Abstract
The mammalian endoplasmic reticulum (ER) harbors disulfide bond-generating enzymes, including Ero1α and peroxiredoxin 4 (Prx4), and nearly 20 members of the protein disulfide isomerase family (PDIs), which together constitute a suitable environment for oxidative protein folding. Here, we clarified the Prx4 preferential recognition of two PDI family proteins, P5 and ERp46, and the mode of interaction between Prx4 and P5 thioredoxin domain. Detailed analyses of oxidative folding catalyzed by the reconstituted Prx4–PDIs pathways demonstrated that, while P5 and ERp46 are dedicated to rapid, but promiscuous, disulfide introduction, PDI is an efficient proofreader of non-native disulfides. Remarkably, the Prx4-dependent formation of native disulfide bonds was accelerated when PDI was combined with ERp46 or P5, suggesting that PDIs work synergistically to increase the rate and fidelity of oxidative protein folding. Thus, the mammalian ER seems to contain highly systematized oxidative networks for the efficient production of large quantities of secretory proteins.
Secretory and membrane proteins are newly synthesized and acquire their native structures in the ER. While many of these proteins undergo disulfide bond formation during folding and assembly, the introduction of a native disulfide bond is frequently not straightforward and includes processes of non-selective oxidation of two cysteines, followed by isomerization1. Accordingly, many organisms have evolved elaborate catalytic systems composed of numerous thiol-disulfide oxidoreductases with distinct functional roles. The ER of higher eukaryotes contains more than 20 members of the PDIs2,3. While the physiological functions of the individual PDIs have not been fully characterized, it is reasonable to postulate that their functions are determined not only by their intrinsic redox and chaperone activities but also by the molecules with which they cooperatively interact4,5.
PDI, a member of PDIs, and ER oxidoreduclin 1 (Ero1) play a central role in disulfide bond formation in eukaryotic cells6. Ero1 is a highly conserved flavoenzyme that generates a disulfide bond de novo in conjunction with bound flavin adenine dinucleotide (FAD) and transfers the disulfide bond to substrates via PDI7,8,9,10. Since Ero1-mediated PDI oxidation yields hydrogen peroxide (H2O2), a source of reactive oxygen species (ROS)11, Ero1 activity is tightly regulated by the redox environment in the ER8. Ero1 is activated only when the ER becomes highly reducing. This feedback regulation is ensured through oxidation/reduction (in the case of yeast Ero1p)12 or internal disulfide rearrangement (in the case of human Ero1α)13,14,15 of redox-sensing regulatory cysteines. In this process, reduced PDI presumably functions as a modulator and substrate of Ero1.
While Ero1 and PDI constitute a major self-regulatory pathway for oxidative protein folding, recent studies report that several other enzymes with significant oxidative activity toward PDIs are present in the mammalian ER4,16. In support of these findings, disruption of two mammalian Ero1 isoforms, Ero1α and Ero1β, only modestly delays oxidative folding of limited substrates, suggesting the existence of Ero1-independent disulfide bond formation pathways in mammalian cells17. Thus, the oxidative folding network in higher eukaryotes may be more complicated and more diversified than previously thought.
Prx4 has recently been identified as an alternative PDI oxidation enzyme. It belongs to the typical 2-Cys Prx family18,19 and irrespective of its redox state, forms a homodecamer within which each dimer constitutes an essential functional unit20,21. During the Prx4 catalytic cycle, a peroxidatic cysteine in one chain reduces H2O2, generating water, and is oxidized to a cysteine sulfenic acid22. This sulfenylated cysteine reacts with a resolving cysteine of the partner chain within the dimer to generate a protein disulfide bond de novo. The concentration of Prx4 in the ER is approximately equivalent to that of PDI and greater than that of Ero1α19. Despite a subtle kinetic defect in oxidative folding of immunoglobulin M, combined loss-of-function mutations in the genes encoding Ero1α, Ero1β and Prx4 profoundly decrease procollagen 4-hydroxyproline content and enhance cysteinyl sulfenic acid modification of ER proteins, resulting in ascorbic acid depletion23.
Since Prx4 is capable of efficiently oxidizing several PDIs, including PDI, P5, ERp46, and ERp57 in vitro19, Prx4 likely cooperates with Ero1α to oxidize PDIs and sustain oxidative folding of secretory proteins in cells. However, the extent to which these two enzymes functionally overlap or complement each other remains to be determined. In the present study, we extensively and quantitatively analyzed activity of Prx4 toward PDIs, which revealed that Prx4 has a strong preference for P5 and ERp46 both in cells and in vitro. Crystallographic and biochemical studies further revealed that Prx4 separately recognizes each Trx domain within PDIs via a specific interaction of its C-terminal extended segment with the peptide-binding groove of the Trx domain. A detailed investigation into oxidative protein folding catalyzed by Prx4 and each PDIs indicated that, while ERp46 and P5 engaged in rapid, but promiscuous, disulfide introduction, PDI was an effective proofreader of non-native disulfide bonds due to its prominent disulfide isomerase activity. By virtue of such distinct functions of PDIs, the rate and fidelity of Prx4-dependent oxidative folding was strikingly increased when PDI was combined with P5 or ERp46. Taken together, the results indicate that PDIs cooperate together to ensure proper and effective oxidative folding of secretory proteins, which likely explains why they are so multiple in the mammalian ER.
Results
Activity of Prx4 and Ero1α toward PDIs in vitro
We first purified recombinant forms of several PDIs, namely PDI, P5, ERp46, ERp57, and ERp72 (supplementary Fig. S1A), all of which are abundant in the mammalian ER and consist of multiple Trx-like domains (Fig. 1A). While PDI, ERp57, and ERp72 possess a U-shaped domain arrangement24,25,26, the full-length structures of P5 and ERp46 remain to be determined. To quantitatively assess Prx4 and Ero1α activity toward these five PDIs, we set up an assay system, wherein Prx4- or Ero1α-catalyzed oxidation of PDIs is coupled to NADPH consumption via the redox cycles of PDIs and glutathione (GSH) (supplementary Fig. S1B). As shown by the decay in absorbance at 340 nm (Abs340), Prx4 showed significant oxidative activity toward all five PDIs while P5 appeared the best substrate of them (supplementary Fig. S1C). The decay curves suggested discrete phases in the oxidation process, which could be explained by the separate oxidation of individual Trx domains at different rates by Prx4 (see supplementary Fig. S2A and S2B). In contrast to Prx4, Ero1α showed much narrower specificity; the enzyme oxidized PDI and, to a lesser extent, ERp46 but oxidized the others to a much lesser extent (supplementary Fig. S1D). Thus, Prx4 likely has the potential to oxidize a broader range of PDIs than Ero1α, as suggested previously19.
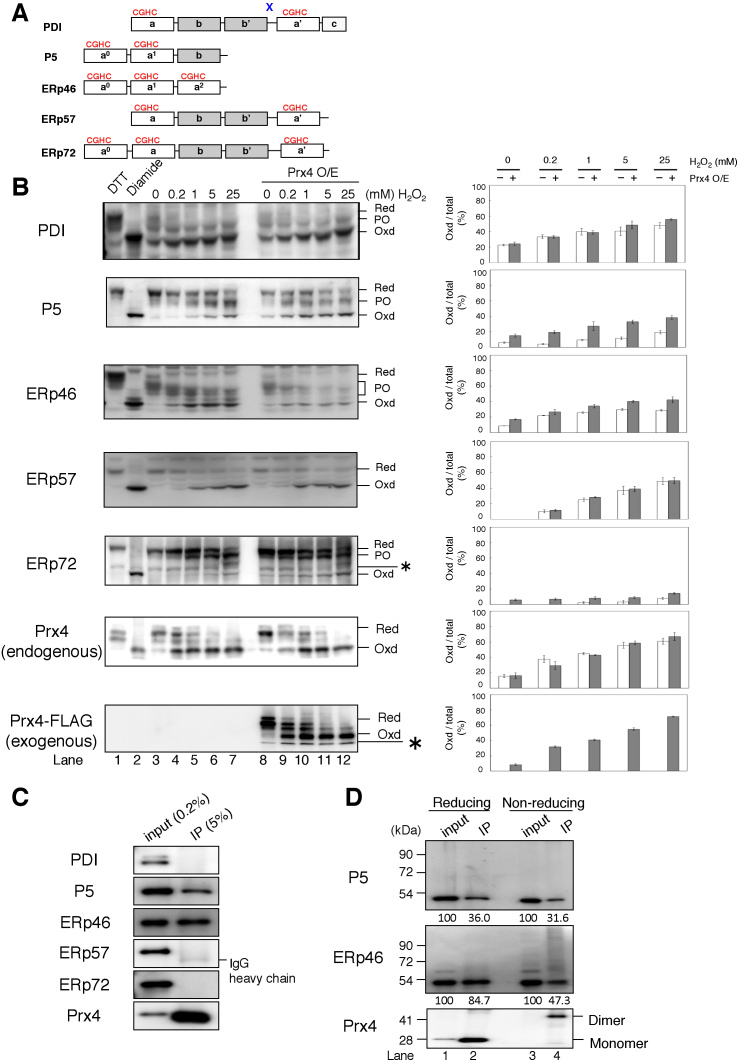
Figure 1. Activity of Prx4 and Ero1α toward PDIs in cultured cells.
(A) Schematic representation of the domain architecture of the five PDIs used in this study. Redox-active Trx domains with a CGHC motif are indicated by a, a′, a0, and a1, and redox-inactive Trx-like domains lacking the motif are indicated by b and b′. The linker region flanked by b′ and a′ in PDI is indicated by “x”. (B) Redox states of PDIs were visualized by immunoblotting the TCA precipitates of wild-type or Prx4-overexpressing HEK293 cells pretreated with various concentrations of H2O2 (left panel), after the alkylation of free cysteines. The representative non-reducing gel images of the three independent experiments are shown. Labels Red, PO, and Oxd denote fully reduced, partially oxidized, and fully oxidized forms of PDIs, respectively. Asterisks indicate non-specific bands. Red and Oxd controls were prepared by pretreatment of the cells with DTT and diamide, respectively. Oxd forms of PDIs are shown as a percentage of total (Red, PO, and Oxd) PDIs in wild-type or Prx4-overexpressing cells (right panel). Band intensities were normalized by taking into account the different reactivities of the antibodies to the reduced and oxidized forms; the Oxd-to-Red ratio of antibody activity was 2.12 for PDI, 1.27 for P5, 1.13 for ERp46, 3.55 for ERp57, 1.45 for ERp72, and 1.29 for Prx4. Also, the antibody activity to PO was assumed to be the same as that to Red. Results in the bar graphs represent the mean ± SD of three independent experiments. Note that an anti-Prx4 antibody used in this study was active against endogenous human Prx4 but not against mouse Prx4-FLAG. The data are cropped to highlight the region of interest and the full-length blots are presented in Supplementary Fig. S3A. (C–D) Co-immunoprecipitation of PDIs with Prx4. Immunoprecipitates obtained using an anti-Prx4 antibody were subjected to reducing (C and D lane 1 & 2) or non-reducing (D lane 3 & 4) SDS-PAGE followed by immunoblotting with antibodies to each PDIs or Prx4. The blotting data are cropped to highlight the bands deriving from each PDIs and Prx4 in (C).
Activity of Prx4 toward PDIs in cultured cells
To examine the activity of Prx4 toward PDIs in cells, we next determined the redox states of endogenous PDIs in wild-type and Prx4-overexpressing HEK293 cells pretreated with H2O2 for the purpose of activation of Prx4 and possibly other H2O2-dependent PDIs oxidation enzymes including glutathione peroxidase 7/8 (GPx7/8)27. While all the examined PDIs but PDI were predominantly in the reduced form at steady state as reported previously28,29 (Fig. 1B left, lane 3), they all showed a significant shift toward oxidized forms upon H2O2 titration (Fig. 1B, lane 4–7). Presumably, externally added H2O2 made the ER environment more oxidizing and hence increased oxidized forms of PDIs in a dose-dependent manner. Direct oxidation by H2O2 and indirect oxidation via H2O2-dependent PDIs oxidation enzymes would also be possible. Indeed, Prx4 overexpression resulted in a further increase in the oxidation of P5, ERp46 and to a lesser extent ERp72 (Fig. 1B, lane 8–12), which is consistent with the high activity of Prx4 toward P5 and ERp46 observed in vitro (supplementary Fig. S1C). By contrast, PDI and ERp57 resisted to further oxidation by overexpressed Prx4 (Fig. 1B). It is also noted that both endogenous (human) and exogenous (mouse) Prx4 were predominantly reduced at steady state but were converted to an oxidized form in a manner that was highly sensitive to H2O2 addition (Fig. 1B and Supplementary Fig. S3A). These results imply that the amount of H2O2 in the ER at steady state is limited and that a slight increase in the H2O2 level is sufficient to convert Prx4 to a form that can oxidize PDIs.
We next investigated the physical interaction between Prx4 and PDIs in cultured cells. Immunoprecipitation with an anti-Prx4 antibody, followed by immunoblotting with an antibody for each PDIs, indicated that endogenous Prx4 binds to P5 and ERp46 exclusively (Fig. 1C). ERp46 not covalently linked to another protein was less detectable under non-reducing conditions than under reducing conditions (Fig. 1D, lane 2 & 4). Instead, several bands of higher molecular weight were clearly observed (Fig. 1D, lane 4), indicating that endogenous ERp46 exists as a population of mixed disulfide species with Prx4. On the other hand, neither PDI, ERp57, or ERp72 was immunoprecipitated with anti-Prx4, suggesting that Prx4 preferentially interacts with P5 and ERp46 under physiological conditions.
Preferential oxidation of P5 and ERp46 by Prx4
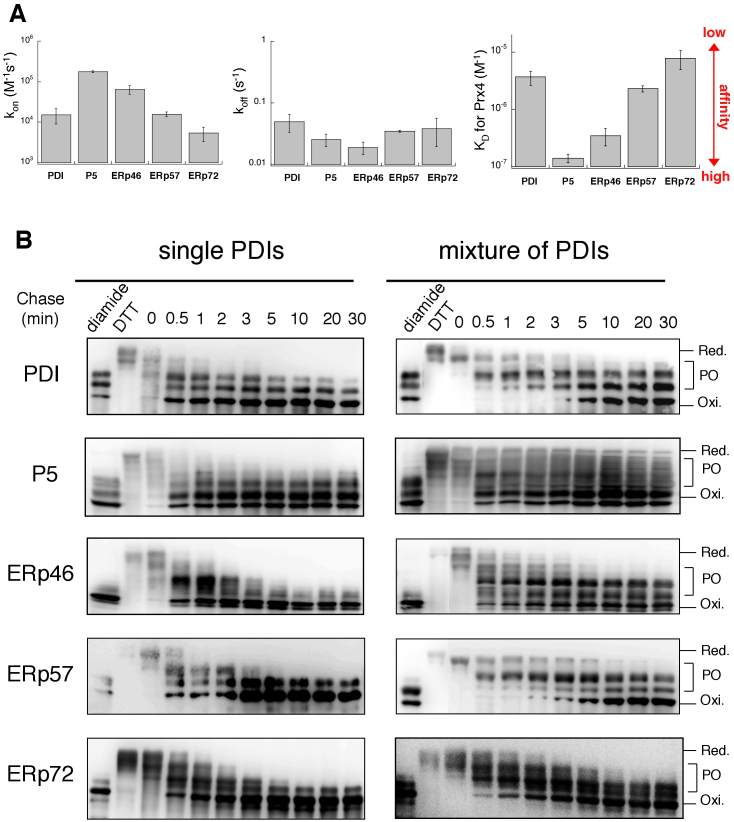
The preferential association of Prx4 with ERp46 and P5 is further supported by the surface plasmon resonance (SPR) analyses. The ‘KD for ERp46 or P5’ value of Prx4 was one or two orders lower than that for other PDIs (Fig. 2A and Supplementary Fig. S4), indicating the particularly high affinity for these two PDIs of Prx4. This observation may suggest that ERp46 and P5 have inhibitory effects on Prx4-catalyzed oxidation of other PDIs. To test this possibility, a mixture of all five PDIs was reacted with Prx4 in vitro, and the time courses of their redox state changes were monitored by immunoblotting with an antibody directed against each PDIs (Fig. 2B right). Notably, fully oxidized forms of ERp46 and P5 were generated at a very early reaction time (~0.5 min of chase), to almost the same extent regardless of the presence of other PDIs (Fig. 2B). On the other hand, full oxidation of the other PDIs, especially PDI and ERp57, were substantially slowed down by the addition of other PDIs (Fig. 2B right); fully oxidized PDI and ERp57 started to appear when the Prx4-catalyzed oxidation of ERp46 and P5 apparently stopped (~5 min of chase). Thus, the presence of reduced ERp46 and P5 significantly inhibited Prx4-catalyzed oxidation of PDI and ERp57. Partially oxidized forms were observed persistently for all the examined PDIs in their mixture. One possible explanation for this would be that the largely decreased molar ratio of H2O2 to PDIs does not allow the full oxidation of all the PDIs involved and that P5 and ERp46 preferentially oxidized by Prx4, in turn, engage in direct oxidation of the other PDIs, resulting in reduction of these two PDIs. Taken together, the results suggested that ERp46 and P5 are oxidized preferentially by Prx4, which is likely explained in terms of the different affinity for each PDIs of Prx4.
Figure 2. Preferential oxidation of ERp46 and P5 by Prx4 in vitro.
(A) SPR analysis of the affinity of PDIs for Prx4. The bar graphs show the association (kon) and dissociation (koff) rate constants and KD values of PDIs for Prx4 (see also Supplementary Fig. S4). Values are the mean ± SD of three independent experiments. (B) Reduced form(s) of a single PDI family member or a subset of PDIs (10 μM each) were reacted with Prx4 (0.2 μM) in buffer (pH 7.5) containing 300 μM H2O2. The reaction was quenched by cysteine alkylation with malPEG2K, and the generated species were resolved by non-reducing SDS-PAGE and visualized by immunoblotting with the indicated antibodies. The representative gel images of the three independent experiments are shown. Labels Red., PO, and Oxi. denote fully reduced, partially oxidized, and fully oxidized forms of PDIs, respectively. The blotting data are cropped to highlight the time course of Prx4-catalyzed oxidation of each PDIs.
Separate recognition of Trx domains of PDIs by Prx4
Cytosolic and mitochondrial members of the 2-Cys Prx family, Prx1, Prx2 and Prx3, accept electrons from thioredoxin to metabolize H2O2. Thus, we hypothesized that Prx4 would recognize the Trx domain of each PDIs separately rather than recognizing the overall structure. To test this hypothesis, we constructed CxxC mutants of P5 and ERp46, in which one CxxC motif was retained and the remainders were replaced with AxxA. We also isolated and purified the Trx domains of P5 and ERp46. Prx4 oxidized isolated P5 a0 at a similar rate to P5 CxxC-AxxA, which contained a CxxC sequence in a0 and an AxxA sequence in a1 (supplementary Fig. S2A). Isolated P5 a1 was oxidized more slowly and had an oxidation rate similar to that of P5 AxxA-CxxC. These results strongly suggest that Prx4 recognizes and oxidizes P5 a0 and a1 independently. Similarly, the isolated a0, a1, and a2 domains of ERp46 were oxidized by Prx4 at almost the same rates as the full-length mutants CxxC-AxxA-AxxA, AxxA-CxxC-AxxA, and AxxA-AxxA-CxxC, respectively (supplementary Fig. S2B). It is thus conceivable that Prx4 discriminates between individual Trx domains, irrespective of where they are located in the whole proteins.
Structural basis for recognition of Trx domains of PDIs by Prx4
We solved the crystal structure of the oxidized form of Prx4 from Mus musculus (mPrx4) at a resolution of 3.3 Å (Table 1). As predicted from the high (96%) identity in the amino acid sequence (supplementary Fig. S5A), mPrx4 forms a decamer that is highly superposable on Prx4 from Homo sapiens (hPrx4) (supplementary Fig. S5B&S5C)20,21. The approximate diameter and height of mPrx4 are 120 Å and 45 Å, respectively. We modeled amino acid residues 79–241 and 247–255, but not those of the C-terminal segments, due to the lack of electron density (supplementary Fig. S5D, left). As is commonly observed for hPrx4 and other typical two-Cys peroxiredoxins30, oxidation of the active-site cysteine residues (Cys127 in one chain and Cys248 in the neighboring chain) induces local unfolding in the C-terminal helical arm, which contains a resolving cysteine (Cys248) and an α6-helix covering the α2-helix of the partner chain. Also, the C-terminal end of the α2-helix, which contains a peroxidatic cysteine (Cys127), is unwound in the oxidized form, exposing the Cys127-Cys248 disulfide on the molecular surface (supplementary Fig. S5D).
Table 1. Data collection and structure determination.
| mPrx4 C54A (full-length) (oxidized form) | P5 a0-Prx4 C-terminal peptide complex | |
|---|---|---|
| Data collection | ||
| Beamline | BL44XU at SPring-8 | BL5A at KEK |
| Space group | P21221 | P212121 |
| Cell dimensions (Å) | a = 87.5, b = 118.7, c = 255.7 | a = 39.0, b = 53.4, c = 133.4 |
| α = β = γ = 90.0° | α = β = γ = 90.0° | |
| Wavelength (Å) | 0.90000 | 1.0000 |
| Resolution range (Å) | 43.73 − 3.30 (3.38 − 3.30) | 41.67 − 2.10 (2.14 − 2.10) |
| No. of total observations | 254,583 | 112,980 |
| No. of unique reflections | 38,676 | 16,851 |
| Completeness (%) | 94.6 (88.7) | 98.9 (92.7) |
| I/σ(I) | 13.9 (4.4) | 26.0 (3.1) |
| Multiplicity | 6.6 (6.5) | 6.7 (5.7) |
| Rmergea | 0.104 (0.450) | 0.070 (0.500) |
| Refinement | ||
| Resolution range (Å) | 43.11 − 3.30 | 41.67 − 2.10 |
| Rworkb | 0.203 | 0.182 |
| Rfreec | 0.255 | 0.245 |
| RMS deviation | ||
| Bond length (Å) | 0.014 | 0.007 |
| Bond angle (°) | 1.8 | 1.0 |
| Ramachandran analysise | ||
| Most favored (%) | 98.3 | 96.4 |
| Allowed (%) | 1.7 | 3.6 |
| Generously allowed (%) | 0.0 | 0.0 |
| Disallowed (%) | 0.0 | 0.0 |
The number in parentheses represent statistics in the highest resolution shell.
aRmerge = ΣΣj|<I(h)> − I(h)j|/ΣΣj|<I(h)>|, where <I(h)> is the mean intensity of symmetry-equivalent reflections.
bRwork = Σ(IIFp(obs)I − IFp(calc)II)/ΣIFp(obs)I.
cRfree = R factor for a selected subset (5%) of the reflections that was not included in prior refinement calculations.
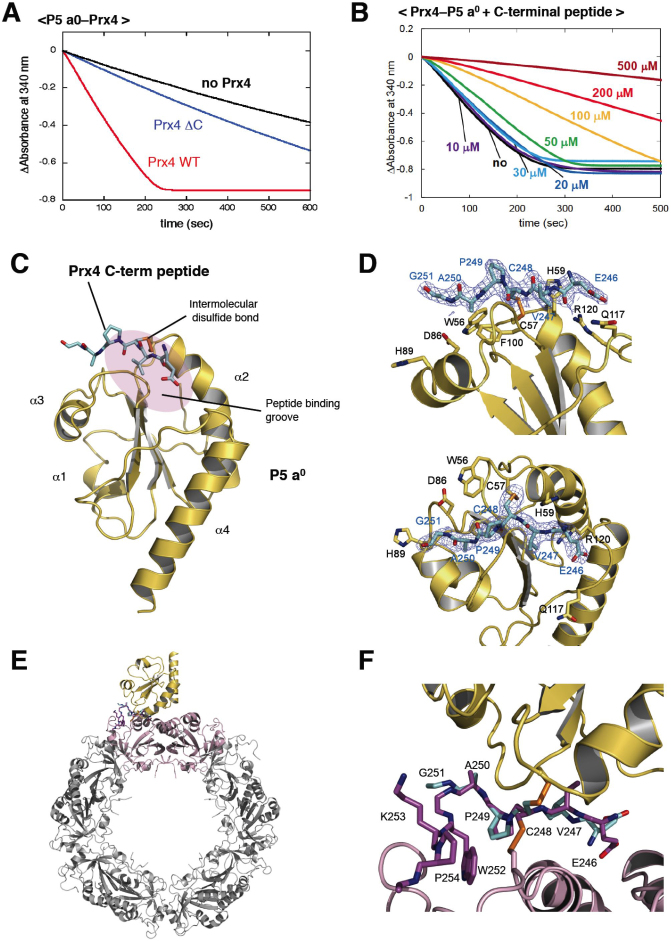
Such redox-dependent conformational changes in Prx4 suggest that the C-terminal segment unstructured upon oxidation plays an essential role in oxidation of PDIs. To examine this, we constructed a C-terminally truncated Prx4 mutant that lacked the residues after Pro249 (Prx4 ΔC). As expected, the mutant grossly lost oxidative activity toward P5 a0 (Fig. 3A) and ERp46 a0, a1, and a2 (data not shown). Also, the titration of the Prx4 peptide composed of residues 244–253 exhibited a striking inhibitory effect on the Prx4-catalyzed oxidation of P5 a0 in the range of 50–500 μM (Fig. 3B). Thus, the C-terminal segment of Prx4 likely plays a key role in the functional interaction with Trx domains of PDIs.
Figure 3. Mode of interaction between Prx4 and Trx of P5.
(A) Activity of the C-terminally truncated Prx4 mutant (Prx4 ΔC) toward P5 a0. The experimental conditions were as described in Materials and Methods. Consumption of NADPH coupled to Prx4 catalysis was detected by monitoring absorbance at 340 nm (Abs340). (B) Inhibition of Prx4-catalyzed oxidation of P5 a0 by the titration of the Prx4 C-terminal peptide. The experiments were carried out as in (a) in the presence of various concentrations (0, 10, 20, 30, 50, 100, 200, 300, 500 μM) of the Prx4 C-terminal peptide. (C) Crystal structure of P5 a0 (yellow) in a complex with the Prx4 C-terminal peptide (cyan) at 2.1 Å resolution. The Prx4 peptide is shown with sticks. Cys57 in the redox-active site of P5 a0 formed a mixed disulfide bond with the resolving cysteine (Cys248) of Prx4, as illustrated by orange sticks. The peptide-binding groove of P5 a0 is shown with a purple oval. (D) Close-up views of the interface of the P5 a0–Prx4 C-terminal peptide complex from two different angles. Electron density map at the 1.0 contour level of the Prx4 C-terminal peptide is indicated with blue mesh. (E) Modeled structure of the P5 a0–Prx4 decamer complex. The Prx4 C-terminal peptide bound to P5 a0 is superposed on the corresponding part of the oxidized Prx4 decamer such that the RMSD of the Cα atoms of Val247, Cys248, Pro249, and Ala250 is minimized. A dimeric unit of Prx4 that interacts with P5 a0 is highlighted in pink. (F) Close-up view of the interface of the P5 a0–Prx4 decamer complex shown in (E).
To gain further insights into the mode of interaction between Prx4 and a Trx domain, we determined the crystal structure of P5 a0 in a complex with the Prx4 C-terminal peptide (His244–Pro263 from mPrx4) at 2.1Å resolution (Table 1). The covalent complex was prepared via an intermolecular disulfide between Cys57 of P5 a0 and Cys248 of the peptide. P5 a0 assumed a prototypical Trx fold composed of a three-stranded central β-sheet surrounded by four α-helices, and the redox-active site was located at the N-terminus of the second α-helix (α2) (Fig. 3C). Importantly, peptide electron density was visible only for the region from Glu246 to Gly251, and, thus, we found it bound a hydrophobic groove of P5 a0 (Fig. 3D). A peptide-binding groove is found near the redox-active cysteine in almost all Trx family proteins31,32,33, which likely underlies the intrinsic broad activity of Prx4 toward PDIs in vitro.
Using the crystal structures of the oxidized form of decameric mPrx4 and the P5 a0–Prx4 peptide complex, we modeled the structure of the Prx4-P5 a0 complex in silico (Fig. 3E). The backbone of the Prx4 peptide is highly superposable on the corresponding part of decameric mPrx4, and only the side chain of Cys248 flips out to form a mixed disulfide with P5 a0 (Fig. 3F). These structural insights indicate that no large conformational changes were induced when P5 a0 formed an intermediate complex with Prx4 via an intermolecular disulfide bond. In other words, the C-terminal region of oxidized Prx4 assumes a conformation readily susceptible to nucleophilic attack by Cys57 of P5 a0.
Oxidative folding of RNase A catalyzed by Prx4 and PDIs
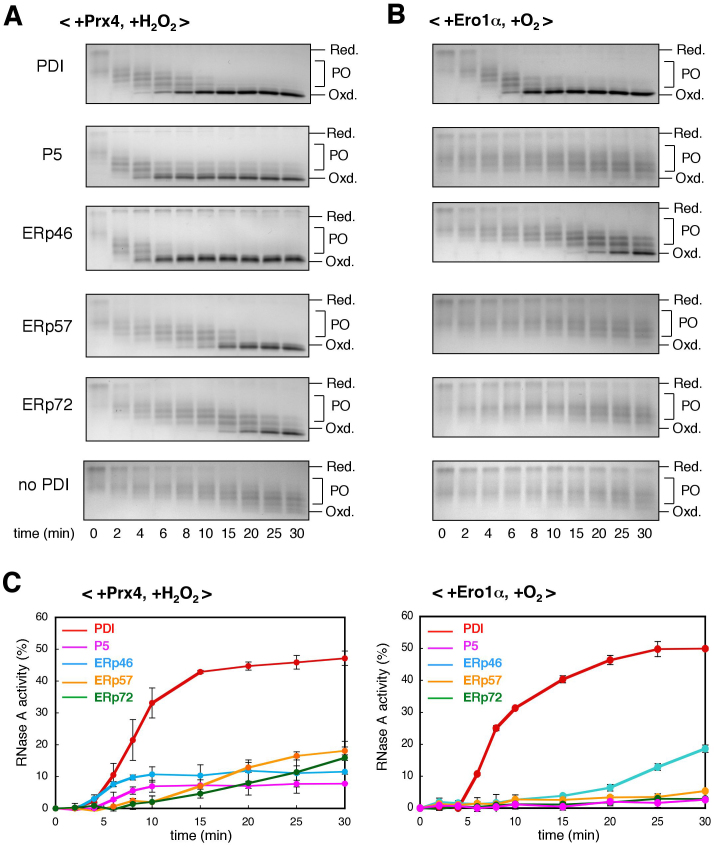
To explore the functional roles of the Prx4-PDIs oxidation pathways, we next monitored, by non-reducing SDS-PAGE, the time course of protein disulfide bond formation in the presence of Prx4 and each PDIs. Fully reduced and denatured RNase A, a model substrate, contains eight free cysteines, whose alkylation largely decreases its electrophoretic mobility. As shown in Fig. 4A and supplementary Fig. S6, Prx4 with any PDIs converted reduced RNase A to a fully oxidized form more rapidly than Prx4 alone (no PDI). Of the PDIs investigated here, P5 and ERp46 were the most active in this respect. Without Prx4, only a small amount of fully oxidized RNase A was generated slowly (supplementary Fig. S7), indicating that PDIs and H2O2 alone were insufficient to oxidize RNase A efficiently. The results are consistent with the observation that Prx4 has broad specificity for PDIs but a particular preference for P5 and ERp46. In contrast, Ero1α showed strong selectivity toward PDIs in full oxidation of RNase A; PDI and, to a lesser extent, ERp46, but not the other PDIs, generated fully oxidized RNase A in an Ero1α-dependent manner (Fig. 4B). Thus, the differences in the activities of Prx4 and Ero1α toward PDIs were reflected in the differences in the efficiencies with which they introduced disulfide bonds into RNase A.
Figure 4. Oxidative folding of RNase A catalyzed by combinations of Prx4 or Ero1α with each PDIs.
(A) Time course of oxidation of reduced and denatured RNase A catalyzed by the Prx4-PDIs combinations. RNase A was refolded for the indicated times by Prx4 and the indicated PDIs in buffer containing H2O2. The reaction was quenched by cysteine alkylation with AMS, and the generated species were subsequently resolved by non-reducing SDS-PAGE. Labels Red., PO, and Oxd. denote fully reduced, partially oxidized, and fully oxidized RNase A, respectively. The representative non-reducing gel images of the three independent experiments are shown. The data are cropped to highlight the region of interest and the full-length gels are presented in Supplementary Fig. S6. (B) Time course of oxidation of reduced and denatured RNase A by the Ero1α-PDIs combinations. Procedures were as described in (A), with the exception that RNase A was refolded in the presence of a constitutively active mutant of Ero1α in buffer saturated with oxygen (no H2O2). The representative non-reducing gel images of the three independent experiments are shown. The data are cropped to highlight the region of interest and the full-length gels are presented in Supplementary Fig. S6. (C) Time course of recovery of RNase A activity measured by the linear increase in Abs295 using cCMP as a substrate45. The initial rate of hydrolysis of cCMP by the same concentration (25 μM) of native RNase A was set to 100%, and all initial rates were expressed as a percentage of this activity. Refolding of RNase A was carried out as described in (A) (left) or (B) (right). Values are the mean ± SD of three independent experiments.
We next monitored the time course of recovery of RNase A activity to see if the Prx4- or Ero1α-dependent PDIs oxidation pathways could promote proper folding through the formation of native disulfide bonds. In the presence of Prx4 or Ero1α, PDI was by far the most efficient in recovering RNase A activity (Fig. 4C). While the RNase A activity was not completely recovered by Prx4-PDI or Ero1α-PDI combination probably due to generation of some misfolded forms under the strong oxidative bias, the time point of reaching the maximum level was almost identical to that of full oxidation. On the other hand, P5 and ERp46 failed to restore RNase A activity efficiently although they rapidly introduced disulfide bonds into reduced RNase A in a Prx4-dependent manner. We conclude that, of the PDIs examined, PDI functions as the most effective catalyst for native disulfide bond formation in concert with Prx4 or Ero1α.
Oxidative folding of bovine pancreatic trypsin inhibitor (BPTI) catalyzed by Prx4 and PDIs
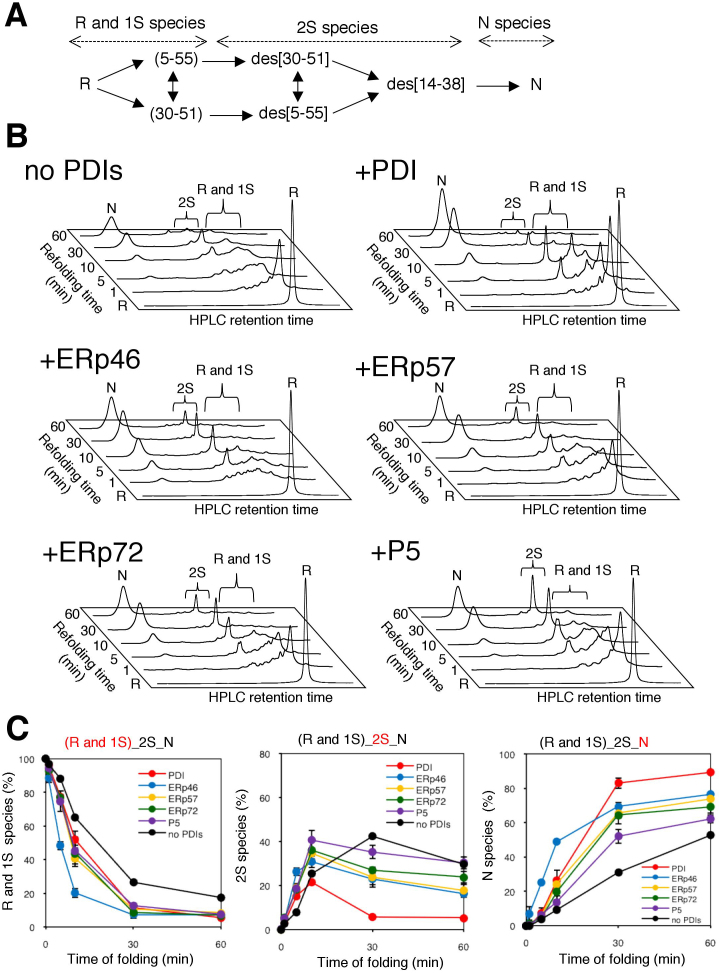
To gain a more general view of the distinct functional roles of PDIs in oxidative folding, we used BPTI as another folding substrate. BPTI possesses three disulfide bonds in its native conformation and accumulates one-disulfide (1S) and two-disulfide (2S) species as folding intermediates (Fig. 5A)34,35. The HPLC profiles shown in Fig. 5B demonstrate the time course of the conformational transitions from R (no disulfide) to 1S and 2S intermediates, and finally to N (three disulfide) species during the catalysis performed by Prx4 and each PDIs. While BPTI can fold correctly within 60 min without the assistance of PDIs, each PDIs significantly increased the efficiency with which the native species was yielded (Fig. 5B).
Figure 5. Oxidative folding of BPTI catalyzed by combinations of Prx4 with each PDIs.
(A) Schematic representation of oxidative folding pathways of BPTI. R and N indicate the reduced and native forms, respectively. 1S and 2S species are ensembles of folding intermediates with corresponding numbers of disulfide bonds. The disulfide pairings in 1S species are shown between parentheses, while the missing disulfides in 2S species are indicated with the prefix ‘des’ (des species). (B) HPLC analysis of oxidative folding of BPTI catalyzed by combinations of Prx4 with each PDIs. R, 1S, 2S, and N species of BPTI were generated by incubation with Prx4 and each PDIs in buffer (pH 7.5) containing H2O2 for the indicated times, and the species were separated by reverse-phase HPLC. (C) Quantitative analyses of the folding intermediates. The occupancies of R + 1S, 2S, and N species were plotted as a function of refolding time. Values are the mean ± SD of three independent experiments.
Of note, the quantitative dissection of each transition step in BPTI folding (Fig. 5C) indicates that the Prx4-ERp46 combination was the most effective in converting R and 1S species to species with more disulfide bonds, corroborating the observation that ERp46 introduces disulfide bonds efficiently. However, significant amount of the 2S species existed even after 60 min incubation in this case. In contrast, the Prx4-PDI combination converted R and 1S species relatively slowly, but yielded more native BPTI than the Prx4-ERp46 combination via the prompt conversion of the 2S form to the native conformation (Fig. 5B & 5C). Thus, PDI contributes more significantly to folding during later steps than during early steps in conversion. The Prx4-P5 combination converted R and 1S species to 2S species at almost the same rate as the Prx4-ERp57 or Prx4-ERp72 combination but accumulated even larger amount of the 2S species throughout the refolding reaction, resulting in the lowest level of conversion to the native species (Fig. 5B & 5C). Thus, P5 was active in acceleration of the early steps of disulfide bond introduction to BPTI, but defective in converting the 2S species to native species compared with PDI. Taken together, the results provide strong evidence for distinct functions of PDIs during Prx4-driven oxidative folding.
Synergistic effects of PDIs on Prx4-driven oxidative protein folding
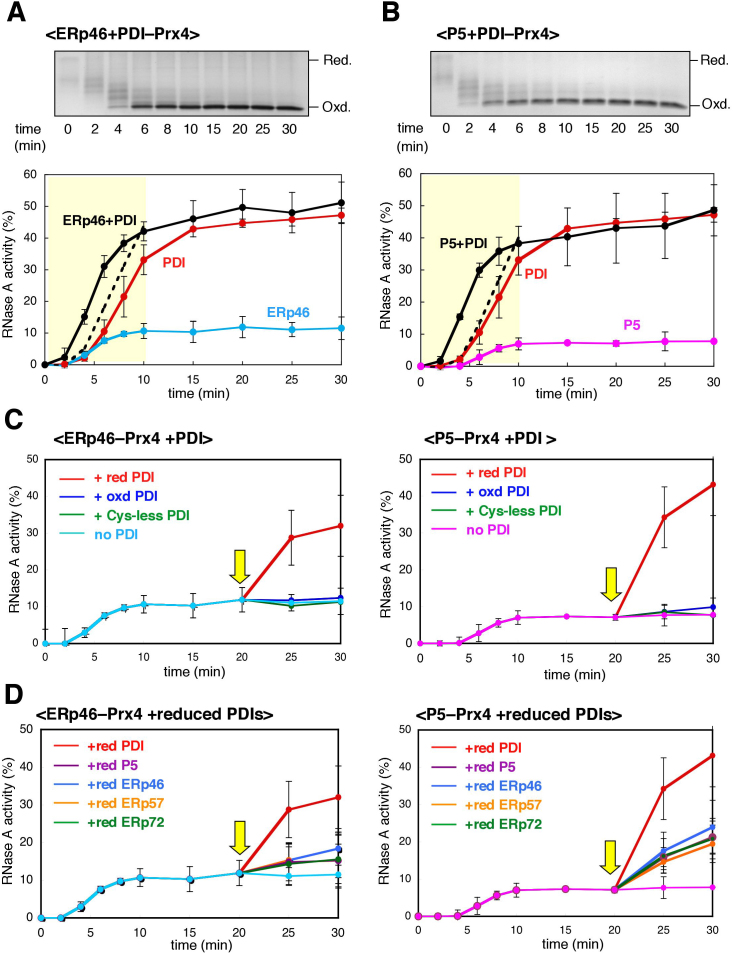
Despite the lack of fidelity, ERp46 rapidly introduced disulfide bonds into RNase A and BPTI in conjunction with Prx4 (Figs. 4 & 5). The Prx4-P5 combination also functioned as an effective disulfide introducer for RNase A (Fig. 4A). These findings suggest that, in the presence of an efficient disulfide isomerase, ERp46 and P5 greatly accelerate native oxidative folding. We therefore investigated the effect of PDI on RNase A refolding catalyzed by Prx4 in the presence of either ERp46 or P5. While Prx4 in the presence of PDI and ERp46 did not significantly affect the rate of conversion to the fully oxidized form (Fig. 6A upper panel), the recovery of RNase A activity was dramatically accelerated by this combination; Prx4 in the presence of ERp46 and PDI restored RNase A activity faster than would have been expected from the simple addition of the curves for Prx4-PDI and Prx4-ERp46 (see the yellow zone in Fig. 6A, lower panel). This was also the case for refolding catalyzed by Prx4 in the presence of P5 and PDI (Fig. 6B). By contrast, the effect was only marginal with Prx4 in the presence of P5 and ERp46 (supplementary Fig. S8), indicating an essential role of PDI in accelerating the native oxidative folding.
Figure 6. Synergistic action of PDIs during Prx4-driven oxidative folding.
(A) Effects of PDI on oxidative folding of RNase A catalyzed by Prx4 and ERp46. RNase A was refolded for the indicated times by Prx4 and ERp46 in the presence of an amount of PDI equimolar to that of ERp46 (2 μM) in buffer containing H2O2. The reaction was quenched by cysteine alkylation with AMS, and the products were subjected to non-reducing SDS-PAGE (upper panel). The representative non-reducing gel images of the three independent experiments are shown. The gel data is cropped to highlight the time course of RNase A oxidation. Time course of recovery of RNase A activity was measured by the change in Abs295, using cCMP as a substrate (lower panel). Values represent the mean ± SD from three independent experiments. The dotted lines indicate the result of the simple addition of the recovery curves for Prx4-PDI and Prx4-ERp46 for the refolding time 0–10 min. (B) Effects of PDI on oxidative folding of RNase A catalyzed by Prx4 and P5. Procedures were as described in (A), with the exception that P5 replaced ERp46 in the reaction mixture. The representative non-reducing gel images of the three independent experiments are shown. The gel data is cropped to highlight the time course of RNase A oxidation. (C) Recovery of RNase A activity by the action of PDI. Reduced and denatured RNaseA was pre-incubated with Prx4 and either ERp46 (left) or P5 (right) in buffer containing H2O2 for 20 min, followed by the addition of reduced, oxidized, or Cys-less PDI (5 μM) (yellow arrow). Values are the mean ± SD of three independent experiments. (D) Effects of reduced PDIs on recovery of RNase A activity. Procedures were as described in (C), with the exception that, after the 20-min pre-incubation, the reduced forms of the indicated PDIs (5 μM) were added to the reaction solution (yellow arrow). Values are the mean ± SD of three independent experiments.
The above findings suggest that, during the Prx4-driven oxidative folding of RNase A, ERp46 and P5 function to rapidly but promiscuously introduce disulfide bonds, whereas PDI serves primarily as a disulfide isomerase. In line with this interpretation, Prx4-catalyzed oxidation of PDI was significantly inhibited in the presence of ERp46 and/or P5, resulting in the prolonged accumulation of reduced form of PDI, a disulfide isomerase active form (Fig. 2). Moreover, addition of reduced PDI to the refolding reaction solution that had been pre-incubated with Prx4 and either ERp46 or P5 for 20 min, dramatically restored RNase A activity to a level close to that achieved de novo by the Prx4-PDI combination (Fig. 6C). This restoration was not observed with oxidized PDI or a redox-inactive (Cys-less) mutant lacking a CxxC motif in both a and a′ (Fig. 6C), suggesting that the disulfide isomerase activity of PDI is essential for the recovery of RNase A activity.
Finally, we examined the effects of adding reduced forms of other PDIs to the reaction solution. RNase A activity was also significantly restored by reduced forms of P5, ERp46, ERp57, and ERp72, but to a much smaller extent than seen with reduced PDI (Fig. 6D). This provides additional evidence for the prominent ability of PDI to isomerize incorrectly formed disulfide bonds. We here note that the cooperation of PDI with P5 was significantly more effective in promoting proper oxidative folding of RNase A than that with ERp46.
Discussion
The present study demonstrated that, while Prx4 is potentially capable of oxidizing a broad range of PDIs in vitro, it has a particular preference for ERp46 and P5 in cells. A likely explanation for this observation is that endogenous PDIs compete with each other for binding to Prx4, resulting in the exclusive formation of Prx4-ERp46 and Prx4-P5 complexes in cells. Accordingly, the SPR analysis revealed the extremely high affinity for ERp46 and P5 of Prx4, and the presence of either of ERp46 or P5 significantly inhibited Prx4 catalysis of other PDIs (Fig. 2). An alternative explanation could be that the formation of stable complexes of PDIs with their partner proteins, for instance, between PDI and prolyl-4-hydroxylase36, or the differential localization of PDIs in the early secretory compartment37, may also contribute to preventing the physical contact of Prx4 with PDI, ERp57, and ERp72 in cells.
Here, we established the structural basis of the interaction between Prx4 and PDIs by showing that a locally unstructured C-terminal segment of oxidized Prx4 binds to the peptide-binding groove of Trx. This mode of interaction is significantly different from that between Ero1α and PDI; Ero1α selectively recognizes PDI via close contact between the protruding β-hairpin of Ero1α and the hydrophobic pocket of PDI b′38, which leads to specific oxidation of a′ in the full-length or the b′a′c fragment of PDI (supplementary Fig. S2C). Thus, Ero1α requires tandemly positioned b′ and a′ for efficient PDI oxidation. In this context, Ero1α can oxidize any isolated Trx domain of PDI and ERp46 with very low efficiency, unlike Prx4 (supplementary Fig. S2C and S2D).
Considering that H2O2 could be generated by Ero1α and metabolized by Prx4, their different specificities for PDIs may have physiological implications. Deregulation of Ero1α oxidation of PDIs would lead to the hyperoxidizing conditions in the ER, which could eventually prevent isomerization or reduction of non-native disulfide bonds and result in ER stress. Also, excessive H2O2 generation mediated by Ero1α would cause mild but significant ER stress, including the induction of immunoglobulin-binding protein (BiP) and homocysteine-induced ER protein (HERP)39. To avoid such imbalanced ER conditions, Ero1α may have acquired the ability to exclusively recognize PDI, in addition to feedback regulation. Unlike Ero1α, Prx4 does not generate but rather scavenges H2O2 using the reducing power conferred by PDIs. If Prx4 had activity only toward PDI and competed with Ero1α for the oxidation of PDI, it would limit Prx4 activity and result in prolonged H2O2 accumulation. The strong preference of Prx4 for ERp46 and P5, which are abundant in the ER but react much less with Ero1α than PDI, seems advantageous to the effective Prx4-mediated reduction of H2O2 in the ER.
The observation that Prx4 is mostly found in the reduced form in cells in the absence of exogenous H2O2 (Fig. 1B) suggests that Prx4 catalysis does not occur primarily at steady state. Nevertheless, the sensitivity of Prx4 to oxidation by H2O2 (Fig. 1B) may have physiological significance. H2O2 can introduce disulfide bonds into PDIs and some folding substrates40 but it has much lower efficiency than Prx4-PDIs combinations in the presence of H2O2 (Fig. 4A and supplementary Fig. S7). Due to the high H2O2 sensitivity of Prx4, the enzyme can efficiently oxidize PDIs, even when the H2O2 concentration is limiting. Conversely, high H2O2 concentrations can compromise peroxidase activity due to irreversible formation of the overoxidized –SO2H form of the peroxidatic cysteine22. The steady-state concentration of H2O2 in the ER increases significantly during the LPS-induced differentiation of B cells to plasma cells, possibly as a result of the activation of Ero1α catalysis41. Thus, it is surmised that Prx4 is well adapted as a primary oxidation enzyme of PDIs in situations where synthesis and oxidative folding of secretory proteins proceed vigorously.
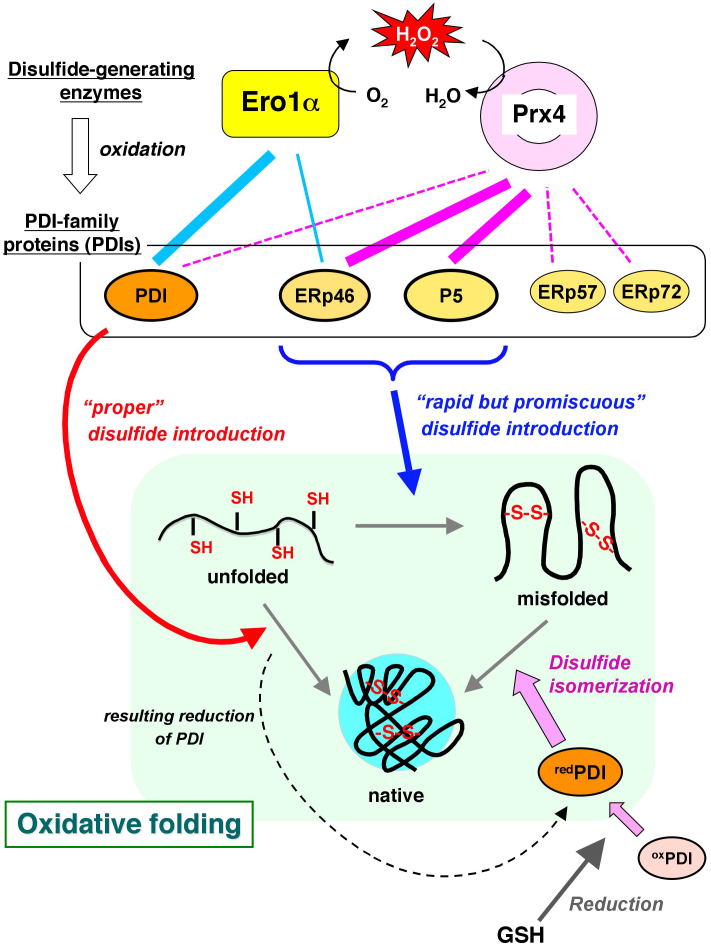
The effective transfer of oxidizing equivalents from Prx4 to ERp46 or P5 led to the rapid introduction of disulfide bonds (Fig. 4A). While speed is often associated with error, that is, the formation of incorrect disulfide bonds, rapid and non-selective introduction of disulfide bonds could be a means of greatly accelerating proper oxidative protein folding if efficient isomerization to native disulfide bonds follows immediately. On the basis of the present findings, non-native disulfide bonds rapidly introduced by ERp46 and P5 can be efficiently proofread by the action of PDI, thereby leading to significantly accelerated production of native proteins. We thus propose a new likely model of the disulfide bond formation network highly systematized in the mammalian ER, where Ero1α and Prx4 have different specificity toward PDIs and consequently each PDIs plays a distinct and significant role during oxidative folding, as illustrated in Fig. 7. Notably, some specialized cells, such as plasma cells and pancreatic β cells, synthesize and secrete large quantities of proteins at an extremely rapid rate. In order to achieve this prodigious task, the cells may have evolved a strategy of a promiscuous but rapid introduction and subsequent proofreading of disulfide bonds.
Figure 7. A new likely model of the disulfide bond formation network that promotes oxidative protein folding in the mammalian ER.
Ero1α and Prx4 function as primary disulfide-generating enzymes with significant oxidative activity toward PDIs in the mammalian ER. On the basis of the present findings, while Ero1α oxidizes PDI and, to a lesser extent, ERp46 selectively, Prx4 has a strong preference for ERp46 and P5 in cells. PDI can efficiently introduce native disulfide bonds and proofread non-native bonds, resulting in native folding (red arrow). By contrast, ERp46 and P5 engage in rapid, but promiscuous disulfide introduction in a Prx4-dependent manner (blue arrow). Rapid, but promiscuous disulfide formation tends to yield misfolded proteins, which can be restored to native conformation through disulfide isomerization by reduced PDI generated as results of disulfide introduction into substrates (broken curve) and reduction by GSH (dark gray arrow). Thus, the cooperation of PDI with P5 and, to a lesser extent, with ERp46 can be one of the possible ways that accelerate proper oxidative protein folding in the ER. The thickness of the lines between disulfide-generating enzymes and PDIs indicates the strength of reactivity of Ero1α (cyan) and Prx4 (magenta) toward PDIs. The broken line denotes interactions between Prx4 and PDIs that were undetected in the cells.
In this context, it is interesting that combined knockdown of PDI and other PDIs significantly hampered the folding of some substrates and induced their degradation in cells42, corroborating our notion that PDIs act cooperatively in oxidative folding. However, the extent of the folding defects observed after the knockdown of PDIs largely varied, relying on combinations of depleted PDIs and substrates of which folding was monitored. It is thus conceivable that endogenous PDIs have different substrate preferences as well as distinct roles in oxidative folding, albeit with some functional overlap. Altogether, we propose that mammalian cells have diversified disulfide bond formation and isomerization pathways to enhance both the rate and fidelity of disulfide bond introduction and thereby accelerate oxidative folding of a wide variety of substrate proteins. Further elucidation of the physiological roles of disulfide bond formation and isomerization networks by global proteomic analyses currently in progress should help advance our current understanding of the mechanisms underlying protein and redox homeostasis in the mammalian ER.
Methods
Plasmid construction and protein preparation
The procedures of plasmid construction and protein expression and purification are described in the Supplementary Information.
Cell culture and transfection
HEK293 cells were cultured in Dulbecco's modified Eagle medium (4.5 g/l glucose) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2/95% air. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Antibodies, reagents, and immunological techniques
Immunoblotting was carried out using a standard procedure and Chemi-Lumi One Super luminol (Nacalai Tesque). Anti-P5 and anti-PDI antibodies were a kind gift from M. Kikuchi. Other antibodies used were anti-ERp46 (Gene Tex products in Fig. 1C and D and laboratory made products in Figs. 1B and 2B), anti-ERp57 (Enzo Life Sciences), anti-ERp72 (Enzo Life Sciences), anti-PDI (Enzo Life Sciences), anti-FLAG M2 (Sigma), anti-FLAG M2 HRP (Sigma), and anti-Prx4 (Abcam).
Analysis of redox states of PDIs in cells
A plasmid for the expression of Prx4 was constructed by inserting cDNA encoding residues 1–274 of mPrx4 into the HindIII-EcoRV site of pCMV-3xFLAG (Sigma). Transfected and control HEK293 cells were treated with 5 mM dithiothreitol (DTT) for 5 min, 3 mM diamide for 5 min, or various concentrations of H2O2 for 10 min. Cells were denatured and precipitated in ice-cold PBS containing 10% trichloroacetic acid (TCA), incubated on ice for 1 h, and centrifuged at 10,000 rpm for 15 min at 4°C. The supernatant was discarded and the pellet was washed twice with ice-cold acetone, dried, and resuspended in reaction buffer (80 mM Tris-HCl, pH 6.8, and 2% SDS) containing 5 mM methoxypolyethylene glycol (average molecular weight, 2000)-maleimide (MalPEG-2K) (NOF Corporation). Samples were left in the dark for 60 min at room temperature. The alkylation reaction was stopped by boiling for 5 min after the addition of an equal volume of 2× Laemmli loading buffer. The gel images were recorded with the LAS-3000 image reader (Fuji Film), and the quantification of each band intensity was done using a software of Multi-Gauge ver. 3.1 (Fuji Film).
Immunoprecipitation
HEK293 cells were lysed in 1% NP-40 lysis buffer (50 mM Tris/HCl, pH 8.0, containing 150 mM NaCl, 1% NP-40, protease inhibitor cocktail [Nacalai Tesque], and 10 μM MG132). Immunoprecipitation was performed with anti-Prx4 antibodies (Abcam) and Protein G-coupled Sepharose beads (GE Healthcare). Beads were washed in 1% NP-40 lysis buffer twice and boiled in Laemmli loading buffer. Immunoprecipitates were subjected to reducing and non-reducing SDS-PAGE and analyzed by immunoblotting.
Surface plasmon resonance
Association (kon) and dissociation (koff) rate constants for the direct binding of PDIs to immobilized mPrx4 were determined by SPR on a Biacore 2000 system (GE Healthcare). Wild-type mPrx4 was coupled to the CM5 sensor chip (GE Healthcare) using amine-coupling chemistry. As a control, one channel was coupled to bovine serum albumin to exclude background binding. Sensorgrams were recorded for three concentrations of each analyte sample at 25°C for a 2 min association phase followed by a 4 min dissociation phase. The running buffer consisted of 20 mM HEPES-NaOH (pH 7.4), 150 mM NaCl, 0.001% Tween-20, 2 mM EDTA, 1 mM GSH, and 0.25 mM GSSG. All analyte samples were diluted in this buffer prior to analysis. Sensorgrams were analyzed by nonlinear regression according to a two-state model using the BIAevaluation 4.1 software. Experiments were replicated three times.
NADPH consumption assay
NADPH consumption coupled to Prx4 or Ero1α catalysis of PDI oxidation was monitored with a HITACHI U-3310 spectrophotometer at a controlled temperature of 30°C27. The oxidation of NADPH by glutathione reductase (supplementary Fig. S1B) accompanies the decrease in absorbance at 340 nm (Abs340). A molar extinction coefficient of 6,200 M−1cm−1 for NADPH was used in the calculations. The Prx4-catalyzed reaction was initiated by addition of H2O2 to a final concentration of 100 μM to a reaction mixture containing 50 mM Tris (pH 7.5), 300 mM NaCl, 200 μM NADPH, 0.05 U glutathione reductase, 300 μM GSH, 10 μM PDIs, and 0.5 μM mPrx4. The Ero1α-catalyzed reaction was initiated by addition of a constitutively active Ero1α mutant (Cys104&131Ala) to a final concentration of 2 μM to a reaction mixture containing 50 mM Tris (pH 7.5), 300 mM NaCl, 200 μM NADPH, 0.1 U glutathione reductase, 500 μM GSH, 10 μM PDIs, and a saturating level of molecular oxygen.
Peptide synthesis
Peptides composed of residues His244-Lys253 (HGEVCPAGWK) and residues His244–Pro263 (HGEVCPAGWKPGSETIIPDP) of Prx4 were synthesized using the Fmoc solid-phase method. The synthetic peptides were purified by reverse-phase HPLC on a C18 column (Cosmosil 5C18-AR-2, 8 × 250 mm; Nacalai Tesque), eluted using a linear gradient of CH3CN in 0.05% TFA from 5% to 60% at a rate of 1%/min and a flow rate of 2.0 ml/min, and monitored at 220 nm. The purity was confirmed by analytical reverse-phase HPLC, and the HPLC fraction was lyophilized and analyzed by MALDI-TOF/MS.
Preparation and crystallization of the P5 a0–Prx4 C-terminal peptide complex
P5 a0 CXXA variant and the Prx4 C-terminal peptide of residues His244–Pro263 were mixed at a 1:5 molar ratio and incubated on ice for 30 min in the presence of 7 mM diamide. The mixed disulfide complex was separated by gel filtration on a Superdex 75 10/300 column pre-equilibrated with 20 mM Tris–HCl (pH 8.1) containing 100 mM NaCl, and concentrated to approximately 7 mg/ml.
The purified complex was vapor-diffused at 20°C under conditions of 20% polyethylene glycol monomethyl ether 2000 (PEG-MME2000), 0.1 M Tris-HCl pH 7.5, and 0.1 M potassium thiocyanate; crystals appeared within 3 days. For cryoprotection, crystals were transferred directly to 12% glycerol containing 20% PEG2000MME, 0.1 M Tris-HCl (pH 7.5), and 0.1 M potassium thiocyanate, and flash-frozen as described above.
Crystallographic analysis
Diffraction data were collected at the Osaka University beamline BL44XU at SPring-8 equipped with an MX225-HE detector (Rayonix), whose acquisition was financially supported by the Academia Sinica and National Synchrotron Radiation Research Center (Taiwan) and at the beamline BL5A at Photon Factory equipped with an ADSC Quantum 315r detector. Data were integrated using HKL2000 software43. The crystallographic parameters are summarized in Table 1. Phase determination was made by molecular replacement using the deposited structures of human Prx4 (PDB code 2PN8) for mPrx4 C54A and P5 a0 (PDB code 3VWW) for the P5 a0–Prx4 C-terminal complex as search models with the program Phaser44. The initial structural models were refined by several cycles of manual rebuilding and refinement with Phenix.refine.
Oxidative folding assay
RNase A from bovine pancreas was reduced and denatured overnight with 7 M guanidinium hydrochloride and 200 mM DTT. The sample was passed through a PD-10 column to remove the denaturing and reducing reagents. Denatured RNase A was incubated at a final concentration of 25 μM together with 0.2 μM Prx4, 2 μM of each PDIs, and 100 μM H2O2 in 50 mM Tris (pH 7.5) and 300 mM NaCl for 0–30 min at 20°C. Aliquots were taken at different time points and 1 mM AMS was added to quench the reaction. Samples were separated by electrophoresis in a 15% non-reducing SDS polyacrylamide gel. Recovery of RNase A activity was measured, after addition of 4 mM cytidine 2′,3′-cyclic monophosphate monosodium salt (cCMP, Sigma) to the aliquots, by monitoring the linear increase in Abs295 on a HITACHI U-3310 spectrophotometer at a controlled temperature of 30°C45.
BPTI was purchased from TAKARA BIO INC. The reduction of BPTI was carried out as previously described46. Briefly, BPTI (10 mg) was dissolved in 1 ml of 0.1 M Tris (pH 8.0) containing 20 mM DTT and 8 M urea, and the solution was allowed to stand for 5 h at 50°C. The denatured/reduced protein was purified by HPLC using an InertSustain™ C18 column (4.6 × 250 mm; GL Sciences Inc.), lyophilized, and stored at −30°C until use. The denatured/reduced BPTI was incubated at a final concentration of 30 μM with 0.1 μM Prx4, 1 μM of each PDIs, and 200 μM H2O2 in 50 mM Tris-HCl (pH 7.5) containing 300 mM NaCl at 30°C. All solutions used in the refolding experiments were flushed with N2 gas, and the reactions were carried out in a sealed vial under an atmosphere of N2. The reaction mixture was quenched with an equivalent volume of 1 M HCl. The mixtures were loaded onto an InertSustain™ C18 HPLC column (4.6 × 150 mm; GL Sciences Inc.), eluted using a linear gradient of CH3CN in 0.05% TFA from 5% to 60% at a rate of 1%/min and a flow rate of 1.0 ml/min, and monitored at 229 nm. The experiments were performed in duplicate. The occupancy of each species at the indicated refolding time was quantified by the software Chrom Merge (TM) version 2.2a (GL Sciences).
Accession numbers
The coordinates and structural factors described in this study have been deposited in the Protein Data Bank with ID code 3VWU for mPrx4 C54A and 3W8J for the P5 a0–Prx4 C-term complex.
Author Contributions
K.I. designed and performed some parts of in vitro experiments and wrote the paper. Y.S. carried out most of experiments using cultured cells. R.K. and M.O. carried out structural and protein folding analyses. M.H. contributed in some in vitro experiments and critical reading of the manuscript. S.M., K.M. and T.H. contributed in plasmid construction and protein purification. M. Suzuki performed crystallographic analysis. M. Saiki carried out peptide synthesis.
Supplementary Material
supplementary information
Acknowledgments
We thank Akiko Sato for technical support. We also thank Roberto Sitia and Masakazu Kikuchi for kindly providing the expression vectors for mPrx4 and several PDIs, respectively. We are also grateful to Junichi Fujii for providing antibodies to mPrx4. This work was supported by Next Generation World-leading Researchers from MEXT to K. Inaba and by the MEXT program “X-ray Free Electron Laser Priority Strategy Program” and Grant-in-Aid for basic science from MEXT to M. Suzuki. The authors declare no conflicts of interest related to this work. This work was also supported by Grant-in-Aid for JSPS Fellows to M. Okumura, M. Hagiwara, and S. Masui.
References
- Arolas J. L., Aviles F. X., Chang J. Y. & Ventura S. Folding of small disulfide-rich proteins: clarifying the puzzle. Trends in Biochemical Sciences 31, 292–301 (2006). [DOI] [PubMed] [Google Scholar]
- Ellgaard L. & Ruddock L. W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Reports 6, 28–32 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatahet F. et al. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxidants & Redox Signaling 11, 2807–50 (2009). [DOI] [PubMed] [Google Scholar]
- Sato Y. & Inaba K. Disulfide bond formation network in the three biological kingdoms, bacteria, fungi and mammals. FEBS J 279, 2262–2271 (2012). [DOI] [PubMed] [Google Scholar]
- Maattanen P., Kozlov G., Gehring K. & Thomas D. Y. ERp57 and PDI: multifunctional protein disulfide isomerase with similar domain architectures but differing substrate-partner associations. Biochem. Cell Biol. 84, 881–889 (2006). [DOI] [PubMed] [Google Scholar]
- Sevier C. S. & Kaiser C. A. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol 3, 836–47 (2002). [DOI] [PubMed] [Google Scholar]
- Araki K. & Inaba K. Structure, mechanism and evolution of Ero1 family enzymes. Antioxid. Redox Signal 16, 790–799 (2012). [DOI] [PubMed] [Google Scholar]
- Sevier C. S. & Kaiser C. A. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochimica et Biophysica Acta 1783, 549–56 (2008). [DOI] [PubMed] [Google Scholar]
- Tavender T. J. & Bulleid N. J. Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum. Antioxidant & Redox Signaling 13, 1177–1187 (2010). [DOI] [PubMed] [Google Scholar]
- Fassio A. & Sitia R. Formation, isomerisation and reduction of disulphide bonds during protein quality control in the endoplasmic reticulum. Histochemistry & Cell Biology 117, 151–7 (2002). [DOI] [PubMed] [Google Scholar]
- Gross E. et al. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci U S A 103, 299–304 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevier C. S. et al. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 129, 333–44 (2007). [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., Riemer J., Christensen B., Sorensen E. S. & Ellgaard L. A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO Journal 27, 2977–87 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker K. M. et al. Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO Journal 27, 2988–97 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K. et al. Crystal structures of human Ero1alpha reveal the mechanisms of regulated and targeted oxidation of PDI. EMBO Journal 29, 3330–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulleid N. J. & Ellgaard L. Multiple ways to make disulfides. Trends. Biochem. Sci. 36, 485–492 (2011). [DOI] [PubMed] [Google Scholar]
- Zito E., Chin K.-T., Blais H. & Ron D. Ero1-beta, a pancreas-specific disulfide oxidase, pomotes insulin biogenesis and glucose homeostasis. Journal of Cell Biology 188, 821–832 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E. et al. Oxidative protein folding by an endoplasmic reticulum localized peroxiredoxin. Mol. Cell 40, 787–797 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavender T. J., Springate J. J. & Bulleid N. J. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO Journal 29, 4185–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., Tavender T. J., Roszak A. W., Cogdell R. J. & Bulleid N. J. Crystal structure of reduced and oxidized peroxiredoxin IV reveals a stable oxidized decamer and a non disulfide-bonded intermediate in the catalytic cycle. J. Biol. Chem. 286, 42257–42266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. F., Wang L., Wang X. F., Sun F. & Wang C. C. Structural insights into peroxidase activity and inactivation of human peroxiredoxin 4. Biochemical Journal 441, 113–118 (2012). [DOI] [PubMed] [Google Scholar]
- Tavender T. J. & Bulleid N. J. Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. Journal of Cell Science 123, 2672–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito E., Hansen H. G., Yeo G. S. H., Fujii J. & Ron D. Endoplasmic reticulum thiol oxidase deficiency leads to ascorbic acid depletion and noncanonical scruvy in mice. Molecular Cell 48, 39–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Xiang S., Noiva R., Lennarz W. J. & Schindelin H. The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites. [erratum appears in Cell. 2006 Mar 10;124(5):1085–8]. Cell 124, 61–73 (2006). [DOI] [PubMed] [Google Scholar]
- Dong G., Wearsch P. A., Peaper D. R., Cresswell P. & Reinisch K. M. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity 30, 21–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G. et al. Structure of the noncatalytic domains and global fold of the protein disulfide isomerase ERp72. Structure 17, 651–9 (2009). [DOI] [PubMed] [Google Scholar]
- Nguyen V. D. et al. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. Journal of Molecular Biology 406, 503–15 (2011). [DOI] [PubMed] [Google Scholar]
- Jessop C. E. & Bulleid N. J. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. Journal of Biological Chemistry 279, 55341–7 (2004). [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C. & Ellgaard L. In vivo reduction-oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxidants & Redox Signaling 10, 55–64 (2008). [DOI] [PubMed] [Google Scholar]
- Wood Z. A., Schroder E., Robin Harris J. & Poole L. B. Structure, mechanism and regulation of peroxiredoxins. Trends in Biochemical Sciences 28, 32–40 (2003). [DOI] [PubMed] [Google Scholar]
- Landerstein R. & Ren B. Protein disulfides and protein disulfide oxidoreductases in hyperthermophiles. FEBS J 273, 4170–4185 (2006). [DOI] [PubMed] [Google Scholar]
- Vivian J. P. et al. Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis. Journal of Molecular Biology 394, 931–43 (2009). [DOI] [PubMed] [Google Scholar]
- Maeda K., Hagglund P., Finnie C., Svensson B. & Henriksen A. Structural basis for target protein recognition by the protein disulfide reductase thioredoxin. Structure 14, 1701–10 (2006). [DOI] [PubMed] [Google Scholar]
- Weissman J. S. & Kim P. S. Reexamination of the folding of BPTI: predominance of native intermediates. Science 253, 1386–93 (1991). [DOI] [PubMed] [Google Scholar]
- Weissman J. S. & Kim P. S. A kinetic explanation for the rearrangement pathway of BPTI folding. Nature Structural Biology 2, 1123–30 (1995). [DOI] [PubMed] [Google Scholar]
- Pihlajaniemi T. et al. Molecular cloning of the beta-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene. EMBO Journal 6, 643–9 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T. & Sitia R. Protein quality control in the early secretory pathway. EMBO Journal 27, 315–27 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Vavassori S., Fagioli C., Sitia R. & Inaba K. Molecular bases of cyclic and specific disulfide interchange between human ERO1alpha protein and protein-disulfide isomerase (PDI). Journal of Biological Chemistry 286, 16261–71 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen H. G. et al. Hyperactivity of the Ero1 oxidase elicits endoplasmic reticulum stress but no broad antioxidant response. Journal of Biological Chemistry 287, 39513–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karala A. R., Lappi A. K., Saaranen M. J. & Ruddock L. W. Efficient peroxide-mediated oxidative refolding of a protein at physiological pH and implications for oxidative folding in the endoplasmic reticulum. Antioxidants & Redox Signaling 11, 963–70 (2009). [DOI] [PubMed] [Google Scholar]
- Bertolotti M. et al. B- to plasma-cell terminal differentiation entails oxidative stress and profound reshaping of the antioxidant responses. Antioxidants & Redox Signaling 13, 1133–44 (2010). [DOI] [PubMed] [Google Scholar]
- Rutkevich L. A., Cohen-Doyle M. F., Brockmeier U. & Williams D. B. Functional relationship between protein disulfide isomerase family members during the oxidative folding of human secretory proteins. Molecular Biology of the Cell 21, 3093–105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. & Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Crysta. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker K. W., Lyles M. M. & Gilbert H. F. Catalysis of oxidative protein folding by mutants of protein disulfide isomerase with a single active-site cysteine. Biochemistry 35, 1972–80 (1996). [DOI] [PubMed] [Google Scholar]
- Okumura M., Saiki M., Yamaguchi H. & Hidaka Y. Acceleration of disulfide-coupled protein folding using glutathione derivatives. FEBS Journal 278, 1137–44 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information