Abstract
The identification of the intestinal stem cell (ISC) markers Lgr5 and Bmi-1 has furthered our understanding of how they accomplish homeostasis in this rapidly self-renewing tissue. Recent work indicates that these markers identify a cycling Lgr5+ ISC which can be replaced by a quiescent Bmi-1+ ISC. Currently, there is little data on how these cells interact to control intestinal crypt homeostasis and regeneration. This interaction likely involves other differentiated cells within the niche as it has previously been demonstrated that the “stemness” of the Lgr5 ISC is closely tied to the presence of their neighboring Paneth cells. To investigate this, we used two conditional mouse models to delete the transcription factor β-catenin within the intestinal crypt. Critically these differ in their ability to drive recombination within Paneth cells and therefore allow us to compare the effect of deleting the majority of active ISCs in the presence or absence of the Paneth cells. After gene deletion, the intestines in the model in which Paneth cells were retained showed a rapid recovery and repopulation of the crypt-villus axis presumably from either a spared ISC or the hypothetical quiescent ISCs. However, in the absence of Paneth cells the recovery ability was compromised resulting in complete loss of intestinal epithelial integrity. This data indicates that the Paneth cells play a crucial role within the in vivo ISC niche in aiding recovery following substantial insult. Stem Cells2013;31:776–785
Keywords: Intestine, Stem cells, Paneth cells, Genetically modified animals
INTRODUCTION
The surface epithelia of the small intestine is a dynamic tissue which undergoes complete self-renewal every 3–4 days [1]. The epithelium comprises two basic repetitive structural units: (a) the crypts of Lieberkuhn which lie at the bottom of the mucosa and are continuous with (b) the villi, finger like projections which protrude into the lumen. This dynamic self-renewal originates from a population of ISCs located within the crypts which function as a stem cell “niche.” The proliferating and differentiating cells migrate upward onto the villi where they eventually die and are shed into the lumen.
For many years the identity of the ISCs was much sought after but the identification of two ISC markers in recent years has allowed progress to be made in the understanding of intestinal biology [2, 3]. The expression profile of these two ISC markers, Bmi1 [3] and Lgr5 [2], suggests that there may be two distinct ISC populations. The Bmi1 expressing cells reside at the +4 position relative to the crypt bottoms. The Lgr5 positive cells are the rapidly cycling columnar base cells (CBCs), with approximately 14 of these cells intermingled with the Paneth cells at the bottom of the crypt. Both these populations fulfill the stem cell characteristics of being self-renewing, multipotent, and essential for crypt maintenance [4]. However, the other stem cell hallmark of quiescence is currently only suggested for Bmi1 cells [5] and is not a feature of the daily cycling CBCs. Based on these markers, it has been proposed that the crypt contains two stem cell populations. The first of these is an active/cycling stem cell population represented by the Lgr5-labeled CBCs [6]. The second is a Bmi1 positive stem cell population at the +4 position which is capable of expanding and renewing the Lgr5 population. Recently, this model has been supported by in vivo and in vitro data showing that, following either diphtheria toxin-mediated or radiation-induced killing of the Lgr5 population, the Bmi1 population can indeed function to replace the Lgr5 population [7, 8].
This concept of two populations of ISCs is further complicated by two recent reports, which demonstrate that self-renewal of the ISCs follows a pattern of neutral drift and elegantly identify a pool of equipotent stem cells that are regulated by its neighbors [9, 10]. This, together with the observation that Lgr5 cells express the highest levels of Bmi1, suggests that Bmi1 and Lgr5 mark overlapping if not identical ISC populations [9, 11]. One of the crucial remaining questions is how these different stem cell compartments interact, especially during the process of crypt regeneration.
It is likely that aspects of the answer to this question will be found by an examination of the interactions between the ISCs and their neighboring cells within the crypt stem cell niche. Indeed the previous suggestion that the “stemness” of the CBCs is closely tied to the presence of their neighboring Paneth cells [9] has now been demonstrated in vitro and in vivo [12]. Evidence from previous studies in which depleting or deleting Paneth cells suggested they were dispensable in intestinal epithelial homeostasis have, upon closer examination shown that the Lgr5 ISCs only exist where they can compete for essential niche signals provided by their specialized daughter Paneth cells [12].
The above studies examined the role of the Paneth cells in relation to presumably normal ISCs. The importance of Paneth cells in a situation where the ISC population is damaged is still unclear. One such circumstance is following loss of function of β-catenin, the conditional deletion of which has been reported to lead to different and conflicting outcomes [13, 14]. The first of these [14] used a tamoxifen (TAM) inducible variant of the Cre recombinase expressed under the control of the villin gene promoter to drive induction of Cre specifically in the intestinal epithelia [13, 15, 16] (vil-Cre-ERT2). Using this system, Fevr et al. [14] demonstrated rapid loss of transit amplifying cells, crypt structures, terminal differentiation of the ISCs, and loss of intestinal homeostasis and function upon deletion of β-catenin. In contrast, a separate study used the promoter element of the rat cytochrome P450A1 (CYP1A1) gene to drive Cre expression in a xenobiotic responsive manner to permit inducible gene deletion in the intestinal epithelia (Ah-cre) [13]. Ireland et al. [13] showed that deletion of β-catenin again reduced cell viability; however, the crypt-villus axis was subsequently rapidly repopulated with unrecombined cells expressing β-catenin and homeostasis was re-established. Given that long-term labeling studies have shown that both the vil-Cre-ERT2 and Ah-cre models induce recombination in the ISCs, the mechanisms underlying these different outcomes are unclear. One possibility is that the two systems differentially drive recombination within the stem cell population, such that in the vil-Cre-ERT2 system a greater proportion of the ISC population is recombined. An alternative possibility is that the vil-Cre-ERT2 model deletes CatnB in differentiated cells, which provide the ISC niche. In the latter scenario, the most obvious candidate here is the Paneth cell, as this has previously been shown to be spared by Ah-cre [13]. To directly address these possibilities, we have confirmed conditions in which both systems deliver the same level of stem cell recombination and have subsequently determined the additional effect of recombining within the Paneth cell population using the vil-Cre-ERT2 system. Analysis of this data indicates that the Paneth cells are critically sensitive to β-catenin loss, and that their loss critically compromises the capacity of intestinal crypts to recover following insult.
MATERIALS AND METHODS
Animal Models
The alleles for the vil-Cre-ERT2[17], Ah-cre [13], floxed β-catenin (CatnBflox) [18], and Rosa26R-lacZ [19] have been described previously. All experiments were performed under the authority of a U.K. project license, which is issued after a local ethical review process. For induction of Ah-cre, the mice received intraperitoneal (i.p.) injections of 80 mg/kg β-napthoflavone (βNF) (Sigma, U.K., http://www.sigma.com) dissolved in corn oil (10 mg/ml) at frequencies stated. For induction of the vil-Cre-ERT2, the mice received i.p. injections of 80 mg/kg TAM unless otherwise stated (Sigma, U.K.) dissolved in corn oil (10 mg/ml) at doses stated. Control experiments were performed by inducing mice which only contained the appropriate Cre transgene and lacZ reporter, referred to as wild type (WT).
Relative Quantitation Analysis
The following methods were all performed according to manufacturer's instructions unless otherwise stated. For analysis of either the levels of CatnB allele recombination or gene expression in the intestinal epithelia, three to five mice from each control and experimental group were harvested. DNA or RNA was extracted either from a 0.5 cm portion of small intestine taken 2–3 cm distally from the stomach and stored at −80°C in RNAlater (Sigma, U.K.) or from crypt epithelia extracted from a whole small intestine [20]. Genomic DNA was extracted using the QiaAMP kit according to manufacturers' instructions and approximately 100 ng was used for quantitative polymerase chain reaction (qPCR). Total RNA was extracted using the RNeasy kit (Qiagen, U.K., http://www.qiagen.com) and DNase treated using the Turbo DNase kit (Ambion, U.K., http://www.ambion.com). Complimentary DNA was transcribed from 1 μg of RNA using random hexamers (Promega, U.K., http://www.promega.com) and the Superscript III (Invitrogen, U.K., http://www.invitrogen.com) kits. For relative quantitation, all samples were run in duplicate on the StepOnePlus PCR machine using Fast Sybr Green master mix (Applied Biosystems, U.K., http://www.appliedbiosystems.com) or Taqman Univeral Mastermix II (Applied Biosystems). The threshold cycle (Ct) values of each gene analyzed were normalized to a reference gene. For qPCR on genomic DNA, Ct values were normalized against the single copy gene ApoB, for expression analysis (qRT-PCR) Ct values were normalized against the ActB gene. Differences between groups were assessed using the 2−ΔΔCT method [21]. One-tailed Mann–Whitney U (M.W.) tests were performed on the ΔCt values to determine significant (p = <.05) differences between groups [22]. Relative quantification was performed using the oligonucleotide sequences indicated in Table 1.
Table 1.
Details of primers and assay used for expression and genomic quantitative analysis
 |
Abbreviation: qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Reporter Visualization and Immunohistochemistry
Whole intestines were mounted and opened along the mesenteric line and stained for LacZ activity [17]. Expression patterns were determined from at least three animals per experimental group using a standard dissecting microscope. The total numbers of stained crypts were counted within the 2–5 cm region distally from the stomach using imageJ software (http://rsbweb.nih.gov/ij/).
For immunohistochemistry (IHC), tissue was fixed in ice cold 10% formalin and processed into wax blocks by conventional means. Section were cut at 5 μm, dewaxed, and rehydrated into phosphate buffered saline. Staining was performed using the Envision+ mouse or rabbit kit (Cat No# K4007 & K4011, Dako, U.K., http://www.dako.com) according to manufacturer's instructions. To detect loss of β-catenin protein, we used a mouse Mab anti-β-catenin antibody (Cat No#610154, BD/Transduction Laboratories, U.K., http://www.bdbiosciences.com) at 1:200, for Paneth cell detection, we used a rabbit pAb anti-lysozyme antibody (Cat No# RB-372, Neomarkers/Labvision, USA, http://www.labvision.com) at 1:200, for proliferation, Ki67 mouse Mab at 1:200 (Cat No# VP-K452, Vector Laboratories, U.K., http://www.vectorlabs.com), and for apoptosis detection, we used a cleaved caspase 3 (Cat No# 9661, Cell Signaling Technologies, U.K., http://www.cellsignal.com) at 1:200. For visualization using immunofluorescence, sections were labeled using Alexafluor 488 and 594 secondary antibodies (Cat No# A11001 and A11072, Invitrogen) at 1:500. Detection of Olfm4 RNA by in situ hybridization was performed using protocols previously described [23–25]. The apoptotic and mitotic index were scored from H&E sections as previously described [26, 27]. Numbers of crypts were counted on 5–10 H&E stained cross-sections from each mouse. Paneth cell number was determined by counting the numbers of cells stained positive for lysozyme in a total of 25 crypts taken from at least three different sections for each mouse.
RESULTS
Identification of a Regime that Delivers Equivalent Levels of Stem Cell Recombination in the Ah-Cre and vil-Cre-ERT2 Systems
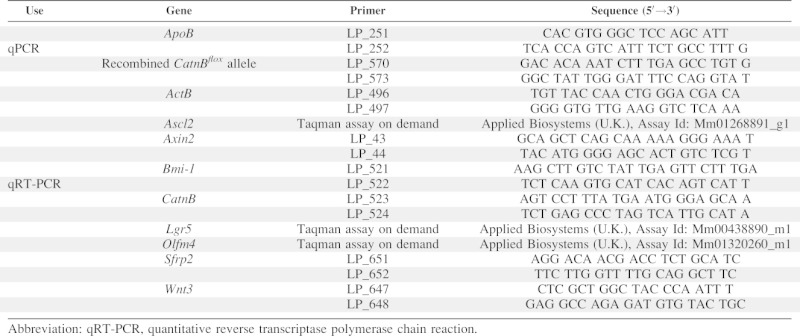
We first defined the level of recombination driven in the Ah-cre system both within the whole intestine and within the stem cell compartment. To define overall recombination levels, we induced Ah-cre CatnBflox/flox mice with 3 × i.p./24 hour injections of βNF and collected tissues at 1-day postinduction (d.p.i). We then quantified the presence of recombined CatnBflox alleles in the genomic DNA from whole intestine as a surrogate marker for the numbers of cells recombined. qPCR analysis showed that for Ah-cre CatnBflox/flox mice average ΔΔCt value was 2.28 in small intestine compared to −0.07 in the liver, a tissue previously shown to have high levels of recombination (Fig. 1A). To determine the efficiency of recombination in the ISC, we induced Ah-cre Rosa26R-lacZ CatnBwt/wt mice with 3 × i.p./24 hour injections of βNF and analyzed intestines at 30 d.p.i. The number of LacZ positive crypts were counted on intestinal whole mounts as a marker of stem cell recombination and established a baseline of an average 9,267 recombined crypts (representing recombined ISCs) (Fig. 1B, 1C).
Figure 1.
Comparison of the specificity and efficiency of Cre/Lox recombination within the intestinal epithelia using the Ah-cre and vil-Cre-ERT2 systems (*, p > .05). (A): Results of quantitative polymerase chain reaction (qPCR) for detection of fold change of the recombined CatnBflox allele in the small intestine using different induction regimes. (B): Quantification of LacZ positive stained crypts 30 d.p.i. using regimes indicated. (C): Whole mount small intestine showing LacZ stained crypts within the small intestine. (D): Results of qPCR for detection of CatnBflox allele within the intestinal crypt epithelia using different induction regimes. Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen.
We next compared the Ah-cre baseline with three different induction regimes in the vil-Cre-ERT2 system: 3 × i.p. injections of TAM (80 mg/kg)/24 hour, 1 × i.p. injection of TAM (80 mg/kg)/24 hour, and 1 × i.p. injection of TAM (40 mg/kg)/24 hour. The average ΔΔCt value for mice recombining using the highest dose was 0.38, indicating a significant 3.53 (p < .404 M.W.)-fold greater amount of the recombined CatnBflox allele compared to the Ah-cre baseline (Fig. 1A). This result is most likely a reflection of the vil-Cre-ERT2 system, which recombines in all epithelial cells of the crypt and villus, in contrast to the Ah-cre system which only recombines in the ISC niche and transit amplifying compartment of the crypt. However, we cannot exclude the possibility that some of the difference observed is due to each Cre system having different recombination efficiency at the CatnBflox allele. The intermediate regime delivered a Ct value of 0.51, which was not significantly different to the highest dose (Fig. 1A). At the lowest dose, the average ΔCt value was 1.94, indicating a 1.1-fold increase in presence of recombined CatnBflox allele compared to the Ah-cre baseline, which was not significantly different (Fig. 1A). For the highest and lowest dose regimes, we then determined the level of ISC recombination (Fig. 1B, 1C). The high regime delivered an average of 8,535 recombined crypts indicating ISC recombination rates equivalent to the Ah-cre baseline. The lowest regime showed a greatly reduced rate with 646 recombined crypts. These experiments therefore established that the high-dose vil-Cre-ERT2 regime delivered the same level of ISC recombination as the Ah-cre regime. Notably, this level of induction results in elevated overall levels of recombination compared to Ah-cre, which presumably reflects the previously reported higher levels of recombination in the villus in the vil-Cre-ERT2 system. In further support of this, analysis of purified crypts from induced mice showed that the higher vil-Cre-ERT2 induction regime delivered equivalent levels of recombination to the Ah-cre baseline (Fig. 1D).
Ah-cre and vil-Cre-ERT2 Driven Deletion of β-Catenin Results in Different Small Intestine Phenotypes
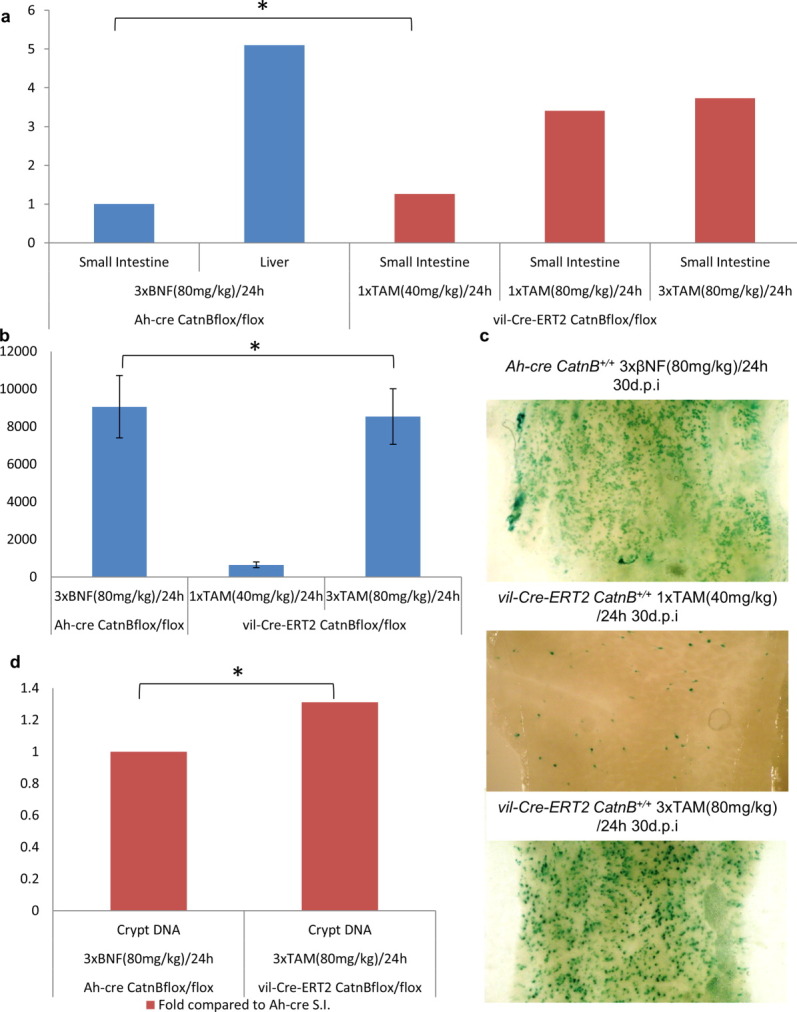
We next assessed whether the different phenotypes previously observed were due to the use of differently targeted CatnBflox alleles. To establish this, we used the Ah-cre and vil-Cre-ERT2 mice to delete the same CatnBflox [18] allele using the regimes characterized above that deliver equivalent levels of ISC recombination. In both models, deletion of CatnB from the intestinal epithelium generated phenotypes similar to the previous reports. Ah-cre mice (3 × i.p. βNF (80 mg/kg)/24 hour) showed no obvious signs of distress or reduction of survival by 30 d.p.i. and partial loss of crypts at 3 d.p.i. compared to WT (Fig. 2A, 2B, 2E). The vil-Cre-ERT2 mice (3 × i.p. TAM (80 mg/kg)/24 hour) became moribund by day 4–5 and had to be euthanized and at 3 d.p.i. mice showed destruction of the crypt/villus axis compared to WT (Fig. 2C–2E).
Figure 2.
Comparison of the Ah-cre and vil-Cre-ERT2 systems for conditionally deleting β-catenin within the small intestine epithelia. (A–D): H&E stained cross-sections of the small intestine, arrow indicate areas of crypt loss. (E): Quantification of the percentage of intestinal crypts lost 30 days after deletion of β-catenin (*, p < .01 M.W.) (F, G): Ki-67 staining indicating proliferating cells at 3 d.p.i. Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen.
Vil-Cre-ERT2 Driven β-Catenin-Deficient Intestines Are Unable to Repopulate like Ah-cre
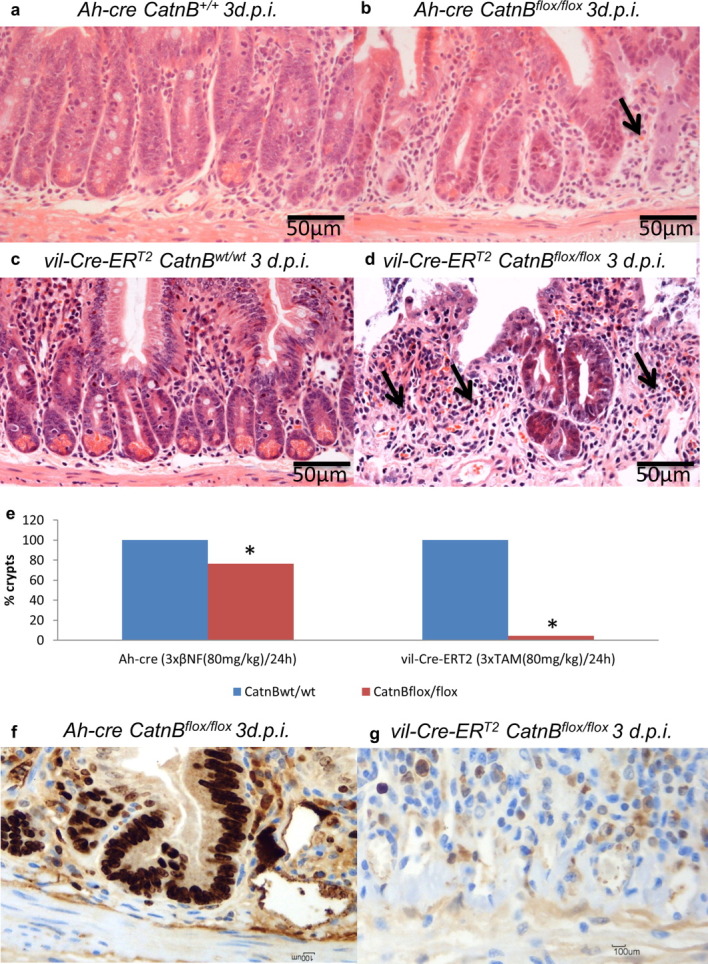
To understand the reasons for the survival of Ah-cre mice in contrast to the vil-Cre-ERT2 mice, we characterized the epithelia after β-catenin deletion. Using the LacZ reporter as a surrogate marker both models showed that after 3 d.p.i. there was a complete loss of all β-catenin-deficient cells compared to controls (data not shown). This was confirmed by expression analysis which demonstrated the deletion of the CatnB gene and alteration of the canonical Wnt pathway target Axin2 (Fig. 3A) compared to appropriate WT controls. For Ah-cre, CatnB expression fold changes were as follows: 1d/0.4*, 2d/0.73, and 3d/1.3, and this trend was reflected by Axin2: 1d/0.43*, 2d/0.24*, and 3d/1.1 (*, p ≤ .01 M.W.). The return to WT expression levels indicated that the epithelium had been repopulated with WT cells. In comparison there was no indication the vil-Cre-ERT2 mice had repopulated as the fold changes were for CatnB: 1d/0.1*, 2d/0.02*, and 3d/0.05* and Axin2: 1d/1.2, 2d/0.18*, and 3d/0.23* (*p ≤ .01 M.W.).
Figure 3.
Characterization of the onset of phenotype when using Ah-cre and vil-Cre-ERT2 systems for conditionally deleting β-catenin in a similar number of intestinal stem cells within the small intestine epithelia. (A): Expression analysis of genes highlighting the phenotype of loss of CatnB in the small intestine epithelia (*, p < .01 M.W.) in comparison to wild-type (WT) controls. Quantification of the cellular phenotype between WT (blue) and CatnBflox (red) mice at each time point (B) crypt height, (C) mitosis, and (D) apoptosis (*, p < .05 M.W.). (E, F): Immunohistochemistry demonstrating pattern of β-catenin loss and regeneration in Ah-cre but not vil-Cre-ERT2 mice (→ indicate cells with nuclear β-catenin; 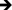 indicates β-catenin deficient cells migrating up on to villus). Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen.
indicates β-catenin deficient cells migrating up on to villus). Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen.
The repopulation in Ah-cre mice compared to vil-Cre-ERT2 was further demonstrated by β-catenin expression patterns (Fig. 3E, 3F). The Ah-cre and vil-Cre-ERT2CatnBflox/flox mice show no apparent loss of β-catenin at 1 d.p.i. compared to a WT mouse. At 2 d.p.i., vil-Cre-ERT2 mice mice show a loss of β-catenin staining in the crypt and villus region. In comparison, the loss is confined to the crypts in the Ah-cre mice, however, these crypts show evidence of β-catenin nuclear localization in single cells near the crypt base. By 3 d.p.i., the β-catenin-deficient crypt cells in the Ah-cre mice have migrated up onto the villus and been replaced with WT cells, in contrast the majority of the crypts in the vil-Cre-ERT2 mice have been lost, however, the few remaining crypts show normal β-catenin localization patterns. These remaining crypts, however, are negative for the proliferation marker Ki-67 and ISC marker Olfm4 unlike the crypts in the Ah-cre mice (Figs. 2F, 2G and 4D, 4E). Further evidence of recovery in the Ah-cre was observed in apoptosis, crypt length, crypt numbers, mitosis, and proliferation levels over 1–3 d.p.i. (Figs. 3B–3D and 4F). In both sets of mice there was a trend of decreasing crypt length by 3 d.p.i., Ah-cre mice showed a 10.8% loss in crypt length compared to a 67.3% (p = <.05) decrease in vil-Cre-ERT2 mice. However, both Ah-cre and vil-Cre-ERT2 mice showed similar increases in apoptosis within the crypts but a reduction in mitosis was only observed in the vil-Cre-ERT2 mice with a 77.8% decrease in mitotic figures. Recovery was also supported by the examination of expression levels of ISC markers and Wnt signaling genes when compared to the appropriate WT controls. Expression levels of the stem cell markers Ascl2, Bmi-1, Lgr5, and Olfm4 recovered to WT levels in Ah-cre CatnBflox/flox by 3 d.p.i. but not the vil-Cre-ERT2CatnBflox/flox mice (Fig. 4A). Whereas, the increase in expression of the Wnt ligand, Wnt3, correlated with a decrease in the Wnt inhibitor Sfrp2 in Ah-cre mice by 3 d.p.i. but not the vil-Cre-ERT2CatnBflox/flox mice (Fig. 4B). This is consistent with an activation of the Wnt pathway to drive repopulation. Further, this recovery of crypt numbers and repopulation in the Ah-cre mice correlates with a significant increase in the proportion of crypts undergoing branching, a surrogate marker for crypt fission, which was absent in the vil-Cre-ERT2 mice (Fig. 4C, 4F).
Figure 4.
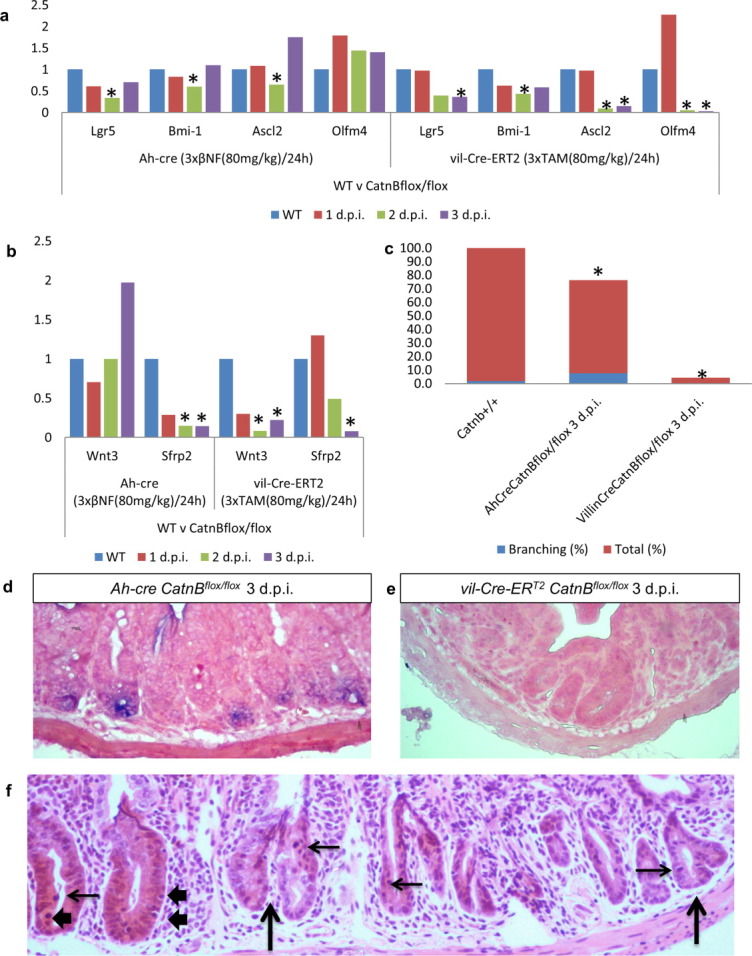
Comparison of the intestinal stem cell (ISC) characteristics using Ah-cre and vil-Cre-ERT2 systems for conditionally deleting β-catenin within the small intestine epithelia. Expression analysis of (A) ISC marker genes and (B) Wnt regulators highlighting the phenotype of loss of CatnB in the small intestine epithelia (*, p < .01 M.W.) in comparison to wild-type (WT) controls. (C): Cellular phenotype indicating % of crypts (red) and the % of branching crypts (blue) (*, p < .01 M.W.) in comparison to WT controls. (D, E): In situ hybridization for expression of the ISC marker Olfm4. (F): H&E stained cross-section of small intestine from Ah-cre CatnBflox at 3 d.p.i.; → indicates mitotic figures, ↑ indicates branching crypts, and 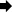 indicates apoptotic bodies. Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen.
indicates apoptotic bodies. Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen.
Vil-Cre-ERT2 but Not Ah-Cre Deletes β-Catenin in the Paneth Cells Leading to Their Death
Using immunofluorescence, we showed that vil-Cre-ERT2 but not Ah-cre recombines within the Paneth cell population (Fig. 5A), as has been previously reported. To define the effects of deleting β-catenin in these cells, we scored the presence of Paneth cells, after IHC staining with an antibody against lysozyme at 2 d.p.i. There was a significant reduction in Paneth cell number between vil-Cre-ERT2CatnBflox/flox and vil-Cre-ERT2CatnB+/+ control mice (p < .048 M.W.) which was not observed with Ah-cre CatnBflox/flox mice (Fig. 5B). The survival of the Paneth cells in the Ah-cre mice was further supported by an increase in expression of the Paneth cell expressed Wnt ligand Wnt3, which was absent in vil-Cre-ERT2 mice (Fig. 4B). We next used IHC to detect cleaved caspase 3 as a marker of apoptosis. After induction for the deletion of β-catenin, Paneth cells positive for cleaved caspase 3 were readily detected in the vil-Cre-ERT2 mice but rarely observed in the Ah-cre mice (Fig. 5C) indicating that β-catenin is critical for the survival of the Paneth cells.
Figure 5.
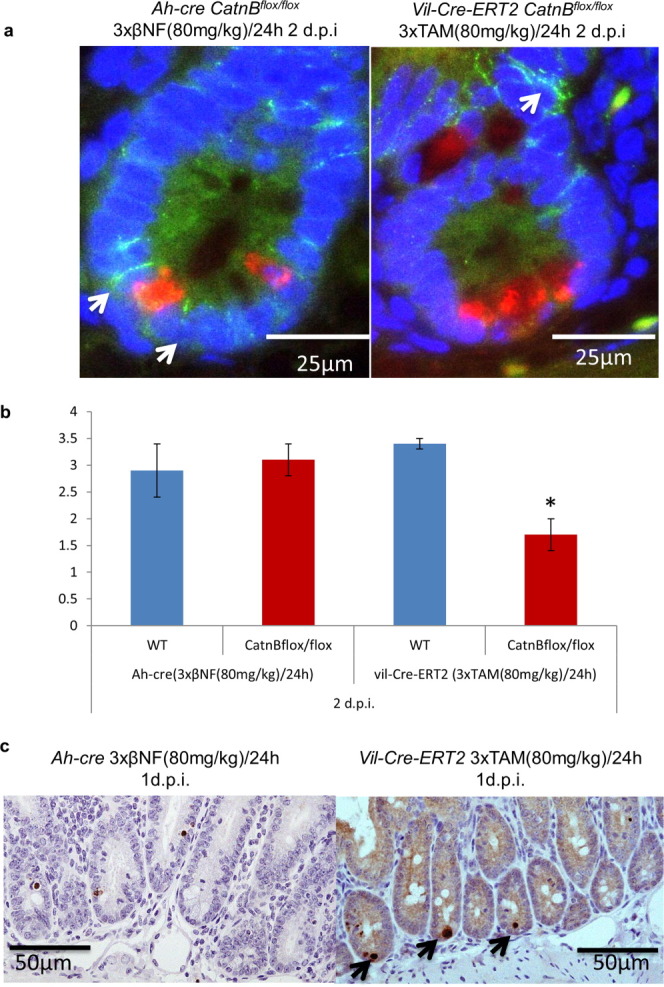
Characterization of the Paneth cells after β-catenin deletion using the Ah-cre and vil-Cre-ERT2 systems. (A): Immunofluorescence to indicate CatnB (green) deletion only occurs in the Paneth cells (red) using the vil-Cre-ERT2 system. (B): Quantification of the number of Paneth cells present 2 days after deletion of β-catenin (*, p < .04 M.W.). (C): IHC staining for cleaved caspase 3 indicating apoptotic cells at the base of the crypt, where the Paneth cells reside, in the vil-Cre-ERT2 but not the Ah-cre mice (→ indicates position of Paneth cells). Abbreviations: βNF, β-napthoflavone; TAM, tamoxifen; WT, wild type.
DISCUSSION
Recently, two populations of cells have been proposed to contain the ISC. One is marked by Bmi-1 expression and is found at position four in the crypt; the second is marked by Lgr5 expression and is located at the base of the crypt. It currently remains unclear how cells transit between these populations, although it has been demonstrated that the Bmi-1 positive population can recapitulate the Lgr5 population. One interpretation of this data is that the Lgr5 population is an “active” stem cell pool, which can be replenished from a more quiescent Bmi-1 pool of cells. This active Lgr5 population appears to be reliant on interactions with the Paneth cells within the crypt, which have been postulated to play a “nurse” role in maintaining stemness. We sought to further understand this interaction by introducing a damaging mutation into the ISC population in the presence or absence of Paneth cells. We selected deletion of β-catenin as the damaging mutation, as previous studies have shown remarkably diverse phenotypic effects of β-catenin loss dependent on the Cre transgene being used. Thus, deletion mediated by vil-Cre-ERT2 leads to catastrophic loss of the epithelium and animal death as opposed to Ah-cre mediated deletion, in which there is rapid repopulation by WT cells and no lethality.
These observations lead to two hypotheses. First, that the difference seen between the two systems is a direct reflection of differing recombination rates within the ISC compartment (either Bmi-1 or Lgr5 positive). Second, that the difference arises as a consequence of differing recombination rates in differentiated cells, such as the Paneth cells. To address these possibilities, we first defined dosing regimens that would deliver equivalent levels of ISC recombination using either Cre system. Having established this baseline, we next ensured that the differences in phenotypes previously observed were not due to technical differences associated with use of different CatnBflox alleles. We therefore performed repeat experiments with the two Cre strains and the same CatnBflox 18 allele. For both Cre transgenes, we observed a similar phenotype to that previously reported in that we observed intestine survival and repopulation using Ah-cre but crypt loss and death using vil-Cre-ERT2. These data show that the phenotypic differences are not attributable to varying levels of functional ISC recombination.
It has previously been shown that the major difference in cell type targeting within the crypt between these two systems is that Ah-cre does not drive recombination within the Paneth cells [13, 17]. However, it does drive recombination in goblet cells, another intestinal epithelium secretory cell, where deletion of CatnB results in a loss of goblet cells [13]. We again confirm here that the Paneth cells are spared in the Ah-cre model and go on to show that loss of β-catenin within the vil-Cre-ERT2 model leads to Paneth cell death. One interpretation of our data is that this is the critical difference between the two models, with loss of Paneth cells having a profound effect on the capacity to regenerate the crypt. This interpretation is supported by recent work demonstrating that when using the vil-Cre-ERT2 model to delete Math-1 all Paneth cells were lost but crypt architecture was otherwise unperturbed, as other Math-1-deficient cells expressed the necessary Wnts [28] whereas other models deleting only Paneth cells show crypt loss [12]. The alternate explanation is that vil-Cre-ERT2 more efficiently targets a potential reserve ISC population. As our data clearly shows that, where repopulation occurs, it always occurs from an unrecombined population, indicating that Ah-cre does indeed spare a “reserve” population of stem cells. The identity of this population is unknown and it is of note that there is no increase in the expression of the Bmi-1 gene, as cells which express this gene have been proposed to contain a “reserve ISC population.” However, the increase in crypt fission in Ah-cre mice does suggest that the reserve population originates from within the ISC niche. Whether this reserve population represents unrecombined ISCs or dedifferentiated ISC daughter cells remains to be seen.
CONCLUSION
In summary, we have explored two models of Cre-mediated intestinal epithelial deletion that have markedly different consequences for intestinal regeneration. We show that the differences we observe do not arise as a consequence of differential recombination rates within the functional ISC. Future work will need to clarify between the three possibilities that are consistent with our data: first, that the Paneth cell is critical as a nurse cell during crypt regeneration; second that the reserve ISC is differentially targeted by the vil-Cre-ERT2 and Ah-cre systems, and finally that both the reserve ISC and Paneth cell compartments are differentially recombined and both contribute to the observed phenotypes.
Acknowledgments
We thank Mark Bishop, Lucie Stocking, Matthew Zverev, and Derek Scarborough for technical assistance; Aliaksei Holik for scientific discussion; Sylvie Robine and D. Louvard for reviewing the manuscript. This work was supported by Cancer Research UK. L.P. would like to thank Drs. Jo Cable and Jo Lello for their kind financial contribution to help complete this work.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicates no potential conflicts of interest.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Cheng H, Leblond C. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anat. 1974;141:461–479. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- 2.Barker N, van Es J, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 3.Sangiorgi E, Capecchi M. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potten C, Loeffler M. Stem cells: Attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 5.Lee G, White L, Hurov K, et al. Response of small intestinal epithelial cells to acute disruption of cell division through CDC25 deletion. Proc Natl Acad Sci USA. 2009;106:4701–4706. doi: 10.1073/pnas.0900751106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scoville D, Sato T, He X, et al. Current view: Intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 7.Tian H, Biehs B, Warming S, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan KS, Chia LA, Li X, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snippert H, van der Flier L, Sato T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Garcia C, Klein A, Simons B, et al. Intestinal stem cell replacement follows a pattern of neutral drift. Science. 2010;330:822–825. doi: 10.1126/science.1196236. [DOI] [PubMed] [Google Scholar]
- 11.van der Flier L, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ireland H, Kemp R, Houghton C, et al. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: Effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–1246. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Fevr T, Robine S, Louvard D, et al. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madison B, Dunbar L, Qiao X, et al. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 16.Kemp R, Ireland H, Clayton E, et al. Elimination of background recombination: Somatic induction of Cre by combined transcriptional regulation and hormone binding affinity. Nucleic Acids Res. 2004;32:e92. doi: 10.1093/nar/gnh090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.el Marjou F, Janssen K, Chang B, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 18.Brault V, Moore R, Kutsch S, et al. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 19.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 20.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 21.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Yuan J, Reed A, Chen F, et al. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillemot F, Nagy A, Auerbach A, et al. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 24.van der Flier L, Haegebarth A, Stange D, et al. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 25.Gregorieff A, Pinto D, Begthel H, et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Marshman E, Ottewell P, Potten C, et al. Caspase activation during spontaneous and radiation-induced apoptosis in the murine intestine. J Pathol. 2001;195:285–292. doi: 10.1002/path.967. [DOI] [PubMed] [Google Scholar]
- 27.Merritt A, Allen T, Potten C, et al. Apoptosis in small intestinal epithelial from p53-null mice: Evidence for a delayed, p53-independent G2/M-associated cell death after gamma-irradiation. Oncogene. 1997;14:2759–2766. doi: 10.1038/sj.onc.1201126. [DOI] [PubMed] [Google Scholar]
- 28.Durand A, Donahue B, Peignon G, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc Natl Acad Sci USA. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]


