvi.73.4.2703-2709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Nagy PD. Tomato bushy stunt virus co-opts the RNA-binding function of a host metabolic enzyme for viral genomic RNA synthesis. Cell Host & Microbe. 2008;3:178–187. doi: 10.1016/j.chom.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Zeng QY, Lu H, Wang XR. Molecular characterization of a glutathione transferase from Pinus tabulaeformis (pinaceae) Biochimie. 2005;87:445–455. doi: 10.1016/j.biochi.2005.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


