Abstract
We have demonstrated that D5 and D2 dopamine receptors exist as heteromers in cells, and determined these receptor interact through amino acids in the cytoplasmic regions of each receptor. Specifically involved in heteromer formation we identified in the carboxyl tail of the D5 receptor three adjacent glutamic acid residues, and in intracellular loop 3 of the D2 receptor two adjacent arginine residues. Any pairing of these three D5 receptor glutamic acids were sufficient for heteromer formation. These identified residues in D5 and D2 receptors are oppositely charged and likely interact by electrostatic interactions.
Keywords: G protein coupled receptors, D5 and D2 dopamine receptor, nuclear localization, protein structure, heteromer, interacting amino acids
1. Introduction
Family A G protein coupled receptors (GPCRs) form heteromers [1-3]. We have reported that dopamine D1-D2 receptor heteromers exist in brain and cultured neurons [4, 5]. These heteromers were subject to conformational changes and separation by agonists [6], the heteromers reformed at the cell surface when the agonist was removed [6]. Identifying specific amino acids involved in GPCR heteromer formation has been hampered by the lack of decisive methodologies. Using our process of inserting a nuclear signal (nls) into a GPCR [7] we have identified residues involved in forming heteromers. We reported that the D1 and D2 heteromers interact by specific residues in the cytoplasmic regions. In intracellular loop 3 (ic3) of the D2 receptor, two arginine residues (274-RR) form an electrostatic interaction with vicinal glutamic residues (404-EE) in the carboxyl tail (c-tail) of the D1 receptor [8]. We also recently identified cytoplasmic residues involved in mu-delta opioid heteromers [9].
Previously we demonstrated heteromerization between the D5 and D2 receptors, our FRET analysis showed D5 and D2 receptors formed a heteromeric complex [10]. The D1 and D5 dopamine receptors share extensive overall homology (80%), however these receptors have negligible homology in their long c-tails. We questioned if D5 and D2 heteromers also form by electrostatic interactions between the D2 ic3 and D5 c-tail. In this report we have determined the specific amino acids in the cytoplasmic regions of D5 and D2 receptors involved in heteromer interactions. We demonstrated that changing the identified cytoplasmic amino acids prevented D5–D2 heteromer formation.
2. Materials and methods
2.1. Fluorescent proteins
cDNA sequences encoding GFP, RFP were obtained from Clontech (Palo Alto, CA), and the receptor constructs generated as described [7].
2.2. Cell culture
HEK cells grown on 60 mm plates in minimum essential medium (MEM), were transfected with 0.5-2 μg cDNA using Lipofectamine (Life technologies, Rockville MD). Dopamine antagonist (+)butaclamol when used, was added to cells and cells visualized by confocal microscopy.
2.3. Microscopy
Live cells expressing GFP, and RFP fusion proteins were visualized with a LSM510 Zeiss confocal laser microscope. In each experiment 5-8 fields, containing 50-80 cells per field were evaluated and the entire experiment was repeated several times (n=3-5).
2.4. DNA Constructs
All the DNA encoding the GPCRs were human origin. Sequences encoding GPCRs were cloned into plasmids pEGFP, as described previously [7 and 11]. The D5 carboxyl tail DNA PCR product, containing no stop codon was subcloned into vector RFP (BD Biosciences) at EcoR1 and Kpn1 and inframe with the start codon of RFP.
2.5. Receptor Constructs
The D5 receptor constructs were prepared using the Quickchange mutagenesis kit (Stratagene) according to the manufacturer’s instructions, and as described [7 and 11]. Receptor DNA was subjected to PCR as previously reported [7]. The reaction mixture consisted of: H2O (32 μl), 10× Pfu buffer (Stratagene) (5μl), dNTP (10mM, 5μl), DMSO (5μl), oligonucleotide primers (100ng, 1μl each), DNA template (100ng), Pfu enzyme (5U). Total volume 50μl. PCR conditions, one cycle at 94 °C for 2 min, 30-35 cycles at 94 °C for 30 sec, 55 °C for 30 sec, 72 °C for 1 min, per cycle, and then one cycle at 72 °C for 5 min. The D2-nls and D1-nls receptor construct was prepared as previously described [7].
3. Results
3.1. Identification of the D5 dopamine receptor amino acids involved in D5-D2 heteromer formation
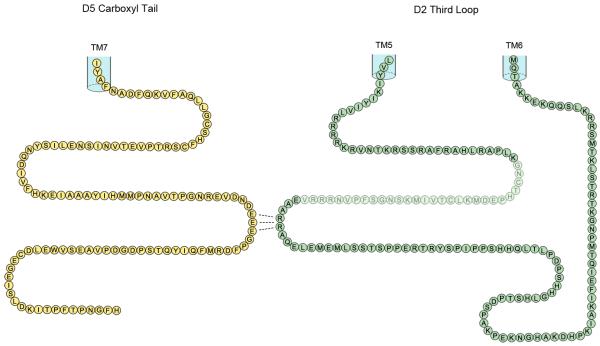
The D5 receptor has an extensive c-tail, extending ~93 amino acids from the palmitoylated cysteine, Fig. 1, (consists of 26% of the total D5 receptor, the D1 receptor c-tail is 95 amino acids in length). There is negligible homology shared between the D1 and D5 receptors throughout their c-tail regions.
Figure 1.
Representation of the cytoplasmic intracellular tail of the D5 dopamine receptor and the cytoplasmic intracellular third loop of the D2 dopamine receptor. The position of the insert of 29 amino acids in the D2 long receptor is indicated by shading.
We incorporated an NLS into the D2 receptor (D2-nls), this did not alter the binding properties, with preserved agonist-detected high affinity and low affinity states, indicative of intact receptor-G protein coupling [7].
We expressed D5 and D2-nls dopamine receptors in cells and demonstrated heteromer formation, Fig. 2, since the D2-nls receptor was able to translocate the D5 receptor to the nucleus. Despite the lack of homology in the extensive c-tails of D1 and D5, in the D5 receptor c-tail there are three adjacent glutamic acids (429–EEE) in a region comparable with the glutamic acid pair (404–EE) located in the D1 receptor, Fig. 1. The D1 receptor glutamic acids (404-EE) were identified as forming hetromers with the D2 receptor [8]. We wished to determine if these three glutamic acids (429-EEE) located in the c-tail of the D5 receptor were involved in forming heteromers with the D2 receptor.
Figure 2.
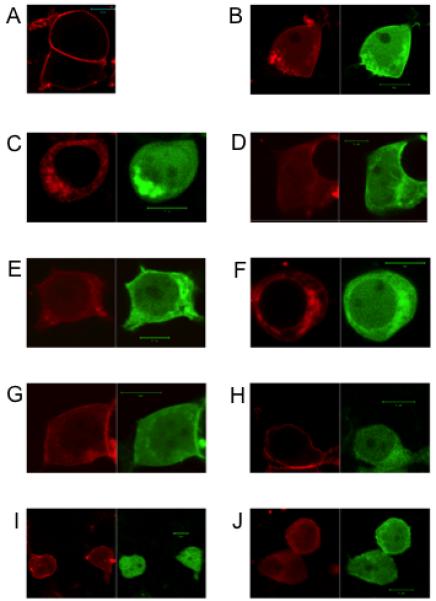
Visualization of expression of D5 and D2-NLS dopamine receptors.
A. D5 (RFP) (red), expressed at the cell surface. B. D5 (RFP) (red) and D2-nls (GFP) (green) co-translocated to the nucleus. C. C1 (D5)(RFP) (red) and D2-nls (GFP) (green) did not co-translocate to the nucleus. D. C4 (D5)(RFP) (red) and D2-nls (GFP) (green) co-translocated to the nucleus. E. C5 (D5)(RFP) (red) and D2-nls (GFP) (green) co-translocated to the nucleus. F. C3 (D5)(RFP) (red) and D2-nls (GFP) (green) did not co-translocate to the nucleus. G. C6 (D5)(RFP) (red) and D2-nls (GFP) (green) co-translocated to the nucleus. H. D5 (RFP) (red) and D2-nls (RR to AA) (GFP) (green) did not co-translocate to the nucleus. I. D5 (RFP) (red) and D1-nls (GFP) (green) co-translocated to the nucleus. J. D5 (RFP) (red) and D1-nls (GFP) (green) co-translocated to the nucleus. Each size bar in figures showing cells indicates length of 10 μm.
We prepared a series of six substitution constructs in the D5 receptor c-tail (Table 1A), and each construct was expressed with the D2-nls receptor. These D5 receptors expressed alone were located predominantly in the cytoplasm. The D5 receptor c-tail constructs C1 (429-EEE to AAA), C2 (429-EEE to EAA) and C3 (429-EEE to EAE) all failed to show D5-D2 heteromerization (Fig. 2 and Table 1A), since the D2 receptor did not translocate these D5 receptors to the nucleus. D5-D2 heteromer formation was observed with D5 receptor constructs C4 (429-EEE to AEE) and C5 (429-EEE to EEA), in which each contained the vicinal –EE residues. Also in C6 construct substitution of the adjacent aspartic acid (DEEE to AEEE) did not affect D5-D2 heteromer formation (Fig. 2 and Table 1A). These experiments indicated a heteromer requirement of at least a pair of glutamic acids (–EE) in the D5 receptor c-tail. Thus like D1 receptor it appears that in the D5 receptor the equivalently located glutamic acid pairs were also involved in heteromerization of D5-D2 receptors. The presence of three glutamic acids (429-EEE) in the D5 receptor, compared to two glutamic acids in (404–EE) in the D1 receptor, would potentially allow two positions for oligomer formation with D2 receptor, utilizing 429-EE or 430-EE.
Table 1 A.
D5 dopamine receptor constructs.
| Heteromer | ||
|---|---|---|
| W/T | … E V D N D E E E G P F D R | Yes |
| C1 | … E V D N D A A A G P F D R | No |
| C2 | … E V D N D E A A G P F D R | No |
| C3 | … E V D N D E A E G P F D R | No |
| C4 | … E V D N D A E E G P F D R | Yes |
| C5 | … E V D N D E E A G P F D R | Yes |
| C6 | … E V D N A E E E G P F D R | Yes |
3.2 Identification of the D2 dopamine receptor amino acids involved in D5-D2 receptor heteromer formation
We wished to determine if the arginines (274–RR) located in ic3 of the D2 receptor were involved in forming heteromers with the D5 receptor. These arginines, identified as being involved in D1-D2 heteromers, were located a distance of 59 amino acids from transmembrane 5 (TM5), Fig. 1. The D2-nls receptor with these arginines substituted (274–RR to AA) and the D5 receptor were co-expressed, Fig. 2. These receptors D5 and D2-nls (RR to AA) did not form heteromers, confirming that both D1 and D5 dopamine receptors utilized the same residues in the dopamine D2 receptor ic3 for heteromer formation.
3.3 Formation of D5-D1 dopamine receptor heteromers
The D1-nls and D5 receptors were co-expressed and formed heteromers, Fig 2, since the D5 receptor was visualized translocating with D1-nls to the nucleus. When D1 receptor-nls was co-expressed with D5 receptor construct, C1 (EEE to AAA), the D1-D5 heteromers remained together indicating that these D5 receptor glutamic acids residues were not involved in forming D1-D5 heteromers.
4. Discussion
There are several accomplishments regarding the oligomeric structures of the D5-D2 dopamine receptors reported. (i) We determined that of three adjacent glutamic acids (429-EEE) in the c-tail of the D5 receptor, any –EE pair was sufficient to form heteromers with the D2 receptor. (ii) We determined adjacent arginines (274-RR) located in ic3 of the D2 receptor, were involved in forming heteromers with the D5 receptor. (iii) We identified single amino acid changes in the D5 receptor that disrupted the D5-D2 heteromers. (iv) We also determined that in the c-tail of the D5 receptor glutamic acid residues (429-EEE) were not involved in D5-D1 receptor heteromer formation. Thus our GPCR-nls incorporation strategy has now enabled elucidation of cytoplasmic structural features of two dopamine receptor families D5-D2, and D1-D2 receptor heteromers [8].
Despite the overall lack of homology in the ~ninety amino acids residues in the c-tails of D1 and D5 receptors, contiguous glutamic acids were located in equivalent positions and each shown to be involved in forming heteromers. The D1 and D2 dopamine receptors form heteromers with D5 receptor using different interacting residues. Although the residues involved in D1-D5 heteromers have not yet been identified, potentially involving TM regions.
Interestingly the rat D5 receptor c-tail contains five contiguous glutamic acids, presumably involved in forming heteromers with the D2 receptor (Table 1B). Thus heteromer formation with these various glycine pairs, (potentially forming heteromers with any of four possible –EE pairs), interact with arginines in ic3 of the D2 dopamine receptor permitting minor variations in the conformation of the D5-D2 heteromer cytoplasmic structures. The mouse D5 receptor has a total of four glutamic acids in a row then alanine and three additional contiguous glutamic acids, (Table 1B), allowing 6 different EE pairings in heteromer formation with the D2 receptor. In comparison the D1 dopamine receptor in human and rodent contains only a single glycine pair (404-EE).
Table 1B.
D5 dopamine receptor cytoplasmic tail sequence in mammalian species.
| Human D5: | E V D N D E E E G P F D |
| Rat D5: | E V G E E E E E G P F D |
| Mouse D5: | E V G E E E E A E E E G P F D |
| Dog D5: | E V D K Q EE S P F D |
Previously using the GPCR-nls strategy we identified discrete cytoplasmic regions in the mu and delta opioid receptors required for oligomer formation. In the carboxyl tail of the delta receptor we identified three glycine residues (-GGG), substitution of any of these residues prevented heteromer formation. In ic3 of both mu and delta receptors we identified three residues (-SVR), substitution of any of these residues prevented heteromer formation.
Thus data from our studies of four heteromer families, dopamine heteromers D5-D2, D1-D2, D1-D5, and mu-delta heteromers, indicate there is not a common mechanism for heteromer formation within Family A GPCRs. Receptor heteromerization arose in the cell to increase complexity, to develop and expand the utility and versatile range of functions of each individual GPCR, thus ways of interacting with other distantly related GPCRs as heteromers were opportunistically adapted. Whereas with closely related GPCRs such as D1 or D5, or mu and delta opioid receptors, these receptors maintained the formations used by homooligomer pairs, in forming heteromers.
In summary, we elucidated precise aspects of the cytoplasmic structure of D5-D2 receptor heteromers. By changing single amino acids in the D5 receptor c-tail we succeeded is disrupting the D5 receptor ability to form heteromers. We can now prepare D5 and D2 receptor expressing cells incapable of forming heteromers. Thus our work on the dopamine and opioid receptors is revealing the nature of the interactions involved in heteromers in the Family A GPCRs.
3 adjacent c-tail D5 receptor glutamic acids form heteromers with the D2 receptor
Adjacent arginines in ic3 of the D2 receptor form heteromers with the D5 receptor
Single amino acid changes in the D5 receptor c-tail disrupt the D5-D2 heteromers
Acknowledgements
This work was partially supported by a Proof of Principle Grant from the Canadian Institutes for Health Research and National Institute on Drug Abuse Grant (DA007223). S.R.G. holds a Canada Research Chair in Molecular Neuroscience.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].George SR, O’Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat. Rev. Drug Discovery. 2002;1:808–820. doi: 10.1038/nrd913. [DOI] [PubMed] [Google Scholar]
- [2].Milligan G. G-protein-coupled receptor heterodimeraztion: contribution to pharmacology and function. Br. J. Pharmacol. 2009;158:5–14. doi: 10.1111/j.1476-5381.2009.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ciruela F, Vallano A, Arnau JM, Sánchez S, Borroto-Escuela DO, Agnati LF, Fuxe K, Fernández-Dueñas V. G protein-coupled receptor oligomerization for what? J. Recept. Signal Transduct. Res. 2010;30:322–330. doi: 10.3109/10799893.2010.508166. [DOI] [PubMed] [Google Scholar]
- [4].Hasbi A, Fan T, Alijaniaram M, Nguyen T, Perreault ML, O’Dowd BF, George SR. Calcium signaling cascade links dopamine D1-D2 receptor heteromer to striatal BDNF production and neuronal growth. Proc. Natl. Acad. Sci. U S A. 2009;106:21377–21382. doi: 10.1073/pnas.0903676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Perreault ML, Hasbi A, Alijaniaram M, Fan T, Varghese G, Fletcher PJ, Seeman P, O’Dowd BF, George SR. The dopamine D1-D2 receptor heteromer localizes in dynorphin/enkephalin neurons: increased high affinity state following amphetamine and in schizophrenia. J. Biol. Chem. 2010;285:36625–36634. doi: 10.1074/jbc.M110.159954. P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].O’Dowd BF, Ji X, Alijaniaram M, Nguyen T, George SR. Separation and reformation of cell surface dopamine receptor oligomers visualized in cells. Eur. J. Pharmacol. 2011;658:74–83. doi: 10.1016/j.ejphar.2011.02.030. [DOI] [PubMed] [Google Scholar]
- [7].O’Dowd BF, Ji X, Alijaniaram M, Rajaram RD, Kong MM, Rashid A, Nguyen T, George SR. Dopamine receptor oligomerization visualized in living cells. J. Biol. Chem. 2005;280:37225–37235. doi: 10.1074/jbc.M504562200. [DOI] [PubMed] [Google Scholar]
- [8].O’Dowd BF, Ji X, Nguyen T, George SR. Two amino acids in each of D1 and D2 dopamine receptor cytoplasmic regions are involved in D1-D2 heteromer formation. Biochem Biophys. Res. Commun. 2012;417:23–8. doi: 10.1016/j.bbrc.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].O’Dowd BF, Ji X, O’Dowd PB, Nguyen T, George SR. Disruption of the mu-delta opioid receptor heteromer. Biochem. Biophys. Res. Commun. 2012;422:556–60. doi: 10.1016/j.bbrc.2012.05.023. [DOI] [PubMed] [Google Scholar]
- [10].So CH, Verma V, Alijaniaram M, Cheng R, Rashid AJ, O’Dowd BF, George SR. Calcium signaling by dopamine D5 receptor and D5-D2 receptor hetero-oligomers occurs by a mechanism distinct from that for dopamine D1-D2 receptor hetero-oligomers. Mol. Pharmacol. 2009;75:843–54. doi: 10.1124/mol.108.051805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].O’Dowd BF, Alijaniaram M, Ji X, Nguyen T, Eglen RM, George SR. Using ligand–induced conformational change to screen for compounds targeting G-protein-coupled receptors. J. Biomol. Screen. 2007;12:175–85. doi: 10.1177/1087057106298287. [DOI] [PubMed] [Google Scholar]