Abstract
The RAD52 epistasis group genes are involved in homologous DNA recombination, and their primary structures are conserved from yeast to humans. Although biochemical studies have suggested that the fundamental mechanism of homologous DNA recombination is conserved from yeast to mammals, recent studies of vertebrate cells deficient in genes of the RAD52 epistasis group reveal that the role of each protein is not necessarily the same as that of the corresponding yeast gene product. This review addresses the roles and mechanisms of homologous recombination-mediated repair with a special emphasis on differences between yeast and vertebrate cells.
Keywords: DT40, reverse genetic study, double-strand break repair, Rad 51 family
Chicken DT40 Cells as an Experimental System to Analyze Homologous Recombination (HR)
Chicken B lymphocyte precursors diversify the variable segments of their Ig genes not only by site-specific V(D)J DNA recombination, but also by a HR process, Ig gene conversion, which occurs in the bursa of Fabricius (recently reviewed in ref. 1). Mature chicken B lymphocytes are also capable of undergoing Ig gene conversion in splenic germinal centers on antigenic stimulation (2). A chicken B lymphocyte line, DT40, transformed with an avian leukosis virus, continuously undergoes Ig gene conversion during in vitro culture (3, 4). Remarkably, targeted integration occurs in these cells with efficiencies that are orders of magnitude higher than those observed in mammalian cells (5). This integration occurs at all loci analyzed including silent loci such as the ovalbumin and α-crystalin loci. Efficient gene targeting is demonstrated in all chicken B lymphocyte lines analyzed, including another avian leukosis virus-transformed cell line, RP9, and a v-rel-transformed cell line 27L2, suggesting that this extraordinary capability might be shared by even some ex vivo chicken B lymphocytes (5). The molecular mechanism responsible for the high targeting efficiencies in chicken B lymphocyte lines is not clear. Conceivably, a common molecule may be responsible not only for Ig gene conversion but also for enhancing gene targeting efficiencies, because both these processes are mediated by HR and are observed only in chicken B lymphocytes but not in chicken non-B cells or mammalian cell lines. Although we have found some molecules to be required for efficient gene targeting in DT40 cells (Table 1), none of them appear to account for the high levels of gene targeting because they are expressed both in DT40 cells and the chicken non-B cell lines.
Table 1.
Cellular phenotypes of null DT40 mutants of genes involved in DSB repair and checkpoints
| Gene, reference | Viability | Genetic instability | DNA damage sensitivity
|
Rad51 focus formation | HR activities | |
|---|---|---|---|---|---|---|
| IR | Crosslinking agents | |||||
| RAD51 (6, 7) | Cell lethal | Extensive* | Defective | |||
| RAD52 (8) | Viable | No | No | No | Yes | Slightly defective† |
| RAD54 (9, 10) | Viable | Yes | Yes | Yes | Enhanced | Defective |
| RAD54B‡ | Viable | No | No | No | Slightly defective† | |
| RAD54/RAD54B‡§ | Viable | Yes¶ | Yes | Yes | Defective | |
| RAD51 paralog‖ (11), (12) | Cell viable‖ | Yes‖ | Yes‖ | Yes‖ | Defective‖ | Defective‖ |
| MRE11 (13) | Cell lethal | Extensive* | Yes | Yes | Defective | |
| KU70 (10) | Viable | No | Yes | Slightly more tolerant** | Yes | Normal |
| ATM (14, 15) | Viable | Yes | Yes | Yes | Slightly delayed | Slightly defective† |
Chromosomal breaks was presumably underevaluated for the reason discussed in the text.
The frequency of gene targeting is reduced by less than 5-fold.
Y. Yamaguchi-Iwai and S.T., unpublished work.
Cells deficient in both RAD54 and RAD54B.
Higher sensitivity than Rad54-deficient cells.
All mutant clones of each of the five Rad51 paralog genes (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) exhibit very similar phenotypes.
M. Takata and S.T., unpublished work.
Besides efficient gene targeting, DT40 cells possess a number of advantages as a tool for reverse genetic studies. First, the relatively invariant character of DT40 cells, during extended periods of cell culture, allows for the performance of sequential gene targeting of up to three genes in a single cell using seven different selection marker genes. Because some DNA repair pathways are complementary to each other, cells deficient in two repair pathways often exhibit an extremely severe phenotype when compared with cells deficient in either pathway alone (10). Using this reasoning we have been able to investigate distinct as well as overlapping roles of independent repair pathways by knocking out multiple genes involved in DNA repair in DT40 cells. Second, the extremely rapid growth rate of DT40 cells, with a doubling time of 8–10 h, makes it easy to perform phenotypic analysis. Third, the absence of functional p53 in DT40 cells offers an additional advantage in the analysis of mutant cells exhibiting genome instability (15). DNA lesions in such mutant cells would elevate the level of p53 product, leading to the decrease in the cloning efficiency of cells, significantly reducing the growth rate, and inducing apoptosis.
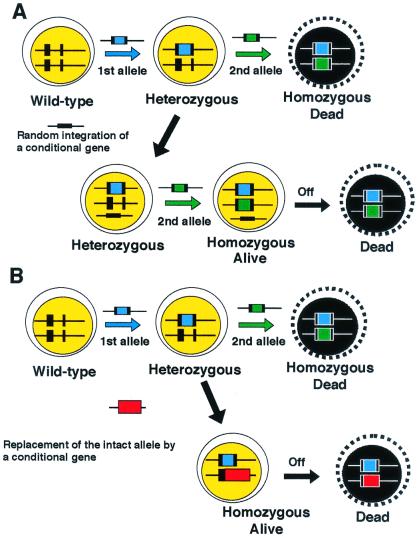
Although HR-deficient yeast mutants are viable, some HR-deficient vertebrate cells show impaired proliferation or even lethality (6, 13) (Table 1). Therefore, we have investigated the effect of HR deficiency in vertebrate cells by generating conditionally mutant cells. Three methods have been used successfully for conditionally suppressing the expression of genes in DT40 cells. As shown in Fig. 1 for RAD51, the structural and functional homologue of Escherichia coli RecA, we generated conditionally null DT40 clones that express a human transgene under the control of a tetracyclin-repressible promoter (6). Although the promoter activity was suppressed by more than 100-fold upon the addition of tetracyclin, possible effects of leaky expression cannot be excluded (16). One way to overcome this disadvantage is to use a chimeric Cre recombinase. The Cre recombinase recognizes loxP sequences and deletes or inverts sequences between two loxP sites depending on the relative orientation of these two loxP sequences. The chimeric Cre recombinase carries a mutated hormone-binding domain of the murine estrogen receptor (17), which binds the antagonist 4-hydroxytamoxifen (OH-TAM). Upon the addition of OH-TAM to the culture media, the chimeric Cre recombinase is translocated into the nucleus where it recognizes loxP sites and recombines the DNA to delete the gene of interest. Cre-mediated recombination works efficiently in DT40 cells; virtually all of the genes flanked by two loxP sequences were deleted within 24 h after the addition of OH-TAM (unpublished work). Although this system allows us to completely inhibit the expression of a gene of interest, Cre-mediated recombination does not occur in a synchronous manner in a population of cells, as observed in the tet repressible promoter system.
Figure 1.
Strategy for generating conditionally gene targeted clones. Ts mutant cDNAs of a gene of interest can be designed based on information from yeast ts mutants, if available. Following the standard protocol to generate ts mutant cells, each mutated cDNA is introduced by targeted integration into the intact endogenous locus of the gene in heterozygous mutant (+/−) DT40 cells. The resulting cells should be homozygous mutant (−/−) cells and express only the mutated protein.
A third method of generating conditionally mutant clones uses temperature sensitive (ts) mutant genes. The physiological body temperature of the chicken ranges from 40.9°C to 41.9°C; the cell culture temperature can vary from 34°C to as high as 43°C without loss of viability. In a procedure designed to generate ts mutants of an essential gene, such as that encoding the kinetochore protein CENP-C (T. Fukagawa, personal communication), the wild-type allele of a heterozygous mutant clone is replaced by mutated cDNA through gene targeting (Fig. 1B), with the result that the cells express only a mutated cDNA instead of the wild-type gene. Each clone transfected with a mutated cDNA is split into two populations, one of which is cultured at 34°C and the other at 43°C. Thus, clones exhibiting a ts phenotype can be identified easily if they survive at 34°C, but die at 43°C. Once appropriate ts mutant clones are identified, it is possible to study the role of the ts genes at each phase of the cell cycle by synchronizing the mutant cells and shifting them to a restrictive temperature only transiently. In comparison to the tet promoter system, which gradually decreases the level of the encoded protein depending on the half-life of its transcripts and the protein, a ts mutated protein may be inactivated rapidly just by shifting the temperature of cell culture. The ts system therefore offers an enormous advantage for the phenotypic investigation of a gene whose mutation is lethal.
Heteroallelic HR Plays an Important Role in DNA Double-Strand Break (DSB) Repair in Yeast but Not in Vertebrate Cells
A chromosomal break is lethal if left unrepaired (18). Two major repair pathways that exist in eukaryotes to tackle DNA DSBs are nonhomologous end-joining (NHEJ) and HR (reviewed in refs. 19–27). NHEJ repairs adjacent broken DNA ends with little or no requirement for extensive sequence homology, whereas the more accurate HR requires an intact template of a homologous sequence either in a homologous chromosome or in a sister chromatid. The primary structures of genes involved in the two pathways are conserved between yeast and vertebrates, including the products of the RAD52 epistasis group genes for HR and the Ku proteins for NHEJ. However, the relative roles of two DSB repair pathways appear to be quite different between yeast and vertebrate cells.
In yeast, HR is preferentially used for DSB repair throughout the cell cycle, except for the G1 phase of haploid cells. Cells deficient in HR, such as rad52 mutants, are extremely hypersensitive to ionizing radiation (IR) (28). On the other hand, NHEJ-deficient cells, such as hdf1 (a homologue of the vertebrate Ku70), show virtually no IR sensitivity even in the G1 phase of haploid cells, whereas cells deficient in both Rad52 and Hdf1 are only slightly more IR sensitive than the rad52 mutants (28). Thus, NHEJ works as a minor backup option for HR in DSB repair. In contrast, murine embryonic stem (ES) clones deficient in either the NHEJ pathway (Ku70−/−) (29, 30) or the HR pathway (Rad54−/−) (31) show elevated radio sensitivity. Furthermore, HR-deficient Rad54−/− adult mice do not exhibit IR sensitivity (32), whereas NHEJ-deficient scid mice, which carry a mutation in the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) gene, are hypersensitive to IR (33). These data indicate that in marked contrast to yeast HR plays a less important role in DSB repair than NHEJ, especially in adult mice. DT40 cells deficient in both DSB repair pathways show a dramatic increase in radio sensitivity when compared with either type of the single mutant clones (10). Likewise, adult mice deficient in both Rad54 and DNA-PKcs are significantly more IR-sensitive than those deficient in Rad54 alone (32). These observations strongly suggest that these pathways act in a complementary fashion.
Diploid yeast cells are more tolerant to IR in the G2 phase than in the G1 phase, indicating that for DSB repair the sister chromatid is the preferred template as compared with the homologous chromosome (34). Likewise, mammalian cells are more tolerant to IR in the late S to G2 phases than in the G1 to early S phases. This increased IR tolerance depends to a large extent on Rad54 (10) and XRCC2 (35, 36), a Rad51-like protein, indicating that HR repairs DSBs using the other intact sister chromatid as a template. Although both yeast and vertebrate mitotic cells repair a broken chromatid through HR using the other intact sister chromatid in the S phase, the usage of interhomologue recombination in DSB repair appears to be quite different between yeast and vertebrates (reviewed in ref. 37). In the G1 phase, wild-type diploid yeast strains are more tolerant to IR than haploid strains, suggesting that chromosomal homologues also can serve as templates for DSB repair (28, 34, 38). In contrast, in the mammalian cells, such heteroallelic recombination, which might result in loss of heterozygosity, occurs only occasionally even after the induction of a DSB (39–43). In agreement with this observation, Rad54 deficiency in DT40 cells elevates IR sensitivity in the S phase but not in the G1 phase (10). These observations suggest that the HR-mediated repair system of vertebrates tends not to perform interhomologue recombination. A simple model to explain this is that HR-mediated repair in the G1 phase requires more intensive homology search between homologous chromosomes in vertebrate cells than in yeast. In contrast, the close proximity of a pair of sister chromatids may allow efficient HR-mediated repair during late S-G2 phases. Probably for this reason, NHEJ plays a major DSB repair role in the G0/1 phases in vertebrate cells (10).
Furthermore, the HR pathway seems to be suppressed in the G0/1 phase in vertebrate cells. It has been observed that neither Rad51 nor Rad54 protein is detectable in the G0 phase even after genotoxic treatments (44), whereas the transcription of the RAD51 and RAD54 genes can be induced by IR in yeast (45, 46). The formation of subnuclear foci of Rad51, which are believed to contain an active and extensively polymerized form of the protein (47–50) (reviewed in refs. 21 and 25), is induced by IR in the S to G2 phases but not in the G1 phase in a rodent cell line (51) (U. Ear, D. Hari, and D. K. Bishop, personal communication). Given that Rad51 plays a central role in an early step of HR-mediated repair, these observations suggest that vertebrate cells are incapable of initiating HR during the G0 and G1 phases. This explains why Rad54-deficient adult mice, where even cells in tissues with rapid turnover are in the G1 phase, show no detectable elevation of IR sensitivity, although deletion of the RAD54 gene in more rapidly cycling murine ES cells causes an increase in IR sensitivity (32). Lack of HR-mediated repair during the G0 and G1 phases, in turn, might avoid the possible interference of NHEJ and HR with one another in competition to reach a DSB end, as has been suggested for the KU and RAD52 genes (52).
Homologous DNA Recombination Is Essential for the Viability of Vertebrate Cells
A wide range of potential insults to genomic DNA is afforded not only by environmental factors, but also by cellular activities per se. Various types of lesions are generated continuously, and estimates of the number of each lesion produced daily per human genome range from a few to several thousand, depending on the type of lesion and the detection technology (53, 54). Spontaneous damage arises in many forms and appears to be efficiently repaired by a variety of repair processes. If the damage is not repaired before the cell progresses to the next stage of the cell cycle, the nature of the damage may alter, resulting in the formation of secondary lesions. For example, if a G1 cell carrying single-strand breaks in its genomic DNA progresses through S phase, the single-strand lesions will be converted to secondary lesions, i.e., DSBs in sister chromatids (reviewed in refs. 55 and 56). Similarly, some types of covalently modified base residues are known to arrest DNA replication, causing a daughter-strand gap that encompasses the damage in the template strand (57). In addition, bacterial studies have indicated that stalled replication forks are actively converted to DSBs as a part of the replication fork restart process (58). Although not yet demonstrated, it seems possible that a similar mechanism of replication restart might occur in higher eukaryotes (reviewed in refs. 55 and 56). Thus, DNA replication at primary lesions in the template strand can result in more severe secondary DNA lesions such as gaps and chromatid breaks.
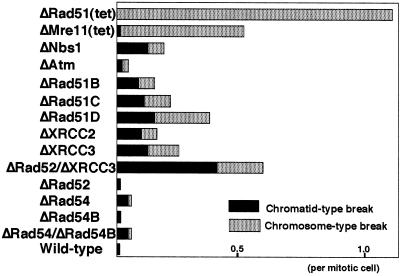
Although HR-deficient yeast cells can proliferate, murine cells deficient in key HR proteins such as Rad51 or Mre11 are nonviable (6, 13, 59, 60). To investigate the essential roles of Rad51 and Mre11, we generated conditionally Rad51- and Mre11-deficient DT40 cells. The depletion of Rad51 or Mre11 causes both the appearance of randomly distributed chromosomal breaks (up to a few breaks per mitotic cell, Fig. 2) and subsequent cell death (6, 13). Furthermore, chromosomal breaks also occur in cells deficient in other genes of the RAD52 epistasis group, including RAD51B/C/D, XRCC2, XRCC3, and RAD54 in DT40 cells (Fig. 2) (10–12) and XRCC2 and XRCC3 in Chinese hamster cell lines (36, 61) (reviewed in ref. 19). These observations suggest that DSBs may occur frequently during the cell cycle and that a defect in HR-mediated DSB repair accounts for the appearance of such chromosomal DSBs. We attribute the lethality of HR defects in vertebrate cells to the important role of HR in maintaining the integrity of chromosomal DNA: the several hundred-fold difference in genome size between vertebrates and lower eukaryotes may account for the contrasting lethality of such deficiency in yeast and vertebrate cells.
Figure 2.
The level of spontaneous chromosomal aberrations in various DT40 mutant clones. Data are presented as macrochromosomal (1–5 and Z) aberrations per mitotic cell.
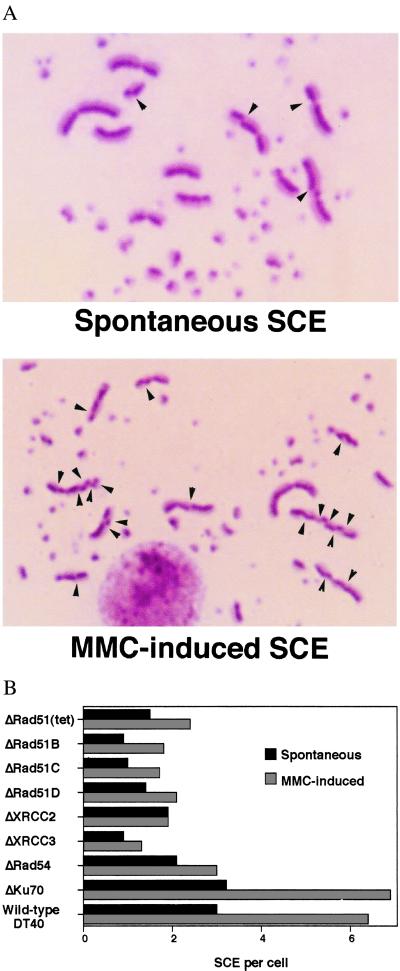
It is likely that HR-mediated DNA repair occurs during DNA replication. This idea is supported by the presence of a number of Rad51 foci at S phase (47, 49). Furthermore, as discussed above, some types of DNA lesions on a template strand are converted to chromatid breaks and daughter-strand gaps by DNA replication. These secondary lesions could stimulate HR with the other intact chromatids and subsequently be repaired by gene conversion. Such gene conversion events might be associated occasionally with sister chromatid crossovers (42, 62). To examine the presence of HR between sister chromatids, we studied the involvement of HR in microscopically visible sister chromatid exchanges (SCEs) by measuring the level of SCE in HR-deficient cells (Fig. 3A). SCE is known to be an S phase-associated repair process and can be induced by treating cells with various mutagens including crosslinking agents and UV before DNA replication (63). Furthermore, both mammalian and chicken cycling cells exhibit up to five spontaneous SCEs per mitosis (7, 64, 65). We showed that HR is indeed responsible for mediating both spontaneous SCE and SCE induced by a crosslinking agent (Fig. 3B), suggesting the presence of HR-mediated repair during the cell cycle (7). Gene conversion events associated with crossover occur frequently during meiosis but only occasionally during mitosis in yeast (66). Given that crossing-over appears to be a relatively infrequent event during mitosis (42, 62, 67) these visible crossing-over events (SCEs) suggest that the level of HR-mediated replication-associated repair may be quite high in vertebrate cells.
Figure 3.
(A) SCE in wild-type DT40 cells. (A) Spontaneous SCE and (B) mitomycin C (MMC)-induced SCE are shown. Arrowheads indicate the sites of SCE. [Reproduced with permission from ref. 7 (Copyright 1999, American Society for Microbiology)]. (B) Reduced levels of SCE in cells deficient in homologous recombination. Cells were labeled with BrdUrd during two cell cycle periods with or without MMC treatment (50 ng/ml) for the last 8 h. Spontaneous and MMC-induced SCEs were scored in the macrochromosomes of metaphase cells. Histograms show the mean value of SCE per cell. The SCE levels of all mutants except ΔKu70 differ significantly (P < 0.002) from wild-type control SCE levels; statistical significance was calculated by the Mann–Whitney nonparametric u test.
The ability to manipulate HR would remove a major bottleneck in various approaches to gene therapy, as well as facilitating further biological research. The recent findings linking HR to DNA replication may have important implications for those interested in genome manipulation by HR. Gene targeting efficiency depends highly on the length of homology of the targeting sequence. Indeed, when this length of homology is increased from 6 kb to 12 kb the efficiency of targeted integration increases by as much as 10-fold in murine ES cells (68). This observation is in marked contrast with gene targeting in Saccharomyces cerevisiae, where fewer than 100 bases of homology are enough for gene targeting (69). Thus, the mechanism of gene targeting in yeast is not necessarily shared by vertebrate cells. If HR is initiated by DSBs or other types of DNA damage in one of the recombining DNA molecules, a fundamental question arises as to whether the ends of the linearized gene targeting construct or DSBs in the genomic DNA initiates HR for gene targeting. In yeast, homologous sequences at the end of the linearized targeting construct appear to invade intact duplex DNA in the genome to initiate HR (69). Thus, “DSBs” in a gene targeting construct might be repaired by interacting with intact homologous sequences in the genome, resulting in targeted integration. An alternative is that a gene targeting construct participates in HR-mediated repair as an intact template DNA. This model is supported experimentally as follows: the remarkable increase in gene targeting frequencies with increasing length of gene targeting constructs (68) can be explained by the increased availability of the longer construct as an intact template DNA for HR-mediated repair. Second, the induction of DSBs in the genome stimulates gene targeting by more than 2 orders of magnitude in rodent cells (70). Third, efficient gene targeting occurs with comparable efficiencies at every locus analyzed including transcriptionally inactive loci in DT40 cells (5). This observation is in agreement with the notion that gene targeting is associated with HR-mediated repair after DNA replication but does not occur during G1, where accessibility to each locus may vary considerably due to difference in high-order chromatin structure.
DSBs arising during DNA replication appear to be repaired in a manner different from DSBs generated by exogenous causes such as IR. HR plays a dominant role over end-joining in repairing spontaneously arising DSBs. Indeed, Rad51-deficient cells are not able to complete even a single cell cycle (6). In contrast, cell lines deficient in end-joining continue to divide and show little chromosomal instability (71–73), whereas only primary cells derived from mice deficient in end-joining exhibit vastly elevated level of chromosomal aberrations (74, 75) (reviewed in ref. 19). Presumably, the HR-mediated repair pathway is intimately associated with DNA replication and ready to act quickly on DNA lesions that arise during DNA replication. On the other hand, the end-joining pathway works as a minor backup option for HR in repairing DNA lesions that arise during DNA replication, although this pathway plays a dominant role in repairing IR-induced DSBs.
The Differing Roles of Rad52 in Vertebrate and Yeast Cells
Rad52 mutants show the most pronounced phenotype among the RAD52 epistasis group mutants in S. cerevisiae (reviewed in ref. 22). Rad52 is essential for an early stage of HR before the appearance of the extensively polymerized form of Rad51 (76–79). Rad52, but not Rad51, also is involved in single-strand annealing (80) and other RAD51-independent forms of recombination (81), which may explain the more pronounced phenotype of the rad52 mutants than that of the rad51 mutants (Table 1). However, Rad52 appears to play a less important role in vertebrate HR compared with Rad51. Although Rad51 deficiency causes lethality in dividing cells, Rad52-deficient mice and cells are viable with no elevated IR sensitivity (8, 82) (Table 1). Additionally, the amino acid sequence identity between the human and chicken Rad52 proteins is only 69%, whereas human and chicken Rad51 share 95% identity (83, 84).
There are a number of possible explanations for the phenotypic differences between S. cerevisiae and vertebrate RAD52. mutants. First, vertebrate Rad52 may not be involved principally in HR but in other DNA metabolic pathways. The roles of homologs are not necessarily the same in different species, as exemplified by the involvement of Mre11 in NHEJ in S. cerevisiae (38, 85) but little involvement of its homolog in Schizosaccharomyces pombe (86). However, Rad52 deficiency causes a slight, marginally significant decrease in gene targeting efficiencies both in murine ES and DT40 cells (8, 82). Immunocytochemical experiments suggest a coordinated response of mammalian Rad52 and Rad51 to DNA damage (50), so that Rad52 indeed plays a role in HR in mammalian cells. Moreover, both the yeast and human Rad52 proteins form ring structures (52, 79, 87) and stimulate DNA strand exchange promoted by Rad51 protein in vitro, further emphasizing the conservation of the roles of human and yeast Rad52 in HR. Second, there may be as yet undescribed orthologs in vertebrates, which might compensate for the absence of Rad52. The number of Rad52 homolog genes varies in various organisms, and two Rad52-like genes are present in both S. cerevisiae (88) and S. pombe (89, 90). Third, the precise mechanism of HR may differ between vertebrate and yeast species, giving rise to the possibility that other analogous molecules may compensate for the lack of Rad52 in vertebrate cells. The two yeast Rad51-like proteins, Rad55 and Rad57, form a heterodimeric complex (91, 92), which is functionally similar to the Rad52 protein in facilitating strand exchange by Rad51 in the presence of replication protein A in vitro (93). This finding suggests that any protein that can substitute for this Rad51-dependent function of Rad52 may be a functional homologue, but not necessarily a structural homologue.
Activities of Rad51-Like Molecules in Yeast and Vertebrates
S. cerevisiae possesses two Rad51-like molecules, Rad55 and Rad57, which form a heterodimer (reviewed in ref. 24). In contrast, mammalian as well as chicken cells possess five Rad51 paralogs (XRCC2, XRCC3, Rad51B, Rad51C, Rad51D) that share ≈20–30% amino acid sequence identity with Rad51 and also with each other (reviewed in ref. 25). Unlike Rad51, none of the vertebrate Rad51 paralogs appears to interact with itself in yeast two-hybrid assays (25, 94), which is also the case for yeast Rad55 and Rad57 (91, 92). Overexpression of yeast Rad51 partially suppresses the DNA repair defect of rad55 and rad57 mutant yeast strains, implying that Rad55 and Rad57 may functionally cooperate with Rad51. This idea is supported by a stable protein–protein interaction between Rad55 and Rad57, and by a transient interaction between Rad51 and Rad55 (91–93). Similarly, yeast two-hybrid studies have suggested that physical interactions may occur in vivo between human Rad51 and XRCC3, XRCC3 and Rad51C, Rad51B and Rad51C, Rad51C and Rad51D, and Rad51D and XRCC2 (25, 94, 95). These observations support the argument that the five Rad51 paralogs may form a functional complex and cooperate with Rad51, in a manner analogous to the yeast Rad55 and Rad57 proteins (reviewed in ref. 19).
All of the Rad51 paralog DT40 mutants show impaired HR, as determined by targeted integration and SCE assays (discussed in the next paragraph). Remarkably, all of the mutant cell lines exhibit very similar phenotypes: spontaneous chromosomal aberrations (Fig. 2), high sensitivity to killing by cross-linking agents (mitomycin C and cisplatin), mild sensitivity to γ-rays, and significantly attenuated Rad51 focus formation during recombinational repair after exposure to γ-rays. Moreover, overexpression of human Rad51 partially corrects the DNA damage sensitivity phenotype in all of the mutants (11, 12). We conclude that the Rad51 paralogs participate in a DNA repair process as a functional unit that facilitates the action of Rad51 in HR. Experiments to confirm this conclusion include ascertaining the epistatic relationship of these genes by generating and characterizing DT40 clones deficient in various combinations of two of the five Rad51 paralogs.
It should be noted that a rodent cell line defective in XRCC2, one of the Rad51-like genes, exhibits not only defective HR but also an increase in IR sensitivity even in the G1 phase in comparison to the wild-type cells (35). Furthermore, other Rad51-like genes, including Rad51C, Rad51D, XRCC2, and XRCC3, all are expressed in some nondividing cells in the brain (36, 96–98), where no Rad51 expression is detectable (99). These observations are inconsistent with the notion that the Rad51 paralogs participate in HR as cofactors of Rad51. Presumably the Rad51-like molecules, but not Rad51, might play a role in intragenic HR in resting cells in mammals.
DT40 Cells Deficient in Both Rad52 and XRCC3, a Rad51 Paralog, Are Not Viable
Biochemical studies have indicated that both the yeast Rad52 protein and the Rad55/57 heterodimer stimulate DNA strand exchange by Rad51 (93). To test the idea that Rad52 and the Rad51 paralogs can partially substitute for each other's function in vertebrate cells, possibly explaining the subtle phenotype of Rad52-deficient cells, we generated a DT40 clone deficient in both Rad52 and XRCC3, one of the five Rad51 paralog genes. Somewhat surprisingly, but in support of our assumption, this double mutant clone was not viable, although cells deficient in either Rad52 (84) or XRCC3 (12) are able to proliferate (unpublished work). This result supports the notion that Rad52 and XRCC3 are indeed complementary to each other for the maintenance of chromosomal integrity in cycling cells. Given the embryonic lethality of mice deficient in XRCC2, Rad51B or Rad51D, the Rad51 paralogs might have taken over an important role of Rad52 in HR during evolution.
The phenotype of Rad51-deficient DT40 cells is much severer even than that of cells deficient in both Rad52 and XRCC3 (Fig. 2, Table 1). It should be noted that the number of chromosomal breaks in Rad51-deficient cells (Fig. 2) was likely underevaluated due to the following reason (6). We were able to measure chromosomal breaks in mitotic cells that had presumably undergone DNA replication when a small amount of Rad51 was still present after the inhibition of the tet-repressible promoter. Upon depletion of Rad51, cells were no longer capable of undergoing even a single cell cycle and exhibited extensive chromosomal aberrations. These observations are in marked contrast with the finding that the number of living cells completely deficient in both Rad52 and XRCC3 gradually decreased over more than 10 cell cycles (unpublished work). These observations support the notion that Rad51 plays a vitally important role in HR in vertebrate cells, whereas both Rad52 and the Rad51 paralogs may work just as cofactors for Rad51. Which important role of Rad52 in HR might have been taken over by Rad51 during evolution? Single-strand annealing (SSA) requires only Rad52 of the RAD52 epistasis group (80), so that Rad52 alone may be able to facilitate homologous paring of even a short stretch of homologous sequences both in vitro and in vivo (100, 101). Active SSA along with other modes of efficient homologous pairing mediated by Rad52 might be useful for unicellular organisms because it would allow rapid repair of some DNA lesions. In contrast, in multicellular organisms, SSA is presumably suppressed because of the following reasons. Higher fidelity of DNA repair would be relatively more important in multicellular organisms than in unicellular organisms, because accumulation of mutations would result in tumorigenesis and malformation in multicellular organisms. In addition, SSA as well as intragenic HR may be mutagenic especially when there are large numbers of various types of repeated sequences in the genome of vertebrates. These reasons could explain why Rad52 plays such an important role in HR in yeast species but not in vertebrate cells.
Other Rad51-Interacting Proteins
The products of the essential mammalian BRCA1 and BRCA2 cancer susceptibility genes are associated with Rad51 in both mitotic and meiotic cells (102–105). However, no structural homologues have been reported in yeast, Caenorhabditis elegans, or Drosophila (106). Brca1 may be involved in two fundamental processes, transcriptional regulation (reviewed in ref. 107) and DNA repair (108–110), implying that Brca1 might stimulate HR by altering the expression of other genes that are directly involved in HR. In contrast, a direct role of Brca2 in HR is suggested by the physical interactions between Rad51 and Brca2 (103–105), impaired Rad51 focus formation caused by a truncating mutation of Brca2 (111), and its lack of involvement in transcriptional regulation. It is noteworthy that human and murine mutant cells in which Brca2 was truncated exhibit phenotypes remarkably similar to those we described for the Rad51-paralog mutants: elevated spontaneous chromosomal aberrations (112), sensitivity to cross-linking agents such as mitomycin C and cisplatin (111), and defective Rad51 focus formation (111, 113). Thus, Brca2 may participate in the formation of a complex involving the Rad51 paralogs to act as a cofactor for Rad51 during HR. The presence of Brca2 homologs in vertebrates but not in yeast (106), and the existence of five vertebrate Rad51 paralogs, compared with only two in yeast (Rad55/57) implies that the activity of Rad51 during HR is regulated in a more complex manner in vertebrate cells than in yeast. It is possible that these Rad51 cofactors might form an interface between Rad51 and cyclin-dependent kinases or between Rad51 and DNA damage checkpoints. Through these interactions, the assembly of Rad51 might be suppressed in the G1 phase and activated by kinases involved in damage checkpoint, as has been suggested for yeast (114).
Conclusions
There are a number of lines of evidence showing that DT40 is not only a valid system for investigating DNA recombination in vertebrate cells, but also is an important system with high relevance to human cells. First, so far no viable mutant cell lines have been reported in any system for several repair proteins, such as Rad51B, Rad51C, Rad51D, Mre11, and Rad51 proteins, so that the conditional approaches used in DT40 have yielded valuable information unobtainable by other means. Second, both murine ES cells and DT40 exhibited the same phenotypes for the already-published mutants of HR genes, including the lethality of Rad51-deficient cells (6, 59), a nearly normal phenotype of Rad52-deficient cells (8, 82), and the elevated radio sensitivity of Rad54-deficient cells (9, 31). These similarities confirm that the DT40 cell line is a reasonable model for the analysis of vertebrate DNA recombination, despite the obvious concerns associated with the use of a transformed cell line, which may have certain cell line-specific characteristics. Gene-targeted DT40 clones have been extensively generated to investigate DNA replication, DNA repair, cell cycle-dependent kinases, cell cycle checkpoints, chromosome-associated proteins, and nuclear and cytoplasmic cell divisions. Because of this large collection of mutant clones, the DT40 system is an extremely useful model to study DNA metabolism, despite the disadvantage represented by an inadequate database of chicken-specific base sequences and the relative dearth of experimental materials.
Acknowledgments
We thank Drs. D. Schild (Lawrence Berkeley National Laboratory, Berkeley, CA), D. Bishop (University of Chicago), D. Pawan (Kyoto University) and M. Lavin (The Queensland Institute of Medical Research, Brisbane, Australia) for critically reading the manuscript. Financial support was provided in part by CREST-Japan Science and Technology (Saitama, Japan), Bayer Yakuhin (Kyoto, Japan), and Grants-in Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan (M.T. and S.T.) and by a grant from the Uehara Memorial Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Naito Foundation. Y.M.Y. received a Japan Society for the Promotion of Science Postdoctoral Fellowship. C.M. is the recipient of an European Molecular Biology Organization Long Term Fellowship and received a Japan Society for the Promotion of Science Postdoctoral Fellowship while in Kyoto.
Abbreviations
- HR
homologous recombination
- ts
temperature sensitive
- DSB
double-strand break
- NHEJ
nonhomologous end-joining
- ES
embryonic stem
- SCE
sister chromatid exchange
- IR
ionizing radiation
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Reynaud C A, Bertocci B, Dahan A, Weill J C. Adv Immunol. 1994;57:353–378. doi: 10.1016/s0065-2776(08)60676-8. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa H, Kuma K, Yasuda M, Furusawa S, Ekino S, Yamagishi H. J Immunol. 1998;160:4232–4241. [PubMed] [Google Scholar]
- 3.Baba T W, Giroir B P, Humphries E H. Virology. 1985;144:139–151. doi: 10.1016/0042-6822(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 4.Buerstedde J M, Reynaud C A, Humphries E H, Olson W, Ewert D L, Weill J C. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buerstedde J M, Takeda S. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 6.Sonoda E, Sasaki M S, Buerstedde J-M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonoda E, Sasaki M S, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Mol Cell Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi-Iwai Y, Sonoda E, Buerstedde J-M, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bezzubova O Y, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J M. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 10.Takata M, Sasaki M S, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata M, Sasaki M S, Sonoda E, Fukushima T, Morrison C, Albala J S, Swagemakers S M, Kanaar R, Thompson L H, Takeda S. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takata M, Sasaki M S, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson L H, Takeda S. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi-Iwai Y, Sonoda E, Sasaki M S, Morrison C, Haraguchi T, Hiraoka Y, Yamashita Y M, Yagi T, Takata M, Price C, et al. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. EMBO J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takao N, Kato H, Mori R, Morrison C, Sonada E, Sun X, Shimizu H, Yoshioka K, Takeda S, Yamamoto K. Oncogene. 1999;18:7002–7009. doi: 10.1038/sj.onc.1203172. [DOI] [PubMed] [Google Scholar]
- 16.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wienands J, Zurn C, Reth M. EMBO J. 1998;17:7304–7310. doi: 10.1093/emboj/17.24.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang L C, Clarkin K C, Wahl G M. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson L H, West M G. Mutat Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 20.Jasin M. Cancer Invest. 2000;18:78–86. doi: 10.3109/07357900009023065. [DOI] [PubMed] [Google Scholar]
- 21.Morrison C, Takeda S. Int J Biochem Cell Biol. 2000;32:817–831. doi: 10.1016/s1357-2725(00)00033-9. [DOI] [PubMed] [Google Scholar]
- 22.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrini J H. Am J Hum Genet. 1999;64:1264–1269. doi: 10.1086/302391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thacker J. Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 25.Thompson L H, Schild D. Biochimie. 1999;81:87–105. doi: 10.1016/s0300-9084(99)80042-x. [DOI] [PubMed] [Google Scholar]
- 26.Jeggo P A. Radiat Res. 1998;150:80–91. [PubMed] [Google Scholar]
- 27.Kanaar R, Hoeijmakers J H, van Gent D C. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 28.Bressan D A, Baxter B K, Petrini J H. Mol Cell Biol. 1999;19:7681–7687. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Jin S, Gao Y, Weaver D T, Alt F W. Proc Natl Acad Sci USA. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang H, Nussenzweig A, Kurimasa A, da Costa Soares V, Li X, Cordon-Cardo C, Li W-H, Cheong N, Nussenzweig M, Iliakis G, et al. J Exp Med. 1997;186:921–929. doi: 10.1084/jem.186.6.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 32.Essers J, van Steeg H, de Wit J, Swagemakers S M, Vermeij M, Hoeijmakers J H, Kanaar R. EMBO J. 2000;19:1703–1710. doi: 10.1093/emboj/19.7.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver D T, Alt F W. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 34.Kadyk L C, Hartwell L H. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheong N, Wang X, Wang Y, Iliakis G. Mutat Res. 1994;314:77–85. doi: 10.1016/0921-8777(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 36.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W, et al. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 37.Haber J E. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 38.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamin M B, Potter H, Yandell D W, Little J B. Proc Natl Acad Sci USA. 1991;88:6652–6656. doi: 10.1073/pnas.88.15.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godwin A R, Bollag R J, Christie D M, Liskay R M. Proc Natl Acad Sci USA. 1994;91:12554–12558. doi: 10.1073/pnas.91.26.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moynahan M E, Jasin M. Proc Natl Acad Sci USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richardson C, Moynahan M E, Jasin M. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richardson C, Jasin M, Dronkert M L, Beverloo H B, Johnson R D, Hoeijmakers J H, Kanaar R, Moynahan M E, Chiu J W, Koller B H. Nature (London) 2000;405:697–700. [Google Scholar]
- 44.Tan T L R, Essers J, Citterio E, Swagemakers S M A, De Wit J, Benson F E, Hoeijmakers J H J, Kanaar R. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 45.Calderon I L, Contopoulou C R, Mortimer R K. Curr Genet. 1983;7:93–100. doi: 10.1007/BF00365632. [DOI] [PubMed] [Google Scholar]
- 46.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 47.Haaf T, Raderschall E, Reddy G, Ward D C, Radding C M, Golub E I. J Cell Biol. 1999;144:11–20. doi: 10.1083/jcb.144.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raderschall E, Golub E I, Haaf T. Proc Natl Acad Sci USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tashiro S, Walter J, Shinohara A, Kamada N, Cremer T. J Cell Biol. 2000;150:283–291. doi: 10.1083/jcb.150.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Maizels N. EMBO Rep. 2000;1:85–90. doi: 10.1093/embo-reports/kvd002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bishop D K, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum R R, Shinohara A. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 52.Van Dyck E, Stasiak A Z, Stasiak A, West S C. Nature (London) 1999;398:728–731. doi: 10.1038/19560. [DOI] [PubMed] [Google Scholar]
- 53.Kunkel T A. Trends Genet. 1999;15:93–94. doi: 10.1016/s0168-9525(98)01664-3. [DOI] [PubMed] [Google Scholar]
- 54.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 55.Flores Rozas H, Kolodner R D. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haber J E. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 57.Wyatt M D, Allan J M, Lau A Y, Ellenberger T E, Samson L D. BioEssays. 1999;21:668–676. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 58.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao Y, Weaver D T. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffin C S, Simpson P J, Wilson C R, Thacker J. Nat Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 62.Bollag R J, Waldman A S, Liskay R M. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- 63.Painter R B. Mutat Res. 1980;70:337–341. doi: 10.1016/0027-5107(80)90023-8. [DOI] [PubMed] [Google Scholar]
- 64.Zwanenburg T S, Natarajan A T. Cytogenet Cell Genet. 1984;38:278–281. doi: 10.1159/000132075. [DOI] [PubMed] [Google Scholar]
- 65.Pinkel D, Thompson L H, Gray J W, Vanderlaan M. Cancer Res. 1985;45:5795–5798. [PubMed] [Google Scholar]
- 66.Paques F, Leung W Y, Haber J E. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dronkert M L, Beverloo H B, Johnson R D, Hoeijmakers J H, Jasin M, Kanaar R. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deng C, Capecchi M R. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leung W, Malkova A, Haber J E. Proc Natl Acad Sci USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang F, Romanienko P J, Weaver D T, Jeggo P A, Jasin M. Proc Natl Acad Sci USA. 1996;93:8929–8933. doi: 10.1073/pnas.93.17.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Darroudi F, Natarajan A T. Mutat Res. 1987;177:149–160. doi: 10.1016/0027-5107(87)90030-3. [DOI] [PubMed] [Google Scholar]
- 72.Kemp L M, Jeggo P A. Mutat Res. 1986;166:255–263. doi: 10.1016/0167-8817(86)90025-8. [DOI] [PubMed] [Google Scholar]
- 73.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 74.Difilippantonio M J, Zhu J, Chen H T, Meffre E, Nussenzweig M C, Max E E, Ried T, Nussenzweig A. Nature (London) 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao Y, Ferguson D O, Xie W, Manis J P, Sekiguchi J, Frank K M, Chaudhuri J, Horner J, DePinho R A, Alt F W. Nature (London) 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 76.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 77.Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 78.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Nature (London) 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 79.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 80.Ivanov E L, Sugawara N, Fishman-Lobell J, Haber J E. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bartsch S, Kang L E, Symington L S. Mol Cell Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rijkers T, van den Ouweland J, Morolli B, Rolink A G, Baarends W M, Van Sloun P P H, Lohman P H M, Pastink A. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bezzubova O, Shinohara A, Mueller R G, Ogawa H, Buerstedde J M. Nucleic Acids Res. 1993;21:1577–1580. doi: 10.1093/nar/21.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bezzubova O Y, Schmidt H, Ostermann K, Heyer W D, Buerstedde J M. Nucleic Acids Res. 1993;21:5945–5949. doi: 10.1093/nar/21.25.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 86.Wilson S, Warr N, Taylor D L, Watts F Z. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stasiak A Z, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West S C, Egelman E H. Curr Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- 88.Bai Y, Symington S. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 89.Ostermann K, Lorentz A, Schmidt H. Nucleic Acids Res. 1993;21:5940–5944. doi: 10.1093/nar/21.25.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suto K, Nagata A, Murakami H, Okayama H. Mol Biol Cell. 1999;10:3331–3343. doi: 10.1091/mbc.10.10.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hays S L, Firmenich A A, Berg P. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson R D, Symington L S. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 94.Schild D, Lio Y-C, Collins D W, Tsomondo T, Chen D J. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 95.Braybrooke J P, Spink K G, Thacker J, Hickson I D. J Biol Chem. 2000;257:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- 96.Dosanjh M K, Collins D W, Fan W, Lennon G G, Albala J S, Shen Z, Schild D. Nucleic Acids Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pittman D L, Cobb J, Schimenti K J, Wilson L A, Cooper D M, Brignull E, Handel M A, Schimenti J C. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 98.Rice M C, Smith S T, Bullrich F, Havre P, Kmiec E B. Proc Natl Acad Sci USA. 1997;94:7417–7422. doi: 10.1073/pnas.94.14.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 100.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugawara N, Ira G, Haber J E. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 103.Chen P L, Chen C F, Chen Y, Xiao J, Sharp Z D, Lee W H. Proc Natl Acad Sci USA. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Katagiri T, Saito H, Shinohara A, Ogawa H, Kamada N, Nakamura Y, Miki Y. Genes Chromosomes Cancer. 1998;21:217–222. [PubMed] [Google Scholar]
- 105.Sharan S K, Bradley A. Genomics. 1997;40:234–241. doi: 10.1006/geno.1996.4573. [DOI] [PubMed] [Google Scholar]
- 106.Rubin G M, Yandell M D, Wortman J R, Gabor Miklos G L, Nelson C R, Hariharan I K, Fortini M E, Li P W, Apweiler R, Fleischmann W, et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Monteiro A N. Trends Biochem Sci. 2000;25:469–474. doi: 10.1016/s0968-0004(00)01632-7. [DOI] [PubMed] [Google Scholar]
- 108.Bhattacharyya A, Ear U S, Koller B H, Weichselbaum R R, Bishop D K. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 109.Moynahan M E, Chiu J W, Koller B H, Jasin M. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 110.Wang Y, Cortez D, Yazdi P, Neff N, Elledge S J, Qin J. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 111.Yu V P, Koehler M, Steinlein C, Schmid M, Hanakahi L A, van Gool A J, West S C, Venkitaraman A R. Genes Dev. 2000;14:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 112.Patel K J, Vu V P, Lee H, Corcoran A, Thistlethwaite F C, Evans M J, Colledge W H, Friedman L S, Ponder B A, Venkitaraman A R. Mol Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 113.Yuan S S, Lee S Y, Chen G, Song M, Tomlinson G E, Lee E Y. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 114.Bashkirov V I, King J S, Bashkirova E V, Schmuckli Maurer J, Heyer W D. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]