Abstract
Eukaryotic initiation factor 2Bε (eIF2Bε) plays a critical role in the initiation of mRNA translation and its expression and guanine nucleotide exchange activity are major determinants of the rate of protein synthesis. In this work we provide evidence that the catalytic epsilon subunit of eIF2B is subject to ubiquitination and proteasome-mediated degradation. Lysates of C2C12 myoblasts treated with proteasome inhibitor were subjected to sequential immunoprecipitations for eIF2Bε followed by ubiquitin. Tandem mass spectrometry (LC-MS/MS) analysis of immunoprecipitated proteins resulted in the identification of five peptides containing ubiquitin (diglycine) modifications on eIF2Bε. The specific lysine residues containing the ubiquitin modifications were localized as Lys-56, Lys-98, Lys-136, Lys-212 and Lys-500 (corresponding to the rat protein sequence). In addition three novel phosphorylation sites were identified including Ser-22, Ser-125, and Thr-317. Moreover, peptides corresponding to the amino acid sequence of the E3 ligase NEDD4 were also detected in the LC-MS/MS analysis, and an interaction between endogenous eIF2Bε with NEDD4 was confirmed by co-immunoprecipitation.
Keywords: mRNA translation, protein synthesis, proteasomal degradation, posttranslational modification
1. Introduction
We recently reported that the catalytic epsilon subunit of the heteropentameric guanine nucleotide exchange factor eIF2B is subject to proteasome-sensitive degradation [1]. To examine the potential ubiquitin modification of eIF2Bε, we analyzed the primary protein sequence with bioinformatics algorithms. Using the UbiPred bioinformatics algorithm [2], 12 of the 27 lysines contained in the rat eIF2Bε protein were identified over the default threshold score of 0.50, with several being close to the highest prediction score of 1.0. In particular, one region contains a cluster of four lysines (Lys-470, Lys-472, Lys-474, Lys-476) with very strong prediction scores and the predictions were conserved in the mouse, rat and human proteins. Using the UbPred prediction algorithm [3], four lysines were the same as those predicted by UbiPred, with three additional potential sites at Lys-493, Lys-500, and Lys-688. Intriguingly, the region immediately N-terminal to the cluster of predicted ubiquitin sites contains a high scoring PEST motif, another potential signal for protein degradation [4]. Based on our previous report and the in silico predictions that several lysines contained within the eIF2Bε protein are putative sites of ubiquitin modification we performed the experiments described herein to verify and characterize such posttranslational modifications. The results demonstrate that eIF2Bε is indeed subject to ubiquitin modification and identify five lysine residues within the rat protein that are modified by ubiquitin as determined by tandem mass spectrometry. The results also identify three novel phosphorylation sites and implicate NEDD4 as the E3 ligase involved in the proteosome-mediated degradation of eIF2Bε.
2. Materials and methods
2.1. Cell culture and reagents
C2C12 myoblasts (ATCC) were maintained in growth medium consisting of Dulbecco’s modified Eagle’s medium (DMEM; Gibco/Invitrogen) containing 25 mM glucose supplemented with 10% fetal bovine serum (FBS; Atlas Biologicals), and 1% penicillin–streptomycin (Invitrogen), at 37°C and 5% CO2. MG-132 (CalBiochem/EMD Biosciences) was prepared as a 10 mM stock solution in DMSO and used at the concentration indicated in the figure legends.
2.2. Plasmids and transfections
The plasmid pFLAG-eIF2Bε in the pcDNA3.1 expression vector (Invitrogen) was generated from the previously cloned rat eIF2Bε cDNA [5] with an N-terminal FLAG-epitope [6]. The plasmid pRK5-HA-Ubiquitin-WT, encoding wild type human ubiquitin C with an N-terminal HA epitope tag [7] was obtained from AddGene, where it was originally deposited by T.M. Dawson (The Johns Hopkins University School of Medicine). Transient transfection of C2C12 myoblasts was accomplished with the Effectene Transfection Reagent (Qiagen) utilizing a modified protocol [8]. Briefly, myoblasts were trypsinized on the day of transfection and treated as a suspension of cells with 1:8 DNA:Enhancer and 1:15 DNA:Effectene ratios (mass:volume), respectively.
2.3. SDS-PAGE and Western blot analysis
Cell lysates or immunoprecipitated proteins were resolved by SDS-PAGE and subjected to Western blot analysis as described previously [9]. Primary antibodies used included: anti-eIF2Bε (generated in house); anti-α-tubulin (Santa Cruz Biotechnology, #sc-32293); anti-DDK (Origene, #4C5); and anti-HA (Santa Cruz rabbit Biotechnology, #sc-805). After overnight incubation with primary antibody, membranes were probed with secondary antibodies (Bethyl Laboratories) in TBST with 5% non-fat dry milk for 1 h at room temperature. Blots were developed with ECL (Pierce/Thermo Scientific) or ECL Plus (GE Healthcare) detection reagents. Images were acquired with a GeneGnome HR imaging system and GeneSnap software (SynGene). In some cases, PVDF membranes were stripped and then re-probed with a different primary antibody.
2.4. Preparation of cell extracts
Unless otherwise noted, cells were lysed with RIPA buffer (Sigma-Aldrich) supplemented with (final concentrations): 1 mM dithiothreitol, 1 mM benzamidine, 0.5 mM sodium vanadate, and 10 μl/ml Sigma Protease Inhibitor Cocktail (Sigma-Aldrich). N-ethylmaleimide (NEM; Sigma-Aldrich) was added to a final concentration of 10 mM immediately before use. Cells were harvested using trypsin, collected by centrifugation at 233×g for 5 min, washed with PBS, recentrifuged, and finally resuspended in lysis buffer. The cell suspension was rocked for 30 min at 4°C followed by centrifugation at 8,200×g for 10 min at 4°C. The cleared lysate was either combined with 2X SDS-PAGE sample buffer or subjected to immunoprecipitation as described below.
2.5. Sequential FLAG(eIF2Bε) and HA(ubiquitin) immunoprecipitation
Immunoprecipitation of FLAG-eIF2Bε covalently modified with HA-Ubiquitin was performed using the FLAG HA Tandem Affinity Purification kit (Sigma-Aldrich). Twenty 10-cm culture dishes were each seeded with 1.5×106 C2C12 myoblasts and simultaneously transfected with pFLAG-eIF2Bε (6.0 μg) and pRK5-HA-Ubiquitin-WT (2.0 μg) and 24 h later the cells were incubated in serum-free DMEM for 16 h. Cells were then treated with MG-132 (10 μM) for 8 hours in serum-free DMEM. Cells were harvested in RIPA buffer supplemented with 10 mM NEM, 10 μM MG-132 and the inhibitors described above. The lysate was sequentially immunoprecipitated using the EZView anti-FLAG M2 affinity resin and the anti-HA agarose affinity resin using the manufacture’s protocol. Proteins were eluted from the HA resin with buffer consisting of 125 mM Tris•HCl, pH 6.8, 4% SDS, 20% (v/v) glycerol, and 0.004% bromophenol blue. Finally, β-mercaptoethanol (5% v/v) was added and the sample boiled at 100°C for 5 min. A small portion of the eluate was used for immunoblot analysis, while the remainder was subjected to electrophoresis on a Criterion SDS-PAGE gel. Proteins were visualized with Bio-Safe Coomassie gel stain (Bio-Rad) and excised bands were sent for analysis at the Taplin Biological Mass Spectrometry Facility at Harvard University (Harvard Medical School, Boston, MA).
2.6. eIF2Bε – Nedd4 co-immunoprecipitation
Cell extracts were prepared as described above, and eIF2Bε was immunoprecipitated from an aliquot of the extract using an anti-eIF2Bε monoclonal antibody [10]. NEDD4 was immunoprecipitated from a separate aliquot of the same cell extract using an anti-Nedd4 antibody (Cell Signaling, #5344). Antibody-antigen complexes were collected using magnetic beads (Qiagen), and the beads were washed thrice with ice-cold buffer (20 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 0.1% β-mercaptoethanol, pH 7.4), resuspended in 1× SDS sample buffer, and then boiled for 5 min. Supernatants were subjected to Western blot analysis using either anti–eIF2Bε or anti–Nedd4 antibody (Cell Signaling, #2740).
3. Results
3.1. Polyubiquitylation of eIF2Bε
C2C12 myoblasts were transfected with a plasmid encoding a FLAG-epitope tagged eIF2Bε (pFLAG-eIF2Bε), serum starved for 16 h and treated with or without the proteasome inhibitor MG-132 for 8 h. In the absence of proteasome inhibitor the expression of FLAG-eIF2Bε (observed as the upper of two bands in Fig. 1A) was nearly identical to that of endogenous eIF2Bε expression. In contrast, with the addition of MG-132 there was a dramatic accumulation of eIF2Bε as well as several additional immunoreactive bands of higher molecular weight (Fig. 1A). The ubiquitin protein is ~8.5 kDa in size [11] and its covalent linkage to substrate proteins results in increasing increments of that mass as the number of ubiquitins on the chain increases. A “smear” of higher molecular weight bands was recognized by the eIF2Bε antibody upon proteasome inhibition (Fig. 1A).
Fig. 1.
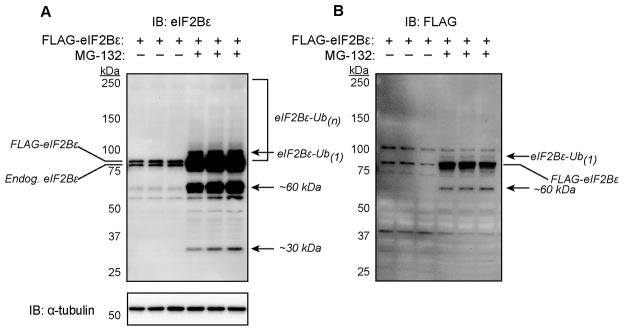
Accumulation of FLAG-eIF2Bε with the proteasome inhibitor MG-132 and appearance of high molecular weight bands. (A and B) C2C12 myoblasts were transfected and treated with MG-132 as described under “Materials and methods”. Equal volumes of the lysates were subjected to immunoblot (IB) analysis with antibodies against eIF2Bε, FLAG epitope, or α-tubulin. The doublet in panel A represents the endogenous (Endog) eIF2Bε and the expressed FLAG-eIF2Bε. Arrows indicate potentially ubiquitinated proteins as well as potential degradation products that accumulate with MG-132 treatment. The experiment was performed with three replicates for each time point and condition; representative blots are shown.
The significance of the faster migrating proteins (i.e., molecular size smaller than full length eIF2Bε; Fig. 1A) recognized by the antibody is unclear, but they are presumably degradation products of eIF2Bε that also accumulate upon proteasome inhibition. In addition to the expected FLAG-eIF2Bε band, at least two additional bands were detected by an anti-DDK(FLAG) antibody. First, a band presumed to be monoubiquitinated eIF2Bε (depicted by the top arrow in Fig. 1B) appeared. Second, the ~60 kDa band was detected, indicating that this smaller protein contained the N-terminus of the FLAG-eIF2Bε protein.
To confirm the ubiquitination of eIF2Bε, cells were transfected with plasmids for the expression of FLAG-eIF2Bε, HA-ubiquitin, or both and then treated with proteasome inhibitor. Immunoprecipitation of FLAG (eIF2Bε) resulted in co-purification of HA-ubiquitin, primarily at molecular weights above the expected size of the eIF2Bε protein (Supplementary Fig. 1). No HA signal was detectable in immunoprecipitate of cells not expressing FLAG-eIF2Bε.
3.2. Tandem affinity purification of eIF2Bε
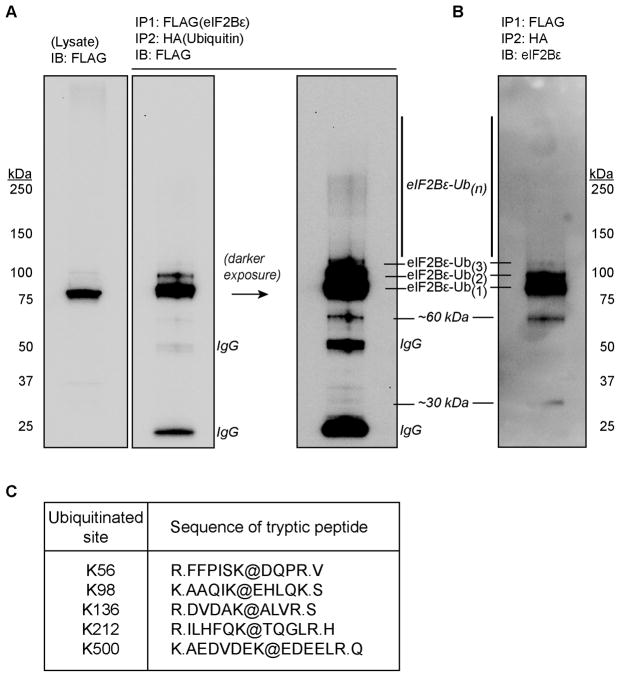
Tandem mass spectrometry is one approach to identify proteins containing ubiquitin modification as well as to localize the modification to specific residues [12]. However, identification of ubiquitin sites can be problematic if the protein is of low abundance and generally enrichment for ubiquitin-conjugated proteins is essential [13]. Thus, the approach for the present investigation was to use sequential immunoprecipitations first to affinity purify eIF2Bε (via FLAG immunoprecipitation), then subsequently to enrich for proteins that were covalently modified by co-expressed HA-Ubiquitin (via HA immunoprecipitation). C2C12 myoblasts were co-transfected with pFLAG-eIF2Bε and pRK5-HA-Ubiquitin and treated with the proteasome inhibitor MG-132 allowing for the accumulation of ubiquitin-conjugated proteins. The co-transfected cells from multiple dishes were trypsinized, pooled, and lysed in a small volume of lysis buffer enabling a large amount of eIF2Bε to be affinity purified. The lysates were subjected to sequential immunoprecipitation, first of FLAG(eIF2Bε) followed by HA-ubiquitin, resulting in successful enrichment of ubiquitin conjugated eIF2Bε as demonstrated by immunoblots utilizing only a small portion (~8%) of the total sample generated. Three distinct bands were recognized by both anti-FLAG (Fig. 2A) and anti-eIF2Bε antibodies (Fig. 2B) at molecular weights greater than the expected size of unmodified FLAG-eIF2Bε. The apparent smearing typical of polyubiquitinated proteins was also present in the double immunoprecipitation upon darker exposure (Fig. 2A, rightmost lane and Fig. 2B).
Fig. 2.
Immunoblots of the FLAG/HA immunoprecipitation used for tandem mass spectrometry analysis and ubiquitin site summary. C2C12 myoblasts were co-transfected with pFLAG-eIF2Bε (6.0 μg) and HA-Ubiquitin (2.0 μg) plasmids and immunoprecipitation was performed as described under “Materials and methods”. (A) SDS-PAGE and immunoblot (IB) with anti-FLAG (anti-DDK) antibody. The first lane represents whole cell lysate (~8% of the lysate represented in the immunoprecipitation lane), the second represents a portion of the final eluate from the anti- HA immunoprecipitation, and the third is a darker exposure of the same IB. (B) Immunoblot with anti-eIF2Bε antibody following sequential FLAG/HA IP. The major higher molecular weight band is presumed to be monoubiquitinated (Ub1) eIF2Bε. Increasingly slower migrating bands are presumed to be eIF2Bε-ubiquitin conjugates with increasing numbers of ubiquitin molecules attached (i.e., Ub2, Ub3…Ub(n)). (C) The remainder of the eluate from sequential FLAG/HA IP was subjected to SDS-PAGE, in-gel trypsin digest, and LC-MS/MS analysis. A summary of eIF2Bε tryptic peptides containing ubiquitinated lysine residues is presented. The lysine residues designated with @ symbol contained a 114.04 Dalton mass shift. Periods denote the sites of cleavage by trypsin.
3.3. Tandem mass spectrometry analysis of eIF2Bε
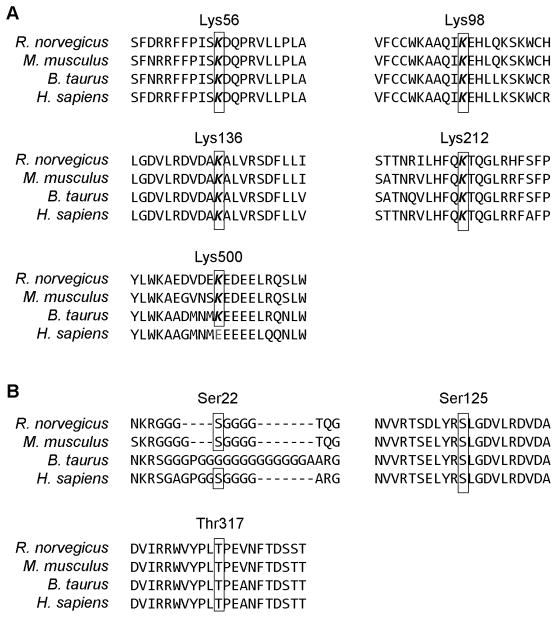
The remaining samples from the FLAG/HA immunoprecipitations were subjected to SDS-PAGE and two major bands (corresponding to ~75–100 kDa and ~100–250 kDa, respectively) were excised, subject to in-gel trypsin digestion, and analysed by LC-MS/MS. The analyses from two such experiments resulted in 67.2% overall coverage of the rat eIF2Bε sequence (481/716 amino acids; Supplementary Fig. 2). Five unique lysine residues containing the -GG modification indicative of ubiquitination were identified in the ~75–100 kDa excised gel slice: Lys-56, Lys-98, Lys-136, Lys-212, and Lys-500 (Figs. 2C). Two of these ubiquitinated peptides (containing Lys-136 and Lys-212) were also positively identified in the gel slice containing the larger molecular size range. Four of the five ubiquitinated lysine residues and the regions flanking them are well conserved (Fig. 3) whereas Lys-500 is not conserved in the human protein. Three of the five identified ubiquitin-modified residues (Lys-56, Lys-98, and Lys-500) were predicted by the in silico algorithms as putative sites of ubiquitin modification while the UbiPred score for Lys-212 (0.49) fell just under the default prediction threshold of >0.50. Although we were unable to confirm ubiquitin modifications at Lys-470–476 due to lack of peptide coverage, we can not rule out that they may be ubiquitinated and this region, due to the presence of a PEST motif, may be of particular interest with regard to eIF2Bε protein stability.
Fig. 3.
Sequence conservation of ubiquitin-modified lysine residues. The amino acid sequences of eIF2Bε from several species are shown. Ten residues are included on either side of the lysine identified as ubiquitinated in LC-MS/MS analysis of rat eIF2Bε. The residue number at the heading of each column corresponds to the rat sequence. Only Lys-500 is not conserved in human eIF2Bε.
The LC-MS/MS analyses also yielded tryptic peptides from the ubiquitin (UBC) protein in the samples with near complete coverage (94.7%). Ubiquitin itself was found to contain diglycine modifications on four of the seven internal lysines: Lys-11, Lys-29, Lys-48, and Lys-63. Although additional ubiquitin-like modifiers are also known to contain a C-terminal (R)GG sequence like ubiquitin (i.e., Rub1, NEDD8, or ISG15) that could potentially leave a diglycine modification on a target protein after trypsin digestion [14], it is unlikely that the eIF2Bε peptides identified in the present investigation were modified by such proteins as no peptides were identified from any of the other ubiquitin-like modifiers known to contain the (R)GG sequence following tryptic digest.
3.4. Interaction of eIF2Bε with NEDD4
In addition to eIF2Bε and ubiquitin, peptides corresponding to the amino acid sequence of two E3 ligases, DDB1 and NEDD4 were detected in the LC-MS/MS analysis. To establish whether or not these proteins interacted with endogenous eIF2Bε, a monoclonal anti-rat eIF2Bε antibody was used to immunoprecipitate eIF2Bε from C2C12 myoblasts, and the immunoprecipitate was subjected to Western blot analysis using anti-DDB1 and anti-NEDD4 antibodies. Although no DDB1 was detected (not shown), NEDD4 was found in the eIF2Bε immunoprecipitate (Fig. 4B). Importantly, no NEDD4 was detected in the absence of anti-eIF2Bε antibody, demonstrating that NEDD4 was binding specifically to eIF2Bε and not non-specifically to the beads used in the immunoprecipitation. As further confirmation of the interaction between NEDD4 and eIF2Bε, NEDD4 was immunoprecipitated from C2C12 myoblasts. As shown in Fig. 4B, eIF2Bε was detected only when anti-NEDD4 antibody was included in the immunoprecipitation reaction. Overall, the results provide strong support for the conclusion that the NEDD4 E3 ligase interacts with eIF2Bε.
Fig. 4.
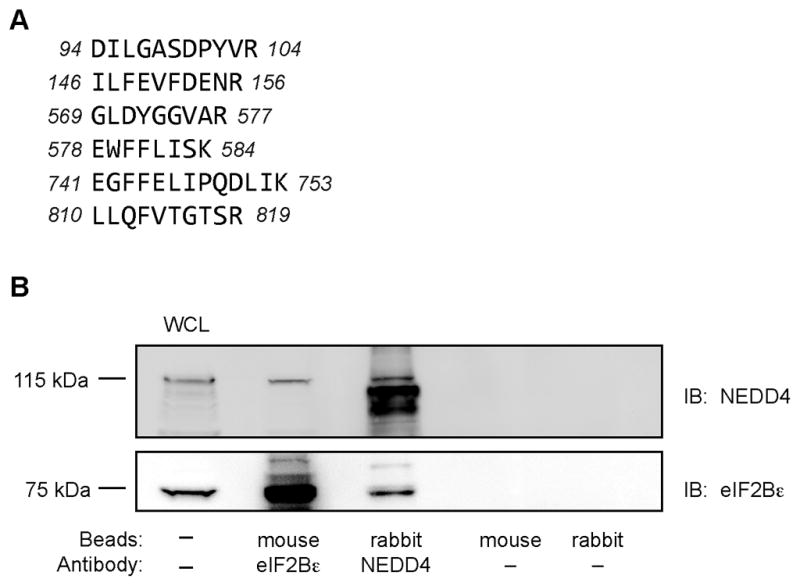
Co-immunoprecipitation of NEDD4 with eIF2Bε. (A) Unique peptides corresponding to mouse NEDD4 identified in two independent MS/MS analyses; several of the peptides shown were identified in both analyses. Total protein coverage by amino acid was 60/887 (6.8%). (B) NEDD4 and eIF2Bε were immunoprecipitated from C2C12 homogenates as described under “Materials and methods”. The immunoprecipitates were subjected to Western blot analysis for either NEDD4 or eIF2Bε. The results are representative of 4 samples that were analyzed.
3.5. Identification of novel phosphorylation sites on eIF2Bε
In addition to the ubiquitin modifications identified, the LC-MS/MS analyses also revealed several phosphorylated residues within the eIF2Bε tryptic peptides. Three phosphorylated serine residues (Ser527, Ser535, and Ser539) were identified that have been previously reported. In addition to these, three additional novel phosphosites were identified in the current analyses: Ser22, Ser125, and Thr317 (Fig. 3B). A fourth phosphosite was contained among the contiguous serine and threonine residues (#325–327) but the exact location of the phosphorylated residue was unclear from the spectra. Each of the novel phosphorylation sites we report here are conserved in the rat, mouse, and human eIF2Bε proteins (Fig. 3B).
4. Discussion
The expression of eIF2Bε appears to be tightly regulated at multiple levels including transcription [15,16], translation of its mRNA [17,18], and as demonstrated herein proteasomal degradation. eIF2Bε, a guanine nucleotide exchange factor that is crucial in controlling global protein synthesis, cellular stress responses, and the regulation of cell growth and proliferation rates, would be expected to be tightly regulated and have multiple checkpoints to control its expression. The literature strongly suggests that eIF2Bε expression is regulated in accordance with the conditions to which the cell or tissue is exposed. In other words, in an anabolic state such as that which occurs in response to resistance exercise, expression of eIF2Bε in skeletal muscle may increase, at least temporarily, to promote or support the anabolic condition [17,18,19,20]. In contrast, in catabolic conditions where muscle protein must be liberated for metabolic purposes rather than deposited/accreted, the activity and/or expression of eIF2Bε is reduced [e.g. 15,19]. Here, we present direct evidence that eIF2Bε is also subject to ubiquitin modification and proteasomal degradation.
A single mutation in any individual eIF2B subunit can lead to its decreased stability as well as affect the stability of the other (non-mutated) subunits [21,22]. It remains unclear whether the eIF2B holoenzyme turns over as a single entity or as individual subunits. One possibility is that disassembly of the complex is required for degradation of individual subunits. An alternative postulate would be that a single subunit may be subject to regulation via degradation and subsequently affects the stability of the remaining subunits in the complex. In support of coordinated degradation of the complex, a recent report [23] concluded that the half life of the five eIF2B subunits is similar. Whether or not newly synthesized subunits enter existing complexes or if individual subunits continuously and dynamically dissociate and reassemble in various conditions is critical in understanding the stability of the complex. Evidence from other multiprotein complexes suggests that availability of additional interacting subunits (presumably in an equimolar ratio) can indeed enhance the stability of individual subunits [e.g. 24].
The present study identified NEDD4 as an eIF2Bε interacting protein. NEDD4 is one of nine members of the NEDD4 family of E3 ligases that are characterized by the presence of a calcium/lipid-binding C2 domain at the N-terminus, a HECT (homologous to E5-AP carboxyl terminus) domain at the C-terminus, and multiple WW protein interaction domains [25,26]. It has been shown to mediate ubiquitylation of a variety of proteins, including several that are involved in cell growth and proliferation. NEDD4 interacts with many of its substrates through the binding of the WW domains to phosphorylated amino acids (tyrosine, serine, or threonine) in the substrate. In this regard, one of the novel phosphorylation sites (Ser125) identified in the present study is located in proximity to one of the ubiquitylation sites (Lys136), making it tempting to speculate that phosphorylation of eIF2Bε might promote its association and ubiquitylation by NEDD4. Future studies will be required to test this hypothesis.
Ubiquitin itself was modified at several residues in the present investigation. Beyond the canonical Lys-48 linked polyubiquitin chains, polyubiquitin chains linked via Lys-11 and Lys-29 of ubiquitin are also implicated in mediating proteasomal degradation [27,28]. Polyubiquitin linkage via Lys-63 has been reported to lead to both lysosomal [27] and proteasomal degradation [29] although others report that Lys-63 linked polyubiquitin chains do not accumulate upon conditional knockout of the 26S proteasome [28], suggesting that Lys-63 may not be involved in proteasomal degradation. At this time, the exact topology and role of any polyubiquitin chains in the context of their physiological role on the eIF2Bε protein remain unknown. Many additional functions (i.e., beyond proteolytic targeting) have been described for “atypical” (mixed-linkage) polyubiquitin chain topologies as well as for monoubiquitination of proteins [30]. Indeed, additional information regarding the function of ubiquitin and other ubiquitin-like modifiers is continuously emerging and the understanding of their function continues to evolve. In light of these novel posttranslational modifications identified on eIF2Bε, future studies are warranted particularly to examine their role in eIF2B assembly, function and stability.
Supplementary Material
Highlights.
Five unique ubiquitylation sites on eIF2Bε were identified by LC-MS/MS
Three novel phosphorylation sites were also identified
The E3 ligase NEDD4 was found to interact with eIF2Bε
The results represent the first evidence on the mechanism of eIF2Bε turnover
Acknowledgments
The authors are grateful to Holly A. Lacko for her expert technical assistance. The studies described here were supported by National Institutes of Health [Grants DK-15658 and DK-13499 (to L. S. Jefferson)] and a grant from The Pennsylvania Department of Health using Tobacco Settlement Funds. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Abbreviations
- DMEM
Dulbecco’s modified Eagle’s medium
- eIF
eukaryotic initiation factor
- HECT
homologous to E5-AP carboxyl terminus
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- NEDD4
neural precursor cell expressed, developmentally down-regulated 4
- NEM
N-ethylmaleimide
- PVDF
polyvinylidene difluoride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tuckow AP, Jefferson SJ, Kimball SR, Jefferson LS. Simvastatin represses protein synthesis in the muscle-derived CC cell line with a concomitant reduction in eukaryotic initiation factor 2B expression. Am J Physiol Endocrinol Metab. 2011;300:E564–570. doi: 10.1152/ajpendo.00383.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinformatics. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, Goebl MG, Iakoucheva LM. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–380. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcìa-Alai MM, Gallo M, Salame M, Wetzler DE, McBride AA, Paci M, Cicero DO, de Prat-Gay G. Molecular Basis for Phosphorylation-Dependent, PEST-Mediated Protein Turnover. Structure (London, England : 1993) 2006;14:309–319. doi: 10.1016/j.str.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Flowers KM, Mellor H, Matts RL, Kimball SR, Jefferson LS. Cloning and characterization of complementary and genomic DNAs encoding the epsilon-subunit of rat translation initiation factor-2B. Biochim Biophys Acta. 1996;1307:318–324. doi: 10.1016/0167-4781(96)00055-3. [DOI] [PubMed] [Google Scholar]
- 6.Fabian JR, Kimball SR, Jefferson LS. Reconstitution and purification of eukaryotic initiation factor 2B (eIF2B) expressed in Sf21 insect cells. Protein Expr Purif. 1998;13:16–22. doi: 10.1006/prep.1998.0860. [DOI] [PubMed] [Google Scholar]
- 7.Lim KL, Chew KC, Tan JM, Wang C, Chung KK, Zhang Y, Tanaka Y, Smith W, Engelender S, Ross CA, Dawson VL, Dawson TM. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escobedo J, Koh TJ. Improved transfection technique for adherent cells using a commercial lipid reagent. Biotechniques. 2003;35:936–938. 940. doi: 10.2144/03355bm06. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher AR, Kimball SR, Dennis MD, Schilder RJ, Jefferson LS. The mTORC1 signaling repressors REDD1/2 are rapidly induced and activation of p70S6K1 by leucine is defective in skeletal muscle of an immobilized rat hindlimb. Am J Physiol Endocrinol Metab. 2013;304:E229–236. doi: 10.1152/ajpendo.00409.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimball SR, Horetsky RL, Jefferson LS. Implication of eIF2B rather than eIF4E in the regulation of global protein synthesis by amino acids in L6 myoblasts. J Biol Chem. 1998;273:30945–30953. doi: 10.1074/jbc.273.47.30945. [DOI] [PubMed] [Google Scholar]
- 11.Schlesinger DH, Goldstein G, Niall HD. The complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry. 1975;14:2214–2218. doi: 10.1021/bi00681a026. [DOI] [PubMed] [Google Scholar]
- 12.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 13.Peng J. Evaluation of proteomic strategies for analyzing ubiquitinated proteins. BMB Rep. 2008;41:177–183. doi: 10.5483/bmbrep.2008.41.3.177. [DOI] [PubMed] [Google Scholar]
- 14.Jeram SM, Srikumar T, Pedrioli PG, Raught B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics. 2009;9:922–934. doi: 10.1002/pmic.200800666. [DOI] [PubMed] [Google Scholar]
- 15.Voisin L, Gray K, Flowers KM, Kimball SR, Jefferson LS, Vary TC. Altered expression of eukaryotic initiation factor 2B in skeletal muscle during sepsis. Am J Physiol Endocrinol Metab. 1996;270:E43–50. doi: 10.1152/ajpendo.1996.270.1.E43. [DOI] [PubMed] [Google Scholar]
- 16.Kubica N, Kimball SR, Jefferson LS, Farrell PA. Alterations in the expression of mRNAs and proteins that code for species relevant to eIF2B activity after an acute bout of resistance exercise. J Appl Physiol. 2004;96:679–687. doi: 10.1152/japplphysiol.00962.2003. [DOI] [PubMed] [Google Scholar]
- 17.Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bε mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem. 2005;280:7570–7580. doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- 18.Kubica N, Crispino JL, Gallagher JW, Kimball SR, Jefferson LS. Activation of the mammalian target of rapamycin complex 1 is both necessary and sufficient to stimulate eukaryotic initiation factor 2Bε mRNA translation and protein synthesis. Int J Biochem Cell Biol. 2008;40:2522–2533. doi: 10.1016/j.biocel.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwee DT, Bodine SC. Age-related deficit in load-induced skeletal muscle growth. J Gerontol A Biol Sci Med Sci. 2009;64:618–628. doi: 10.1093/gerona/glp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fluckey JD, Knox M, Smith L, Dupont-Versteegden EE, Gaddy D, Tesch PA, Peterson CA. Insulin-facilitated increase of muscle protein synthesis after resistance exercise involves a MAP kinase pathway. Am J Physiol Endocrinol Metab. 2006;290:E1205–1211. doi: 10.1152/ajpendo.00593.2005. [DOI] [PubMed] [Google Scholar]
- 21.Richardson JP, Mohammad SS, Pavitt GD. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol Cell Biol. 2004;24:2352–2363. doi: 10.1128/MCB.24.6.2352-2363.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leng X, Wu Y, Wang X, Pan Y, Wang J, Li J, Du L, Dai L, Wu X, Proud CG, Jiang Y. Functional analysis of recently identified mutations in eukaryotic translation initiation factor 2Bε (eIF2Bε) identified in Chinese patients with vanishing white matter disease. J Hum Genet. 2011;56:300–305. doi: 10.1038/jhg.2011.9. [DOI] [PubMed] [Google Scholar]
- 23.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 24.Keppler BR, Archer TK. Ubiquitin-Dependent and -Independent Control of Subunit Stoichiometry in the SWI/SNF Complex. J Biol Chem. 2010 doi: 10.1074/jbc.M110.173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearwin-Whyatt L, Dalton HE, Foot N, Kumar S. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. BioEssays. 2006;28:617–628. doi: 10.1002/bies.20422. [DOI] [PubMed] [Google Scholar]
- 26.Yang B, Kumar S. Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 2010;17:68–77. doi: 10.1038/cdd.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedford L, Layfield R, Mayer RJ, Peng J, Xu P. Diverse polyubiquitin chains accumulate following 26S proteasomal dysfunction in mammalian neurones. Neurosci Lett. 2011;491:44–47. doi: 10.1016/j.neulet.2010.12.064. [DOI] [PubMed] [Google Scholar]
- 29.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28:359–371. doi: 10.1038/emboj.2008.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.