Abstract
The human β-globin genes are regulated by a locus control region (LCR) and are expressed at extremely high levels in erythroid cells. How transcriptional fidelity of highly expressed genes is regulated and maintained during the cell cycle is not completely understood. Here, we analyzed the association of transcription factor USF, the co-activator CBP, topoisomerase I (Topo I), basal transcription factor TFIIB, and RNA polymerase II (Pol II) with the β-globin gene locus at specific cell-cycle stages. The data demonstrate that while association of Pol II with globin locus associated chromatin decreased in mitotically arrested cells, it remained bound at lower levels at the γ-globin gene promoter. During early S-phase, association of CBP, USF and Pol II with the globin gene locus decreased. The reassociation of CBP and USF2 with the LCR preceded reassociation of Pol II, suggesting that these proteins together mediate recruitment of Pol II to the β-globin gene locus during S-phase. Finally, we analyzed the association of Topo I with the globin gene locus during late S-phase. In general, Topo I association correlated with the binding of Pol II. Inhibition of Topo I activity reduced Pol II binding at the LCR and intergenic regions but not at the γ-globin gene promoter. The data demonstrate dynamic associations of transcription factors with the globin gene locus during the cell cycle and support previous results showing that specific components of transcription complexes remain associated with highly transcribed genes during mitosis.
Keywords: globin gene locus, cell-cycle, transcription factor, mitosis, RNA polymerase II
Introduction
Transcription is regulated at many different levels, including modification of chromatin structure, recruitment of transcription complexes and alteration of transcription elongation properties [Orphanides and Reinberg, 2002; Juven-Gershon and Kadonaga, 2010]. Recent data suggest that moderately expressed genes are expressed in spikes for only short time periods during the cell cycle [Chubb et al., 2010; Suter et al., 2011]. In contrast, highly expressed genes are transcribed for longer time periods. This suggests differences in transcription complex assembly and activity during the cell cycle between highly expressed genes and genes that are expressed at lower levels. Understanding of how transcription competence of specific gene loci is maintained during the cell cycle is limited. Mitotic chromatin has long been viewed as rigid and relatively inaccessible. However, Martinez-Balbas et al. [1995] demonstrated that DNase I hypersensitive sites associated with gene promoters remain accessible during mitosis. Furthermore, Nishino et al. [2012) recently demonstrated that mitotic chromatin fibres are flexible and dynamic and not organized as regular 30nm fibres. Other studies have shown that components of the basal transcription machinery, including TATA-binding protein (TBP) and TFIIB, as well as specific transcription factors and chromatin modifying activities, remain bound at mitotic chromatin and may thus bookmark genes for rapid re-expression after exit from mitosis [Christova and Oelgeschlager, 2002; Chen et al., 2005; Blobel et al., 2009; Zaidi et al., 2010]. In general, Pol II transcription is repressed during mitosis; however, some data suggest the possibility that highly transcribed genes may maintain Pol II binding and even low levels of transcription during mitosis [Parsons and Spencer, 1997].
The S-phase represents a stage in the cell-cycle in which the process of transcription is affected by DNA synthesis [Chakalova et al., 2005]. Although many studies have shown that transcription takes place during S-phase, the process of replication is inhibitory to transcription. Current models suggest that transcription takes place in specialized domains in the nucleus, called transcription factories, while DNA synthesis takes place in other domains, called replication factories [Wei et al., 1998; Chakalova et al., 2005; Sutherland and Bickmore, 2009, Hiratani et al., 2009]. According to this model, active genes are relocated to replication factories sometime during S-phase and are then moved back to transcription factories. DNA binding proteins are likely involved in regulating the association of gene loci with transcription and replication factories. In general, open and transcriptionally active chromatin replicates early during S-phase while inaccessible heterochromatin replicates late [Peng et al., 2008].
Both transcription and replication of chromosomal templates lead to torsional stress of the DNA and results in positive and/or negative supercoiling [Vos et al., 2011; Durand-Dubief, 2011]. For example, transcription of longer genes leads to overwinding of DNA (positive supercoiling) in front of the RNA polymerase and underwinding of DNA (negative supercoiling) behind the RNA polymerase. Topoisomerases (Topos) are involved in relaxing these supercoils to allow continuation of transcription and replication. While Topo II catalyzes a double strand break through which another double strand of DNA is transferred, Topo I changes the linking number by introducing a nick into one strand of the double helix and passing the other strand through. Previous studies have shown that Topo I and II are involved in transcription by Pol II [Kretzschmar et al., 1993; Collins et al., 2001; Sperling et al., 2011]. Consistently, the association of Topo I and II with chromatin correlates with the binding of Pol II [Khobta et al., 2006; Capranico et al.; 2007, Bermejo et al, 2009]. Topo I but not Topo II activity is inhibited by the cancer drug camptothecin [Pommier, 2009]. Camptothecin arrests the Topo I/DNA complex, which then blocks the propagation of replication and transcription. Inhibition of Topo I activity causes reduction in transcription of specific genes and alterations in Pol II binding to chromatin.
The human β-globin genes are expressed at extremely high levels in erythroid cells and regulated by a locus control region (LCR), which consists of five DNase I hypersensitive sites (HS1 to HS5) [Bulger and Groudine, 1999; Levings and Bungert, 2002]. The LCR associated HS sites recruit transcription factors, chromatin modifying activities and transcription complexes [Liang et al., 2008]. The recruitment of Pol II to promoters involves transcription factors that interact with basal promoter elements, such as the TATA box, downstream promoter elements (DPE), and initiator [Juven-Gershon and Kadonaga, 2010]. A well understood pathway involves the binding of TFIID (TATA binding protein, TBP, plus TBP associated factors, TAFs) to the TATA box or initiator and the subsequent recruitment of TFIIB, which then mediates the association of Pol II [Thomas and Chiang, 2006]. How transcription complexes are assembled at enhancers and LCRs is not understood but could involve co-factors like CBP/p300 [Rada-Iglesias et al., 2011]. We previously demonstrated that transcription factor USF is critical for the recruitment of transcription complexes to the LCR and that it associates with the co-activator CBP/p300 [Crusselle-Davis et al., 2006; Crusselle-Davis et al., 2007; Liang et al., 2009].
In this study we analyzed the recruitment of the co-regulator CBP, Pol II, basal transcription factor TFIIB, USF2, and Topo I with the β-globin gene locus at specific stages during the cell cycle in K562 cells. K562 cells are human erythroleukemia cells that express high levels of the γ-globin genes and low levels of the adult β-globin gene. We found that Pol II and TFIIB remain associated at relatively high levels with the γ-globin gene promoter but at much lower levels with HS2 during M-phase. Furthermore, like USF, the interaction of CBP with HS2 precedes reassociation of Pol II during S-phase. Finally, we demonstrate that Topo I binding to the globin gene locus correlates with the binding of Pol II and that inhibition of Topo I activity by camptothecin reduces the association of Pol II with LCR element HS2 but not with the γ-globin gene promoter.
Materials and Methods
Cell Culture
Human erythroleukemia (K562) cells were cultured in RPMI 1640 containing L-glutamine supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The cells were grown at 37°C with 5% ambient CO2 levels and maintained at a density between 1.5 × 105 and 2.5 × 105 cells/mL.
Cell Cycle Synchronization and Mitotic Arrest
The cell cycle synchronization of K562 cells at the border of G1/S-phase was achieved by double thymidine block as described previously by Vieira, et al. (2004). In brief, cells were incubated in media containing 2 mM thymidine for 16–17 h, then allowed to recover in fresh media for 10–12 h, followed by a second round of incubation in media containing 2 mM thymidine for an additional 16–17 h. Cells were either harvested for ChIP and other analyses at specific time points after release from the block, or subsequently arrested in M-phase. Additionally, S-phase synchronized K562 cells were also subjected to treatment with camptothecin (Cpt), an inhibitor of DNA topoisomerase I (Topo I). For cells treated with Cpt, G1/S-phase synchronized cells were allowed to recover in fresh media for 5 h, and then incubated in media containing 20 μM Cpt for 1 h before being harvested for ChIP analysis.
In order to arrest K562 cells in M-phase, cells synchronized in S-phase by double thymidine block were released from the block and allowed to recover in fresh media for 5 h. Then, the cells were incubated in media containing 60 ng/mL of nocodazole for 12 h and harvested for ChIP analysis (Parsons and Spencer, 1997).
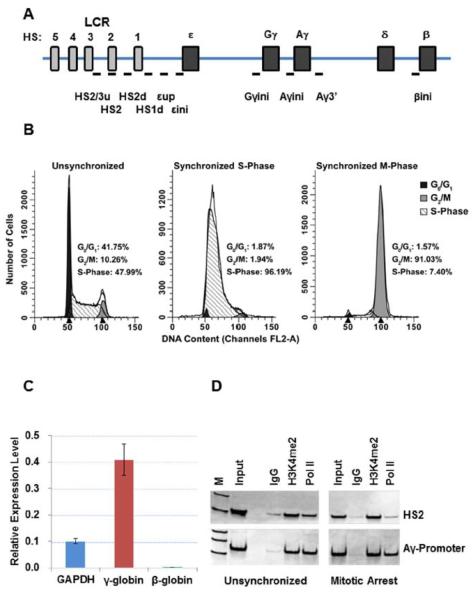
For each synchronization or arrest, aliquots of the blocked/arrested cells were collected for flow cytometry analyses in order to confirm proper cell cycle synchronization and/or arrest. Cells were fixed in 100% ethanol, treated with RNase, and stained with propidium iodine before being subjected to flow cytometry. The cell cycle stage was determined using ModFit LT software. Figure 1 depicts a representative flow cytometry profile for these experiments.
Figure 1.
A. Diagram of the human β-globin gene locus indicating the position of genes and LCR HS sites. Small bars indicate the position of PCR-amplicons analyzed in this study. These regions are as follows: HS2/3/u, two sets of primers in the linker region between HS3 and HS2; HS2, core region of HS2; HS2d, a region between HS2 and HS1; HS1d, a region downstream of HS1; εup, a region between HS1 and ε-globin; εini, a region overlapping the transcription initiation site of the ε-globin gene; Gγini, a region overlapping the transcription start site of the Gγ-globin gene; Aγini, a region overlapping the transcription start site of the Aγ-globin gene; Aγ3', a region at the 3'end of the Aγ-globin gene, βini, a region overlapping the transcription start site of the β-globin gene. B. Flow cytometry analysis of unsynchronized, synchronized at the G1/S-phase border, and M-phase arrested K562 cells. The cells were arrested at the G1/S-phase border (labeled as Synchronized S-Phase) using the double thymidine method, then released from the block and arrested in M-phase using nocodazole (labeled as Synchronized M-Phase). C. Analysis of GAPDH, γ-globin, and β-globin gene expression by reverse transcription (RT) qPCR. The data represent means +/− standard error of the means of results from two experiments. D. ChIP analysis of of the interaction of H3K4me2 and Pol II with HS2 and the Ay-globin promoter (Aγini). ChIP enriched DNA was amplified with the indicated primers and electrophoresed on TBE gels followed by staining with SYBR green.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed using modified protocols originally described by Leach, et al. [2003] and Vieira, et al. [24]. K562 cells were cross-linked in 1% formaldehyde and quenched with 0.125 M Glycine, then washed twice with 1× Phosphate Buffer Solution (PBS) containing protease inhibitors (Roche). Cell nuclei were swelled and then lysed, followed by sonication on ice. Sonication conditions were optimized to shear chromatin into fragments of approximately 500 bp in length.
The sonicated chromatin was pre-cleared by incubation with Protein A sepharose beads (GE Healthcare) for two hours, and then dilutions of the pre-cleared sonicated chromatin were incubated with specific antibodies overnight. The immune complexes were captured using Protein A sepharose beads. Following a series of washes, the DNA was eluted off the beads and purified using a QIAprep Spin Miniprep purification kit (Qiagen). The following antibodies were used in these experiments: rabbit IgG (eBioscience), RNA Polymerase II (CTD45H8, Upstate Biotechnology Inc.), serine-2 phosphorylated RNA Polymerase II (MMS 129R,Covance), and TFII-B (C-18, sc-225), TFIID (TBP, sc-273), CBP (A-22, sc-369), BRG1 (H-88, sc-10768), and USF2 (N18, sc-861), Topo I (C-15, sc-5342) (all from Santa Cruz Biotechnology).
Quantitative PCR (qPCR)
The ChIP samples were analyzed by quantitative Polymerase Chain Reaction (qPCR) using the Bio-Rad MyiQ system. The qPCR conditions were as follows: 95°C for 5 min, followed by 40 cycles of 94°C for 30 s, 59°C for 20 s, and 72°C for 30 s. Additionally, a melting curve was performed from 60°C to 95°C (reading every 0.5°C) to ensure primer specificity. Final quantitation analysis was performed using the standard curve method, with standard curve samples generated from 10-fold serial dilutions of the input sample. Results were normalized to IgG levels. Statistical analysis of the qPCR data was performed using Student's t-test, and significance was concluded if p < 0.05. Primers relevant to the β-globin gene locus included the following: a region between LCR HS 3and HS2 (HS2/3), a region upstream of HS2 (HS2u), the HS2 core region (HS2), a region downstream of HS2 (HS2d), a region downstream of HS1 (HS1d), a region between HS1 and ε-globin (εup), the ε-globin promoter (εini), the Gγ-globin gene promoter (Gγini), the Ay-globin promoter (Aγini), a region 3' to the Aγ-globin gene (Aγ3'), and the β-globin gene promoter (βini). Primers for the promoter region of the Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) were also used in control experiments. The sequences of the primers listed above are detailed in Table 1.
Table 1.
List of qPCR primers used for ChIP
| Primer | Sequences |
|---|---|
| Human HS2 core | US 5'-CGCCTTCTGGTTCTGTGTAA-3' |
| DS 5'-GAGAACATCTGGGCACACAC-3' | |
| Human Gυ-Globin Gene Promoter | US 5'-CCTTCAGCAGTTCCACACAC-3' |
| DS 5'-CTCCTCTGTGAAATGACCCA-3' | |
| Human 3' Aυ-Globin Gene | US 5'-CCATGATGCAGAGCTTTCAA-3' |
| DS 5'-TTTGCTCATCAAAACCCACA-3' | |
| Human βini | US 5'-GTCAGGGCAGAGCCATCTAT-3' |
| DS 5'-AACGGCAGACTTCTCCTCAG-3' | |
| Human EKLF | US 5'-TTCACCGGAGGACAGAGC-3' |
| DS 5'-CCTCCCCAGTAAACAGCAAC-3' | |
| Human GAPDH | US 5'-GCCAATCTCAGTCCCTTCC-3' |
| DS 5'-CCTACTTTCTCCCCGCTTTT-3' | |
| Human Necdin promoter | US 5'-GTGTTATGTGCGTGCAAACC-3' |
| DS 5'-CTCTTCCCGGGTTTCTTCTC-3' | |
| HS2d | US 5'-GGAAGGCATGAAAACAGGAA-3' |
| DS 5'-CCGTATGTGAGCATGTGTCC-3' | |
| HS1d | US-5'CCTCCCTACCACCTTTAGCC-3' |
| DS-5'GCAGAGCCCACATTTTCTTC-3' | |
| εup | US-5'AGAGAGCCCCAGGCAATACT-3' |
| DS-5'GGGGTGATTCCCTAGAGAGG-3' |
For gene expression analysis RNA was isolated from K562 cells, reverse transcribed and the cDNA was subjected to qPCR as described previously (Crusselle-Davis et al., 2007).
Results
The human erythroleukemia cell line K562 expresses high levels of the γ-globin genes and low levels of the adult β-globin gene (Fig. 1C) [Dean et al., 1983]. A diagram of the β-globin gene locus is shown in Fig. 1A. We analyzed the interaction of specific proteins with the β-globin locus at different time points during S-phase and during M-phase. To arrest cells in M-phase, S-phase synchronized cells were released and incubated in the absence of thymidine and in the presence of nocodazole, which arrests cells in early M-phase due to deficient spindle formation. The double thymidine block in combination with nocodazole is widely used to analyze specific events during S- or M-phase. Aliquots of unsynchronized, S-phase synchronized, and M-phase synchronized cells were subjected to flow cytometry and the profiles are shown in Fig. 1B. The data show that the double thymidine treatment resulted in a cell population in which more than 95% of the cells were in S-phase. The combined double thymidine and nocodazole treatment enriched M-phase cells to more than 90%. For S-phase analysis, K562 cells were subjected to double thymidine block to arrest cells at the G1/S-phase border. Aliquots of the cells were taken at different time points after removal of thymidine and subjected to ChIP analysis.
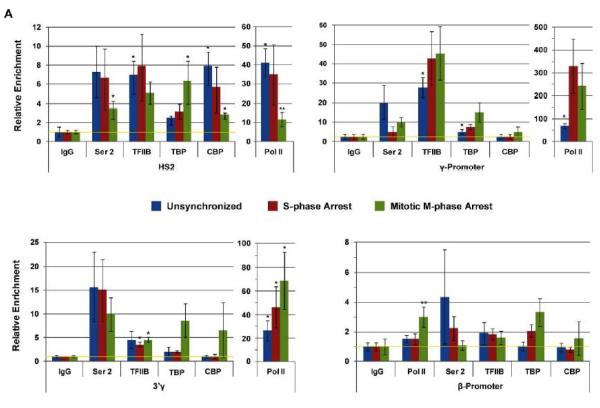
We first examined the association of RNA polymerase II (Pol II) with LCR element HS2 and with the Aγ-globin promoter in unsynchronized and mitotically arrested K562 cells. As a control we examined association of H3K4me2, a histone modification associated with active chromatin, with the two regions. The data show that while Pol II partially dissociates from the LCR during M-phase it remains associated with the Ay-globin gene promoter (Fig. 1D). To further examine the association of transcription factors and Pol II with the globin gene locus during S- and M-phase we performed ChIP experiments and subjected the precipitated DNA to qPCR. In these experiments we analyzed the interactions of Pol II, Pol II phosphorylated at serine 2 (Pol II S2P), TFIIB, TBP, and CBP (Fig. 2). We focused on these proteins because previous studies have shown that components of the basal transcription machinery, mainly TBP, remain associated with specific genes during M-phase [Parsons and Spencer, 1997; Chen et al., 2002; Christova and Oelgeschlager, 2002; Chen et al., 2005; Blobel et al., 2009; Zaidi et al., 2010]. The data show that all proteins examined were associated with LCR element HS2 in unsynchronized and G1/S-phase synchronized cells. The binding of TBP occurred at low levels at the LCR HS2. We observed a decrease in binding of Pol II, Pol II S2P, and CBP, with LCR HS2 during mitosis. The binding of TBP and TFIIB at LCR HS2 was similar in unsynchronized, S-phase arrested, and M-phase arrested cells. At the Aγ-globin promoter the binding of Pol II increased in G1/S-phase synchronized cells and remained at a significant level in M-phase cells. The binding of TFIIB at the Aγ-globin promoter was relatively constant during the different cell cycle stages. The binding of Pol II S2P, TBP and CBP was low at this site. The interaction pattern at the 3' end of the γ-globin gene was similar compared to the promoter but the efficiencies of the binding of the factors were lower compared to the promoter region. Finally, association of all the proteins with the adult β-globin gene promoter, which exhibits low activity in K562 cells, was very inefficient (Fig.2, compare the scale at the β-promoter with that of the other regions examined). The summary data show that Pol II associates with LCR HS2 and largely dissociates from this site during M-phase. In contrast, binding of Pol II remains high in both G1/S-phase and M-phase synchronized cells at the γ-globin promoter.
Figure 2.
Analysis of Pol II and transcription factor binding at the human β-globin gene locus in unsynchronized, synchronized (at the G1/S-border), and M-phase arrested cells. K562 cells were subjected to double thymidine block at G1/S-phase and nocodazole mediated arrest in early M-phase. Aliquots of the cells were taken from unsynchronized, G1/S-phase synchronized (indicated as S-phase arrest) and M-phase arrested (indicated as Mitotic arrest) cultures. The cells were subjected to ChIP assays using antibodies specific for Pol II, Pol II phosphorylated at serine 2 (indicated as Ser 2), TFII B, TBP, and CBP. Precipitation with IgG antibodies served as negative controls. The DNA was isolated from the immunoprecipitates and subjected to qPCR using primers specific for LCR element HS2, the Gγ-promoter region, the Aγ-3'end, and the β-globin promoter region. The data represent means +/− standard errors of the means of three independent experiments with PCR performed in duplicate. The data are shown relative to the IgG control, set at 1. The yellow line indicates IgG background levels.
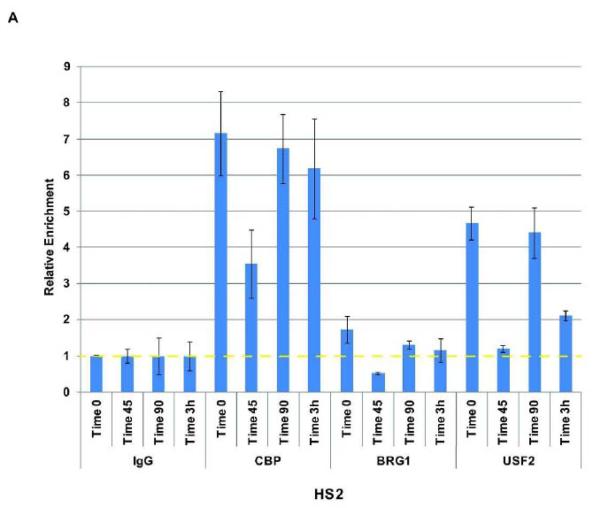
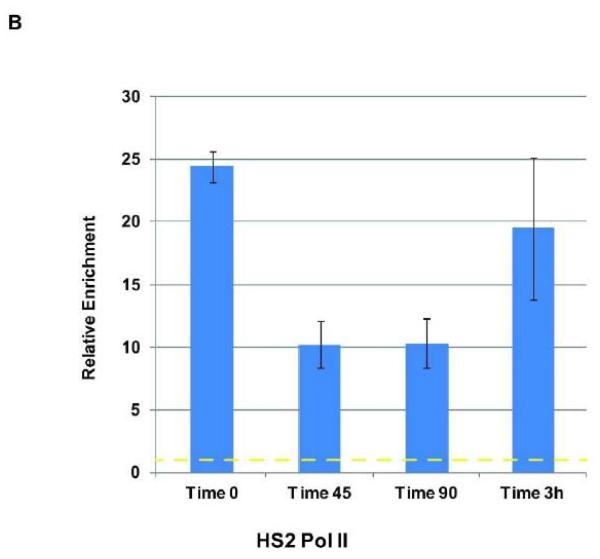
We previously demonstrated that Pol II dissociates from the β-globin gene locus during early S-phase in K562 cells [Vieira et al., 2004]. Transcription factor USF2, which is implicated in the recruitment of transcription complexes in the β-globin gene locus, also dissociated from the globin locus but re-associated with LCR element HS2 and the γ-globin gene promoter before recruitment of Pol II [Vieira et al., 2004]. The dissociation of USF and Pol II correlated with ongoing replication at the early S-phase stage [Vieira et al., 2004]. The previous interpretation was based on chromatin immunoprecipitation experiments followed by a non-quantitative PCR. Here, we repeated these experiments using quantitative PCR (qPCR) and also included the analysis of co-factors CBP and BRG1 (Fig. 3). We have shown previously that USF interacts with CBP in erythroid cells [Crusselle-Davis et al., 2007]. Because CBP is also interacting with components of the basal transcription machinery, it may be involved in recruiting Pol II to the LCR. BRG1 is an ATPase dependent chromatin remodeling factor that has been implicated in LCR function [Kim et al., 2007]. The data largely confirm the previous results and demonstrated that Pol II, USF2 and CBP dissociate from locus control region HS2 in early S-phase (45 min after release from the double thymidine block) and reassociate at later time points (Fig. 3 A and B). While USF2 and CBP were already bound at LCR HS2 at the 90-minute time point, the binding of Pol II was still reduced and revealed maximal levels only at the 3-hour time point. The recovery of BRG1 associated LCR HS2 fragments was very inefficient. The data suggest that the reassociation of Pol II after completion of globin locus replication is mediated by USF2 and its co-factor CBP. We note that the results shown in Fig. 3 are reproducible but reflect the data from a single synchronization/ChIP/qPCR experiment. We observed similar patterns of interaction in separate experiments; however, the level of binding and the exact timing of dissociation and reassociation varied somewhat in these complex experiments.
Figure 3.
Analysis of Pol II and transcription factor interaction with LCR element HS2 at different time points during S-phase. K562 cells were subjected to double thymidine block to arrest cells at the G1/S-phase border at which time point an aliquot was taken (Time 0). After the release from the block aliquots of cells were taken at different time points (45 min, 90 min, and 3 hrs, as indicated). The cells were subjected to ChIP analysis using antibodies specific for CBP, BRG1, USF2 (A), and Pol II (B). Precipitation with IgG antibodies served as negative controls in these experiments and the data were compared to the IgG samples which were set at 1. The bars represent the results of a single ChIP experiment with the qPCR performed in triplicate. Data are represented as the means +/− standard error of the means. Independent experiments yielded qualitatively similar data. The dotted yellow lines represent IgG background levels.
There is increasing evidence showing that enhancers recruit transcription complexes and are transcribed [Kim et al., 2010; Koch and Andreau, 2011]. This has been shown to be the case for the LCR associated HS sites a long time ago [Tuan et al., 1992]. In fact, transcription complexes associate with LCR HS sites in undifferentiated cells and produce non-coding transcripts [Vieira et al., 2004, Levings et al., 2006]. It has been shown that topoisomerases (Topo) I and II are involved in the process of transcription [Vos et al., 2011]. Both enzymes can relieve the supercoil tension arising from transcription elongation and have also been implicated in the process of transcription initiation. We analyzed the recruitment of Pol II and Topo I at various positions along the β-globin gene locus in K562 cells. To analyze the interaction of Topo I with the globin gene locus, we trapped its chromatin interactions using the Topo I inhibitor camptothecin (Cpt), which leads to a covalent interaction of Topo I with DNA [Pommier, 2009]. Furthermore, we analyzed interaction of Topo I and Pol II with the globin gene locus in synchronized cells 5 hours after the release from the double thymidine block. The rationale for this was that replication could interfere with Topo I and Pol II binding.
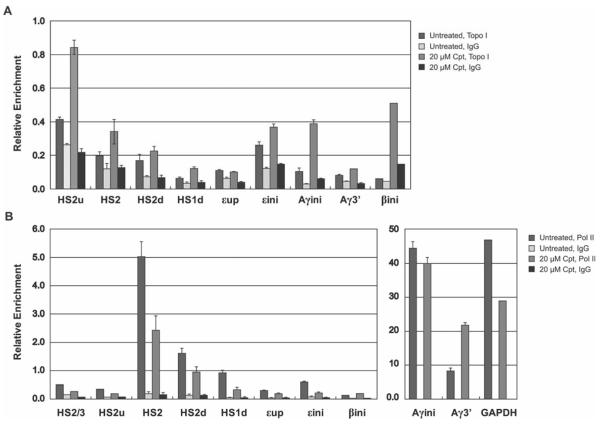
The data demonstrate that Topo I binding was detectable at LCR HS2 and at the transcribed Aγ-globin gene, both at the 5' and 3' end, after treatment with camptothecin (Fig. 4). We observed a gradual decrease in Topo I binding in a region downstream of the LCR HS2 core. The pattern of Topo I binding was very similar to the binding of Pol II. Pol II efficiently interacted with HS2 and with the Aγ-globin gene promoters. We also observed a gradual decrease in Pol II binding with the HS2 downstream region. The occupancy of Pol II at the γ-globin gene promoter was much higher compared to HS2. In contrast, Topo I appeared to bind more efficiently at LCR HS2. Furthermore, the binding of Pol II decreased at HS2 in the presence of camptothecin but remained unaltered at the γ-globin gene promoter. Interestingly, we detected a small but reproducible increase in Pol II binding at the adult β-globin gene promoter in the presence of camptothecin. This was accompanied by an increase in β-globin gene transcription (data not shown). The data reveal a correlation between Topo I and Pol II associations in the globin gene locus and are largely consistent with previous data suggesting that Topo I is involved in Pol II recruitment to specific sites in the genome [Vos et al., 2011].
Figure 4. Analysis of Topo I and Pol II binding with specific region in the β-globin gene locus in camptothecin treated K562 at late S-phase.
Quantitative PCR results of ChIP assays of Topo I (upper graph) and Pol II (lower graph) binding to the β-globin locus in synchronized K562 cells, untreated (no drug) or treated with 20μM of camptothecin (Cpt) for 1hour. Cells were cross-linked with formaldehyde and the chromatin was isolated, fragmented, and precipitated with antibodies against Topo I, Pol II, or the unspecific IgG antibody. DNA was purified from the precipitate and analyzed by quantitative PCR using primers specific for the indicated regions in the LCR, intergenic region, and promoters of ε-globin, γ-globin, and β-globin in the β-globin locus (the amplicons are indicated in the diagram shown in Fig. 1A: HS2/3, LCR HS2 and HS3 linker region; HS2u, HS2 5' flanking region; HS2, HS2 core; HS2d, HS2 downstream region; HS1d, HS1 downstream region; εup: ε-globin gene upstream region; εini, ε-globin promoter; Aγini, Aγ-globin promoter; Aγ3', Aγ-globin gene 3' region; βini, β-globin promoter; GAPDH, GAPDH promoter region.). Bars represent the relative enrichment over the input DNA. Error bars represent standard deviation from two independent experiments.
Discussion
The vertebrate globin genes are expressed at extremely high levels in erythroid cells [Stamatoyannopoulos, 2005]. High-level expression is mediated by transcriptional and post-transcriptional mechanisms. Transcription of the globin genes is activated by multiple DNA elements including, promoters, proximal enhancers, and locus control regions. Recent reports demonstrated that the globin loci associate with transcription foci, also called transcription factories, in erythroid cells [Ragoczy et al., 2006; Osborne et al., 2004]. In the β-globin gene locus the association with transcription foci is dependent on the LCR. If transcription takes place in transcription foci, how is this process maintained during the cell cycle? The replication of the globin locus during S-phase and the condensation of chromatin during M-phase likely disrupt associations with transcription foci. In this study, we analyzed the association and dissociation of specific transcription factors, RNA polymerase II (Pol II) and Topoisomerase (Topo) I with the β-globin gene locus at different cell cycle stages in the human erythroleukemia cell line K562.
The analysis of transcription factor association with the LCR at specific time points during S-phase revealed that USF, CBP, and Pol II dissociated from HS2 during early S-phase. USF and CBP re-associated with the globin locus before Pol II is recruited to the globin gene locus after replication. We previously demonstrated that the dissociation of Pol II from the globin locus coincides with replication [Vieira et al., 2004]. Thus, these data confirm and extend our previous observations. We propose that USF and CBP are involved in the reassociation of Pol II with the globin gene locus during S-phase. USF associates with LCR element HS2 and with the globin gene promoters and inactivation of USF diminished recruitment of Pol II to the β-globin gene locus [Crusselle-Davis et al., 2006, Liang et al., 2009]. Furthermore USF interacts with the co-activators CBP/p300 [Crusselle-Davis et al., 2007]. These proteins have been shown in many studies to aide in the recruitment of transcription complexes [Orphanides and Reinberg, 2002]. The fact that USF is not an erythroid-specific protein but expressed in many cell types suggests that one function of USF in general may be to regulate Pol II recruitment at specific cell-cycle stages. CBP may aid USF in this process.
The analysis of transcription factor association with the globin locus at the G1/S-phase border and during M-phase revealed that the levels of Pol II were relatively high at LCR element HS2 in unsynchronized and G1/S-phase synchronized cells and declined to low levels at M-phase. The low level association of Pol II during M-phase is likely due to the fact that although the nocodazole mediated arrest in M-phase was very efficient (see Fig. 1B) a small number of cells (less than 10%) was still in S/G2 phase. Nevertheless, the situation was different at the γ-globin gene promoter and at the 3' end of the Aγ-globin gene. Pol II levels were low in unsynchronized cells and increased at the G1/S-phase border. Importantly, we did not observe a dramatic decrease in Pol II binding at the promoter and at the 3' end of the γ-globin gene in M-phase arrested cells. Again, even though the nocodazole mediated arrest was not 100%, the fact that we did not observe a significant decrease in Pol II, and as a matter of fact, TFIIB and TBP, binding at the γ-globin promoter suggest that these proteins remain associated with the γ-globin gene during M-phase. These data are consistent with previous studies showing that TBP remains associated with active promoters in M-phase [Chen et al., 2002, Christova and Oelgeschlager, 2002]. However, most previous studies demonstrated that Pol II dissociates from mitotic chromatin. It is possible that Pol II remains associated with genes that are highly expressed, like the globin genes in erythroid cells. In fact, a previous report showed that Pol II remains associated with the γ-actin gene in HeLa cells [Parsons and Spencer, 2007]. The continuous binding of Pol II at the 5' end of the γ-globin gene could ensure rapid re-initiation of transcription during early G1. It is unlikely the γ-globin gene is expressed during M-phase. This is supported by our unpublished observation that the levels of H3 tri-methylated at lysine residue 36 (H3K36me3) were decreased in M-phase compared to unsynchronized cells and G1/S-phase synchronized cells (Shermi Liang, unpublished). H3K36 methylation is an indicator for active elongation [Buratowski and Kim, 2010]. Furthermore, the levels of serine 2 phosphorylated Pol II were very low at the β-globin gene locus during M-phase. This suggests that Pol II remains bound at the promoter in M-phase but is not engaged in productive transcription. The persistent binding of Pol II at the γ-globin promoter is likely mediated by TBP and TFIIB. However, a previous report demonstrated that the erythroid-specific transcription factor NF-E2 also binds to mitotic chromatin [Xin et al., 2007]. We and others have shown that NF-E2 interacts with LCR HS sites and with the globin gene promoters [Liang et al, 2008]. Furthermore, NF-E2 has been shown to be required for the efficient recruitment of Pol II to the adult β-globin gene promoter but not to LCR element HS2 [Johnson et al., 2001, Zhou et al., 2010]. Thus, NF-E2 could assist basal transcription factors in maintaining Pol II binding at the γ-globin promoter during mitosis. In this respect, it is important to note that NF-E2 has been shown to interact with a TBP associated factor (TAF) [Amrolia et al., 1997].
Topoisomerases have long been known to play a role in the process of transcription [Vos et al., 2011]. Topo I and Topo II are involved in relaxing negative and positive supercoils upstream and downstream of the transcribing polymerase and have been implicated in transcription complex formation at promoters. We detected relatively high levels of Topo I at the active γ-globin gene promoter and at LCR element HS2, sites that also recruit Pol II with high efficiency. Thus, our data confirm previous results and further suggest that Topo I not only regulates the activity of Pol II at promoters but also at enhancer and locus control regions. It is interesting to note that inhibition of Topo I led to a reduction in the association of Pol II with LCR HS2 and HS2 downstream regions but not with the γ-globin gene promoter. In fact, we detected an increase in the accumulation of Pol II at the 3' end of the γ-globin gene. While there are many possible interpretations for these results, the data could suggest that Topo I is particularly important for the recruitment of Pol II to LCR HS2. Reitman and Felsenfeld [1990] previously assayed the recruitment of Topo II to the chicken β-globin gene locus and found that it is associated with almost all of the DNase I HS sites analyzed. The data suggested that topoisomerases preferentially act at nucleosome-free regions. This is consistent with our data showing that in the human β-globin gene locus, Topo I preferentially associates with LCR HS2 and the γ-globin promoter region.
We treated late S-phase cells for only one hour with 20μM camptothecin in these experiments to minimize negative effects of replication and DNA damage/repair on Pol II recruitment. Higher concentrations of camptothecin and longer incubation times will affect other processes that indirectly influence recruitment of Pol II to chromatin (e.g. transcription coupled DNA repair) [Vos et al., 2011].
The finding that Topo I is recruited together with Pol II to LCR HS2 and that Pol II recruitment to this site is impaired in the presence of Topo I inhibitors is interesting as identifying proteins that are differentially required for the recruitment of Pol II to the LCR or to the globin gene promoters could aid the functional analysis of the role of Pol II recruitment to the LCR.
Acknowledgements
We thank the members of the UF ICBR Flow Cytometry core for help with flow cytometry. We thank our colleagues in the Bungert laboratory, particularly Blanca Ostmark, for assistance and encouragement. This work was funded by grants from the NIH (RO1 DK083389, RO1 DK52356 to J.B.). Michael Rosenberg was supported in part by an NIH summer supplement for undergraduate students (RO1 DK52356S1).
Funding: NIH RO1DK052356, NIH R01DK052356S1, and NIH RO1DK083389 to J.B.
Footnotes
This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: [10.1002/jcb.24542]
References
- Amrolia PJ, Ramamurthy L, Saluja D, Tanese N, Jane SM, Cunningham JM. The activation domain of the enhancer binding protein p45NF-E2 interacts with TAFII130 and mediates long-range activation of the alpha- and beta-globin gene loci in an erythroid cell line. Proc Nat Acad Sci USA. 1997;94:10051–10056. doi: 10.1073/pnas.94.19.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Capra T, Gonzales-Huici V, Fachinetti D, Cocito A, Natoli G, Katou Y, Mori H, Kurokawa K, Shirahige K, Foiani M. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S-phase transcription. Cell. 2009;123:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–77. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Kim T. The role of cotranscriptional histone methylations. Cold Spring Harb Symp Quant Biol. 2010;75:95–102. doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capranico G, Ferri F, Fogli MV, Russo A, Lotito L, Baranello L. The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie. 2007;89:482–489. doi: 10.1016/j.biochi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P. Replication and transcription: shaping the landscape of the genome. Nat Rev Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromatin. Mol Cell Biol. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Dundr M, Wang C, Leung A, Lamond A, Misteli T, Huang S. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol. 2005;168:41–54. doi: 10.1083/jcb.200407182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T. Association of human TFIID-promoter complexes with silenced mitotic chromatin. Nat Cell Biology. 2002;4:79–82. doi: 10.1038/ncb733. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Liverpool TB. Bursts and pulses: Insights from single cell studies into transcriptional mechanisms. Curr Opin Genet Dev. 2010;20:478–484. doi: 10.1016/j.gde.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Collins I, Weber A, Levens D. Transcriptional consequences of topoisomerase inhibition. Mol Cell Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusselle-Davis VJ, Vieira KF, Zhou Z, Anantharaman A, Bungert J. Antagonistic regulation of beta-globin gene expression by helix-loop-helix proteins USF and TFII-I. Mol Cell Biol. 2006;26:6832–43. doi: 10.1128/MCB.01770-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusselle-Davis VJ, Zhou Z, Anantharaman A, Moghimi B, Dodev T, Huang S, Bungert J. Recruitment of coregulator complexes to the beta-globin gene locus by TFII-I and upstream stimulatory factor. Febs J. 2007;274:6065–73. doi: 10.1111/j.1742-4658.2007.06128.x. [DOI] [PubMed] [Google Scholar]
- Dean A, Ley TJ, Humphries RK, Fordis M, Schechter AN. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Nat Acad Sci USA. 1983;80:55155519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand-Dubief M, Svensson JP, Persson J, Ekwall K. Topoisomerases, chromatin and transcription termination. Transcription. 2011;2:66–70. doi: 10.4161/trns.2.2.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Takebayashi S, Lu J, Gilbert DM. Replication timing and transcriptional control: beyond cause and effect—part II. Curr Opin Genet Dev. 2009;19:142–149. doi: 10.1016/j.gde.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and downstream promoter. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Kadonaga J. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol Cell Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khobta A, Ferri F, Lotito L, Montecucco A, Rossi R, Capranico G. Early effects of topoisomerase II along transcribed genes in human cells. J Mol Biol. 2006;357:127–138. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- Koch F, Andrau JC. Initiating RNA polymerase II as hallmarks of enhancer activity and tissue-specificity. Transcription. 2011;2:263–268. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar M, Meisterernst M, Roeder RG. Identification of human topoisomerase I as cofactor for activator-dependent transcription by RNA polymerase II. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach KM, Vieira KF, Kang SH, Aslanian A, Teichmann M, Roeder RG, Bungert J. Characterization of the human beta-globin downstream promoter region. Nucl Acids Res. 2003;31:1292–1301. doi: 10.1093/nar/gkg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings PP, Bungert J. The human beta-globin locus control region. Eur J Biochem. 2002;269:1589–99. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- Levings PP, Zhou Z, Vieira KF, Crusselle-Davis VJ, Bungert J. Recruitment of transcription complexes to the beta-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. Febs J. 2006;273:746–55. doi: 10.1111/j.1742-4658.2005.05107.x. [DOI] [PubMed] [Google Scholar]
- Liang S, Moghimi B, Yang TP, Strouboulis J, Bungert J. Locus control region mediated regulation of adult beta-globin gene expression. J Cell Biochem. 2008;105:9–16. doi: 10.1002/jcb.21820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang SY, Moghimi B, Crusselle-Davis VJ, Lin IJ, Rosenberg MH, Li X, Strouboulis J, Huang S, Bungert J. Defective erythropoiesis in transgenic mice expressing dominant-negative upstream stimulatory factor. Mol Cell Biol. 2009;29:5900–10. doi: 10.1128/MCB.00419-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Dey A, Rabindran SK, Ozato K, Wu C. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995;83:29–38. doi: 10.1016/0092-8674(95)90231-7. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Nishino Y, Eltsov M, Yasumasa Y, Ito K, Takata H, Takahashi Y, Hihara S, Frangakis AS, Imamoto N, Ishikawa T, Maeshima K. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parsons GD, Spencer CA. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol. 1997;17:5791–5802. doi: 10.1128/mcb.17.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. Topoisomerase inhibitors: chemistry, biology and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol Cell Biol. 1990;10:2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Jeong KS, Kitada T, Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc Natl Acad Sci USA. 2011;108:12693–12698. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hemat. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Molina N, Gatfield D, Schneider K, Schibler U, Naef F. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Tuan D, Kong S, Hu K. Transcription of the HS2 enhancer in erythroid cells. Proc Natl Acad Sci USA. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Vieira KF, Levings PP, Hill MA, Crusselle VJ, Kang SH, Engel JD, Bungert J. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J Biol Chem. 2004;279:50350–7. doi: 10.1074/jbc.M408883200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L, Zhou GL, Song W, Wu XS, Wei GH, Hao DL, Lu X, Liu DP, Liang CC. Exploring cellular memory molecules marking competent and active transcriptions. BMC Mol Biol. 2007;8:31–41. doi: 10.1186/1471-2199-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Li X, Deng C, Ney PA, Huang S, Bungert J. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the beta-globin gene locus. J Biol Chem. 2010;285:15894–905. doi: 10.1074/jbc.M109.098376. [DOI] [PMC free article] [PubMed] [Google Scholar]