SUMMARY
Translation factor eIF5A, containing the unique amino acid hypusine, was originally shown to stimulate methionyl-puromycin synthesis, a model assay for peptide bond formation. More recently, eIF5A was shown to promote translation elongation; however, its precise requirement in protein synthesis has remained elusive. Here we use in vivo assays in yeast and in vitro reconstituted translation assays to reveal a specific requirement for eIF5A to promote peptide-bond formation between consecutive proline residues. Addition of eIF5A relieves ribosomal stalling during translation of three consecutive proline residues in vitro, and loss of eIF5A function impairs translation of polyproline-containing proteins in vivo. Hydroxyl radical probing experiments localized eIF5A near the E site of the ribosome with its hypusine residue adjacent to the acceptor stem of the P-site tRNA. Thus, eIF5A, like its bacterial ortholog EFP, is proposed to stimulate the peptidyl-transferase activity of the ribosome and facilitate the reactivity of poor substrates like proline.
INTRODUCTION
Ribosomes catalyze protein synthesis with the assistance of tRNAs and translation factors. There are three tRNA binding sites on the ribosome: a centrally located peptidyl-tRNA binding P site, an aminoacyl-tRNA binding A site, and an exit E site that binds deacylated tRNA following its transfer from the P site. Translation initiation factors aid in the assembly of an 80S ribosome in eukaryotes in which the initiator methionyl-tRNA (Met-tRNAiMet) is bound in the P site base-paired to the start codon on the mRNA. To extend the polypeptide, the eukaryotic elongation factor eEF1A (EFTu in bacteria) delivers aminoacyl-tRNA to the A site in a codon-dependent manner. Following accommodation of the tRNA, the amino acid attached to the A-site tRNA is juxtaposed to the peptidyl portion of the P-site tRNA in the active site (peptidyl transferase center [PTC]) of the large (60S) ribosomal subunit. Peptide bond formation links the extended polypeptide to the A-site tRNA, leaving a deacylated tRNA in the P site. Next, elongation factor eEF2 (EFG in bacteria) promotes translocation of the tRNAs and mRNA such that the A site is vacant and ready to accept the next aminoacyl-tRNA (reviewed in Schmeing and Ramakrishnan, 2009). A common misconception is that the ribosome is a monolithic machine that catalyzes all peptide bonds at equivalent rates regardless of the amino acid. In fact, certain residues including the imino acid proline are poor substrates for peptide bond formation (Wohlgemuth et al., 2008; Pavlov et al., 2009). Recently, it was shown that the translation elongation factor EFP is essential for translation of polyproline sequences by bacterial ribosomes (Doerfel et al., 2013; Ude et al., 2013); however, it is currently unclear how eukaryotic ribosomes manage to synthesize peptide bonds with poor substrates.
In addition to the canonical elongation factors eEF1A and eEF2, eIF5A has also been linked to translation elongation. eIF5A was initially discovered and characterized based on its ability to stimulate methionyl-puromycin synthesis (Kemper et al., 1976; Schreier et al., 1977; Benne and Hershey, 1978), a reaction analogous to the synthesis of the first peptide bond. The aminoacyl analog puromycin reacts with Met-tRNAiMet bound in the P site of 80S initiation complexes. Based on its ability to stimulate methionyl-puromycin synthesis, eIF5A was initially thought to function as a translation initiation factor, stimulating formation or reactivity of 80S initiation complexes. However, since the assay involves formation of a peptide bond between puromycin and methionyl-tRNA, puromycin reactivity also reports on the peptidyl transferase activity of the ribosome, a key component of translation elongation. Depletion of eIF5A in vivo or inactivation of a temperature-sensitive mutant of yeast eIF5A impaired translation elongation and stabilized polysomes in the absence of cycloheximide (Saini et al., 2009) and increased the average ribosomal transit time in vivo (Gregio et al., 2009; Saini et al., 2009). Moreover, addition of eIF5A resulted in a two-fold stimulation in the rate of tripeptide synthesis using a reconstituted yeast in vitro translation system. Taken together, these data revealed a role for eIF5A in translation elongation. However, it is difficult to rationalize the essential requirement for eIF5A in yeast with the modest two-fold stimulation of tripeptide synthesis, suggesting that eIF5A may have a more specialized and critical requirement in translation elongation.
eIF5A is of particular interest because it is the only protein that contains the modified amino acid hypusine and because eIF5A and hypusine have been linked to tumorigenesis and cancer (Silvera et al., 2010; Scuoppo et al., 2012). The hypsuine modification is present in all archaea and eukaryotes that have been examined, and it is formed by the transfer of an n-butylamine moiety from spermidine to the ε-amino group of a specific lysine side chain (Lys51 in yeast eIF5A), followed by addition of a hydroxyl group. The hypusine modification is essential for eIF5A function: deoxyhypusine synthase, which catalyzes the first step in hypusine formation, is essential for yeast viability and derivatives of eIF5A lacking hypusine fail to stimulate in vitro methionyl-puromycin (Park, 1989; Park et al., 1991) and tripeptide synthesis (Saini et al., 2009). Interestingly, bacterial EFP and eIF5A are orthologs, and in some bacteria a lysine side chain in EFP, corresponding to the site of hypusine modification in eIF5A, is post-translationally modified by the addition of a β-lysine residue (Navarre et al., 2010; Yanagisawa et al., 2010; Roy et al., 2011; Peil et al., 2012). Like eIF5A, EFP was found to stimulate methionyl-puromycin synthesis, and this activity was dependent on the β-lysine modification (Park et al., 2012). Earlier studies revealed that the impact of EFP on dipeptide synthesis varied for different aminoacyl analogs (Glick et al., 1979; Ganoza and Aoki, 2000), suggesting that EFP, and by extension eIF5A, may facilitate the reactivity of certain amino acids in peptide bond synthesis. Consistent with these findings, recent reports showed that EFP enhances the synthesis of proteins containing stretches of consecutive proline residues (Doerfel et al., 2013; Ude et al., 2013).
RESULTS
eIF5A Stimulates Translation through Polyproline Sequences in vivo
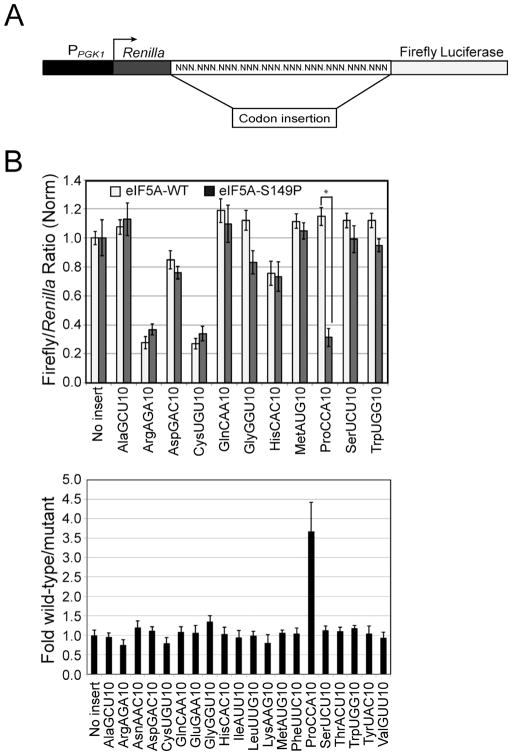
To further define the role of eIF5A in translation elongation and to determine whether eIF5A, like EFP, stimulates translation of specific amino acid motifs, we monitored the expression of a set of dual-luciferase reporters in isogenic yeast strains expressing wild-type eIF5A or the temperature-sensitive eIF5A-S149P mutant (Zuk and Jacobson, 1998; Saini et al., 2009). The dual-luciferase reporters, developed by Beth Grayhack and colleagues to examine codon bias in translation (Letzring et al., 2010), express a single mRNA in which the 5′ Renilla luciferase and 3′ firefly luciferase open reading frames (ORFs) are joined in-frame by sequences encoding repeats of 10 identical codons for each of the 20 amino acids (Fig. 1A). For the initial analysis, the inserted sequences repeated the optimal codon for each amino acid (Letzring et al., 2010). As shown in Fig. 1B (upper panel) and as previously observed (Letzring et al., 2010), the ratio of firefly to Renilla luciferase activity varied depending on the repeated codon. While the ratios for most constructs were similar to the no insert control, low ratios were observed for the ArgAGA and CysUGU reporters (Fig. 1B, upper panel); whereas, high ratios were observed with GluGAA and PheUUC codon insertions (see Fig. S1D). These eIF5A-independent effects might reflect codon or aminoacyl-tRNA abundance or impacts of the inserted amino acids on luciferase activity in the bifunctional Renilla-firefly luciferase fusion protein.
Figure 1. eIF5A Stimulates Translation of Polyproline Motifs in vivo.
(A) Schematic of Renilla-firefly luciferase reporter construct. Codon repeats were inserted in-frame between the Renilla and firefly luciferase ORFs (Letzring et al., 2010).
(B) Dual luciferase reporter constructs containing 10 repeats of the indicated codon were introduced into isogenic yeast strains expressing wild type eIF5A or temperature-sensitive eIF5A-S149P. (Top panel) Following growth at semi-permissive 33°C, the firefly-to-Renilla luciferase ratio for each construct was normalized to the ratio obtained from reporters with no insert between the ORFs. (Bottom panel) The fold difference in luciferase ratios between cells expressing wild-type eIF5A and eIF5A-S149P was quantitated and then normalized to the values obtained from the no insert control. *Statistical significance for ProCCA(10) was measured by student’s t-test with a p-value <0.05. Error bars were calculated as propagated standard deviations (SD) for three independent transformants.
If eIF5A stimulates the translation of specific amino acids, then the ratio of firefly to Renilla luciferase activity is expected to decrease when these reporters are analyzed in the strain containing eIF5A-S149P when grown at the semi-permissive temperature (33°C). As shown in Fig. S1A, the slow-growth phenotype of the eIF5A-S149P mutant at 30°C is exacerbated at 33°C, and the mutant strain fails to grow at 37°C. The impaired growth at 33°C is marked by reduced levels of eIF5A (Fig. S1B) and by retention of polysomes in the absence of cycloheximide (Fig. S1C), indicative of a general translation elongation defect in the strain. Analysis of all 20 luciferase reporter constructs revealed that only the Pro codon insertions revealed a strong dependence on eIF5A (Fig. 1B, upper panel). For the ProCCA reporter, the ratio of firefly to Renilla luciferase in the strain expressing wild-type eIF5A was ~3.7-fold greater than the ratio observed in the strain expressing eIF5A-S149P (Fig. 1B, lower panel), whereas this normalized ratio ranged from 0.75 (ArgAGA) to 1.35 (GlyGGU) for reporters containing any of the other 19 codon insertions.
To test whether the impaired expression of firefly luciferase from the construct containing the ProCCA codon repeats was specific to mutation of eIF5A, two other translation elongation factors were evaluated. No significant differences in firefly to Renilla luciferase ratios were observed when constructs containing proline or alanine codon insertions were examined in strains expressing temperature-sensitive mutants of translation elongation factors eEF2 or eEF3 (Fig. S2A–B). Thus, polyproline peptide bond-formation shows a unique dependence on eIF5A. Alternatively, this result could reflect a specific requirement for eIF5A to promote peptide bond formation by Pro-tRNA. Consistent with this hypothesis, reporters containing 10 repeats of the Pro codons CCA, CCG or CCU displayed a strong requirement for eIF5A, whereas no Ala codon insertions conferred a dependence on eIF5A (Fig. S2C). While these data are not definitive, they suggest that the amino acid proline rather than the tRNA likely determine the requirement for eIF5A.
To define the number of consecutive proline residues needed to impose a requirement for eIF5A, the dual-luciferase reporters were modified to contain one, two, three, four, six, eight or ten consecutive ProCCA or PheUUC codons. As shown in Fig. 2, luciferase ratios for the Phe codon insertion constructs were the same in the wild type and eIF5A-S149P mutant strains (fold wild-type/mutant = ~1.0). Likewise, insertion of one or two proline codons did not significantly impact luciferase ratios in the eIF5A mutant strain compared to the wild-type control. In contrast, insertion of four proline codons resulted in reduction of the luciferase ratio in the eIF5A-S149P mutant strain. (Some reduction may be evident with insertion of three Pro codons as well). Insertion of six, eight or ten proline codons further exacerbated the defect, and the normalized ratio of firefly to Renilla luciferase in the strain expressing wild type eIF5A was ~3–4.5-fold greater than the ratio observed in cells expressing eIF5A-S149P. These results indicate that at least four (or perhaps three) consecutive proline codons are needed to impose an eIF5A-dependency on protein synthesis.
Figure 2. Translation of Three or More Consecutive Proline Codons Reveals eIF5A Dependency.
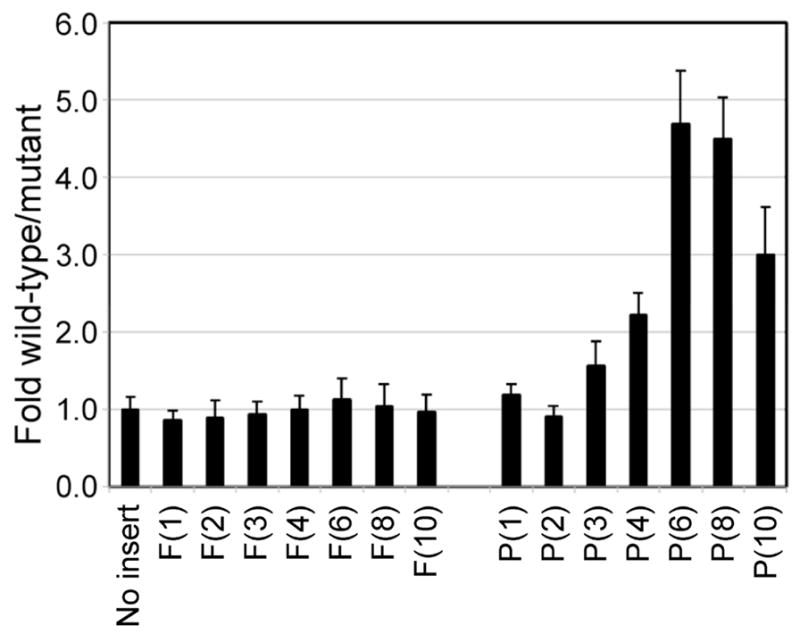
Dual luciferase reporters containing 1, 2, 3, 4, 6, 8, or 10 consecutive PheUUC (F) or ProCCA (P) codons were assayed in wild type or eIF5A-S149P mutant strains and the fold differences in luciferase ratios were quantitated and normalized to the no insert control as described in Fig. 1 (error bars = propagated SD).
Expression of Yeast Polyproline-containing Proteins Requires eIF5A in vivo
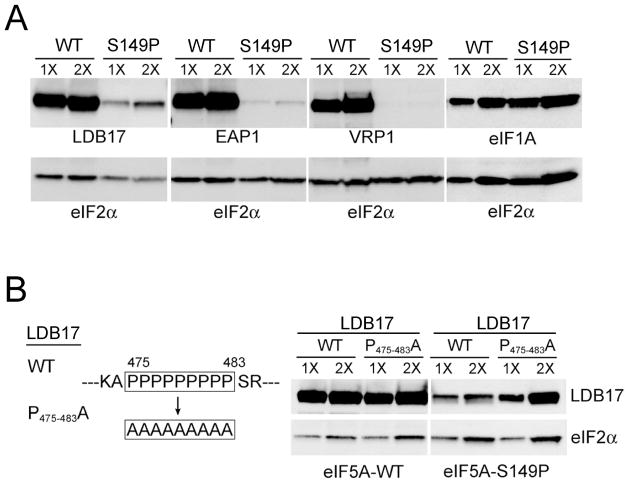
Analysis of the Saccharomyces cerevisiae genome identified 549 proteins (out of 5886 open reading frames) that contain polyproline motifs with at least three consecutive proline residues. To test whether expression of yeast proteins containing polyproline motifs is dependent on eIF5A, selected plasmids from the Yeast ORF Collection (Open Biosystems) were introduced into isogenic strains expressing wild type eIF5A or the temperature-sensitive mutant eIF5A-S149P. Transformants were grown at the semi-permissive temperature of 33°C to partially inactivate eIF5A-S149P and in galactose medium to induce the GAL1 promoter used to drive ORF expression. Protein expression was monitored by Western analysis using antibodies to detect the HA-tag incorporated at the C-terminus of each ORF (Gelperin et al., 2005). As shown in Figure 3A, expression of Ldb17 (9 Pro motif), Eap1 (two 6 Pro, one 3 Pro motif), and Vrp1 (multiple poly-Pro sequences including one 9 Pro, one 8 Pro, one 6 Pro, four 5 Pro, three 4 Pro, and two 3 Pro motifs) were dramatically reduced in the eIF5A mutant strain relative to the wild- type eIF5A strain and the loading control eIF2α (no poly-Pro motifs). Stable expression of Tif11 (eIF1A, no poly-Pro motifs) from a Yeast ORF Collection plasmid in the wild type and mutant eIF5A strains indicates that the eIF5A-sensitive expression of the poly-Pro proteins is not due to impacts on the expression system (e.g. the GAL1 promoter, growth at 33°C) (Fig. 3A). In addition, substituting Ala in place of the nine Pro residues in the C-terminal 9 Pro motif restored Ldb17 expression in the eIF5A-S149P mutant (Fig. 3B), directly linking eIF5A function to the synthesis of poly-Pro motifs.
Figure 3. Expression of Polyproline-containing Proteins Requires eIF5A in vivo.
(A) Plasmids expressing HA-tagged forms of the yeast proteins LDB17, EAP1, VRP1 or eIF1A under the control of the yeast GAL1 promoter were introduced into isogenic strains expressing wild-type eIF5A or eIF5A-S149P. Cells were grown at semi-permissive 33°C in galactose medium, broken with glass beads in the presence of 10% trichloroacetic acid (TCA), and two different amounts of each extract differing by a factor of 2 were loaded in successive lanes and subject to immunoblot analysis using monoclonal anti-HA or polyclonal anti-yeast eIF2α antiserum.
(B) The experiment in panel A was repeated using an LDB17 construct in which Ala codons were substituted for the nine Pro codons in the polyproline motif.
eIF5A Plays an Essential Role in Polyproline Peptide Synthesis
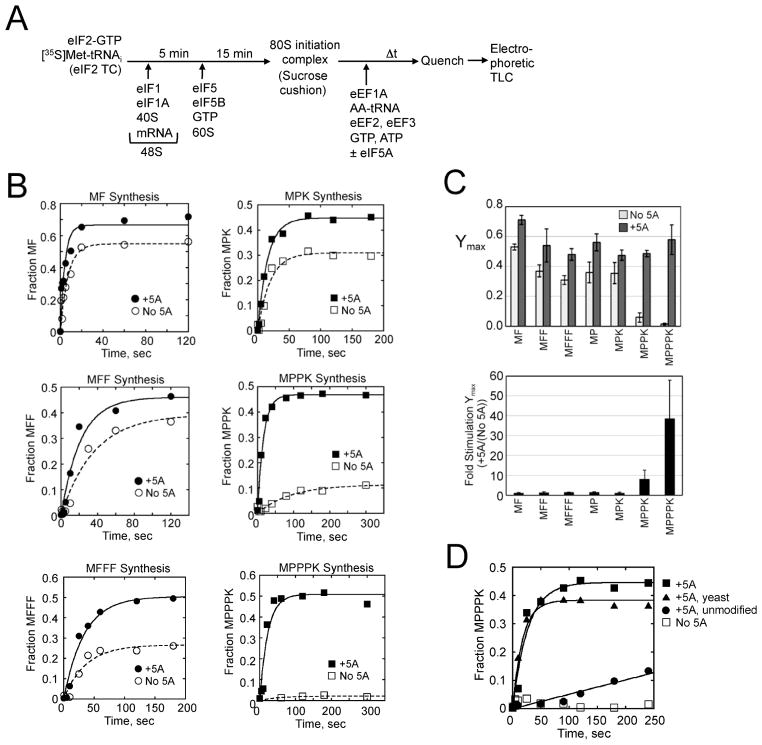
An in vitro reconstituted yeast translation assay was used to directly examine the eIF5A requirement for polyproline synthesis. As shown in Figure 4A, minimal translation initiation (48S) complexes encoding polyproline or polyphenylalanine were assembled using unstructured model mRNAs to avoid the requirement for initiation factors that function in mRNA recruitment. Following ribosomal subunit joining and assembly of an 80S initiation complex with [35S]Met-tRNAiMet in the P site of the ribosome, the complex was pelleted through a sucrose cushion to remove initiation factors and unbound Met- tRNAiMet. Next, elongation factors and the necessary aminoacyl-tRNAs were added to the purified 80S complexes in the absence or presence of excess recombinant eIF5A. The recombinant eIF5A was prepared from E. coli that co-expresses the hypusine formation enzymes (Fig. S3A), and the presence of hypusine in the recombinant eIF5A was confirmed by Electrospray Ionization (ESI) QTOF MS analysis (Figure S3B, see Experimental Procedures). Peptide synthesis was monitored by electrophoretic thin-layer chromatography (TLC) (Youngman et al., 2004; Eyler and Green, 2011).
Figure 4. eIF5A Stimulates Synthesis of Polyproline Peptides.
(A) Scheme for in vitro reconstituted translation elongation assay.
(B) Fractions of MF, MFF, MFFF (left column) or MPK, MPPK, and MPPPK (right) synthesis in elongation assays (Fig. S3D–E) performed in the absence (open symbols) or presence of eIF5A (closed symbols) were plotted and fit to a single exponential equation.
(C) Summary of maximum fractions of peptide synthesis (Ymax, top) and fold stimulation of Ymax by adding eIF5A (bottom) calculated from the data in panel B. Error bars are (upper) SD from at least three independent experiments and (lower) calculated propagated SD.
(D) Effect of eIF5A hypusine modification on peptide synthesis. Fraction of MPPPK synthesis (Fig. S3F) in reactions lacking eIF5A, containing unmodified eIF5A (no hypusine), or containing hypusinated eIF5A prepared from E. coli (+5A, see Experimental Procedures) or purified from yeast (+5A, yeast) was plotted and fit to a single exponential equation.
Synthesis of Met-Phe (MF), MFF, and MFFF peptides progressed well in the absence and presence of eIF5A (Figs. 4B and S3D–E) with less than two-fold stimulation in both the observed rate constant and the fraction of maximal yield (Ymax) for formation of the peptides in the presence of eIF5A (Fig. 4C). These results are consistent with the previously reported ~2-fold stimulation of MFF synthesis upon adding eIF5A to the reconstituted system (Saini et al., 2009). In preliminary experiments, the proline-containing peptides Met-Pro (MP), MPP and MPPP failed to resolve as discrete spots on the TLC (see Figs. S3C, E, and F). However, incorporation of a lysine residue at the C-terminus of the proline peptides enabled resolution of the peptides by TLC and facilitated their quantitation. The fraction of maximum peptide yield for Met-Pro-Lys (MPK) peptide synthesis was stimulated ~1.3-fold by adding eIF5A (Ymax = 0.36 in the absence of eIF5A and 0.48 in the presence of eIF5A) (Figs. 4B–C, and S3E [top panels]). Thus the presence of a single Pro residue conferred a modest eIF5A dependency for peptide synthesis. In contrast, synthesis of the MPPK peptide containing two prolines was significantly impaired in the absence of eIF5A (Ymax = 0.06 ±0.03). An 8.3-fold stimulation of Ymax was observed upon adding eIF5A (Ymax = 0.49 ±0.02) (Figs. 4B- C, and S3E). The large difference in reaction endpoints for the proline-containing peptides in the presence versus the absence of eIF5A suggests that competing reactions are likely occurring (e.g. peptidyl-tRNA drop-off). Since the observed rates reflect both peptide bond formation and these competing reactions, we have limited our analysis to the reaction endpoint differences and not observed rates. Remarkably, no detectable formation of the MPPPK peptide containing three consecutive prolines occurred in the absence of eIF5A during the time course of experiments. The addition of eIF5A efficiently restored MPPPK synthesis stimulating the Ymax at least 39-fold (Ymax = 0.58 ±0.1) (Fig. 4C). Thus, consistent with the results of the in vivo assays, eIF5A is required for synthesis of peptides containing consecutive Pro residues.
To assess the importance of the hypusine modification on eIF5A, MPPPK synthesis was analyzed using different forms of the factor. As shown in Figs. 4D and S3F, no MPPPK synthesis was detected in the absence of eIF5A and very little synthesis was detected in assays that included unmodified eIF5A prepared from E. coli (see Experimental Procedures). In contrast, hypusine-modified eIF5A, prepared either from yeast or from E. coli co-expressing the hypusine modification enzymes (see Experimental Procedures), readily stimulated MPPPK synthesis (Ymax = 0.38 ±0.02 for yeast eIF5A; and Ymax = 0.45 ±0.02 for recombinant eIF5A). Thus, the hypusine modification of eIF5A is necessary for efficient polyproline synthesis in vitro.
eIF5A Prevents Ribosome Stalling on Consecutive Proline Codons
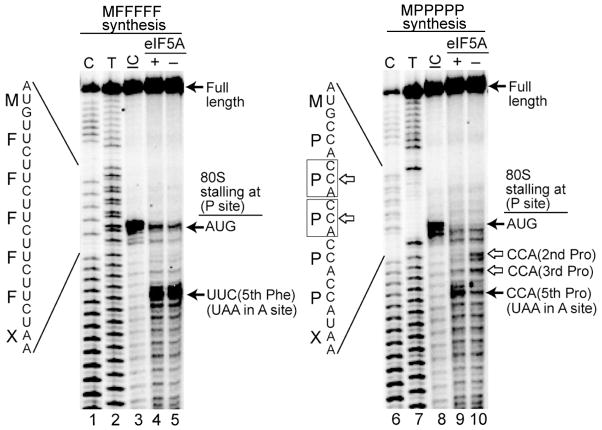
The in vitro peptide synthesis assays revealed defects in synthesizing MPPK and MPPPK, suggesting that ribosomes stall when translating polyproline sequences. To directly detect ribosome stalling, toeprinting assays were performed to determine the position of the ribosome on mRNAs encoding MPPPPP and MFFFFF in the reconstituted in vitro translation system described above. A [32P]-labeled primer was annealed to the 3′ end of the mRNA and extended by reverse transcription. The ribosome blocks reverse transcriptase 15–16 nucleotides downstream of the first nucleotide of the P-site codon. As shown in Fig. 5 (lanes 3 and 8), toeprinting confirms that the AUG start codon is positioned in the P site in 80S initiation complexes. When elongation factors and Phe-tRNAPhe were added to the ribosomal complexes translating the MFFFFF mRNA, this toeprint was diminished and a new toeprint was observed, corresponding to the final Phe codon in the P site (Fig. 5, lanes 4–5). The position of this toeprint is consistent with the ribosome translating to the end of the ORF and arresting with the stop codon in the A site. (No release factors were included in the translation reaction). Consistent with the robust synthesis of the MFFF peptide in the presence and absence of eIF5A (Figs. 4B–C and S3D), production of the MFFFFF complex in the toeprinting assays was unaffected by the presence or absence of eIF5A (Fig. 5, lanes 4–5).
Figure 5. eIF5A Prevents Ribosome Stalling on Consecutive Proline Codons.
Reconstituted peptide synthesis assays were performed in the absence or presence of eIF5A using mRNAs encoding the peptides MFFFFF (left panel) or MPPPPP (right panel). The position of the 80S ribosome was determined by reverse transcription of the mRNA template using a [32P]-labeled primer, and C and T sequencing reactions were run alongside. Reactions lacking elongation factors were performed to identify 80S initiation complexes (IC) on the AUG codon (lanes 3 and 8). The identity of the 80S toeprint signals is indicated on the right; and the sequences of the mRNA and the corresponding amino acids are shown on the left with the sites of ribosome stalls at the 2nd and 3rd proline codons boxed.
Toeprinting analysis of elongation complexes revealed ribosomal stalling on the MPPPPP mRNA. In reactions lacking eIF5A (Fig. 5, lane 10), toeprints were observed corresponding to the ribosome stalling with the second or third Pro codon in the P site. Importantly, addition of eIF5A diminished the abundance of these stalled complexes and increased the yield of ribosome complexes with the final Pro codon in the P site and the stop codon in the A site (Fig. 5, lane 9). These results indicate that in the absence of eIF5A the P-site tRNA is linked to an MPP or MPPP peptide and that a Pro codon, and presumably Pro-tRNA, are in the A site. The positions of the stalled ribosome complexes are consistent with the in vivo assays (Fig. 2) that showed that at least three consecutive proline residues are necessary to observe an eIF5A-dependency, and indicate that peptide bond formation between diprolyl-tRNA and Pro-tRNA is particularly dependent on eIF5A. Moreover, these results are consistent with reports of E. coli ribosomes stalling on tri-proline motifs with the peptidyl-tRNA linked to consecutive C-terminal prolines and the third Pro codon in the A site of the ribosome (Tanner et al., 2009; Woolstenhulme et al., 2013).
eIF5A Binds Near the E and P sites of the 80S Ribosome
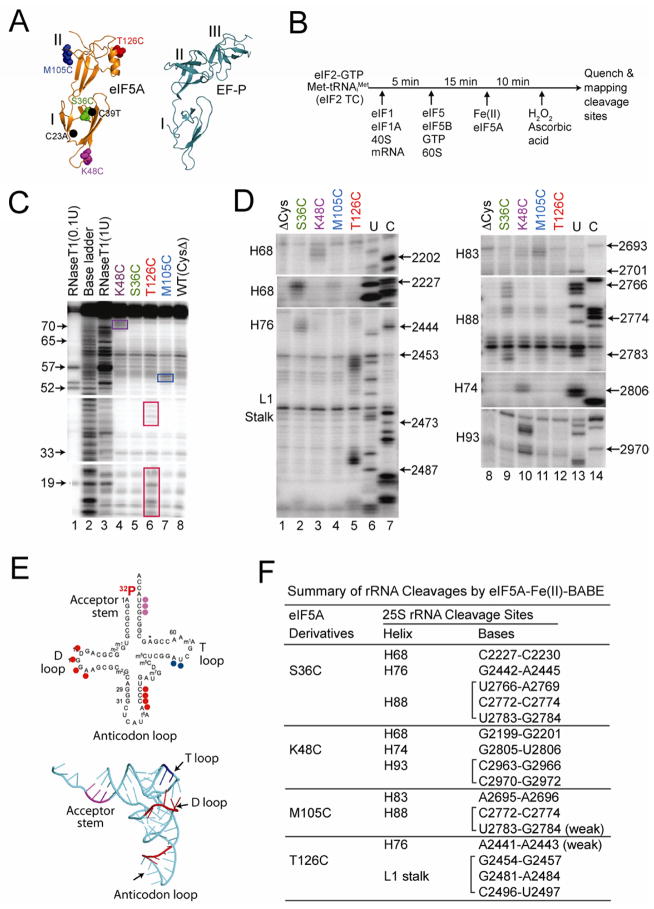
Directed hydroxyl radical mapping was used to identify the binding site for eIF5A on the yeast 80S ribosome. It is noteworthy that EFP, which contains an extra C-terminal domain not found in eIF5A, binds between the P and E sites and contacts both the large and small subunits of the bacterial 70S ribosome (Blaha et al., 2009). A cysteine-less derivative of yeast eIF5A was generated by mutating the native Cys23 and Cys39 residues to Ala and Thr, respectively. Yeast expressing the eIF5A-C23A, C39T mutant (eIF5A-ΔC) as the sole source of eIF5A grew as well as cells expressing the wild type protein (Fig. S4A), indicating that the mutations do not affect eIF5A function. Next, single Cys residues were introduced at four surface-exposed sites generating eIF5A-ΔC-S36C, eIF5A-ΔC-K48C, eIF5A-ΔC-M105C, and eIF5A-ΔC-T126C (Fig. 6A). All four mutant proteins supported yeast cell growth at wild type rates (Fig. S4A), suggesting that the mutations did not interfere with the essential hypusine modification of eIF5A or with the function of the protein on the ribosome. The eIF5A-ΔC and four single Cys mutant proteins were purified from yeast, derivatized with Fe(II)-1-(p-bromoacetamidobenzyl)- EDTA [Fe(II)-BABE], which links the ferrous iron to the Cys residue, and then added to assembled 80S complexes containing 5′ end-labeled Met-[32P]tRNAiMet in the P site (see Fig. 6B).
Figure 6. Directed Hydroxyl Radical Probing of eIF5A Binding to 80S Ribosomal Complexes.
(A) Ribbon representation of T. thermophilus EFP (right panel, pdb 3HUW, (Blaha et al., 2009)) and yeast eIF5A (left panel, pdb 3ER0) showing the protein domains (Roman numerals), the positions of the C23A and C39T mutations (black dots) that removed the native Cys residues in eIF5A, and the sites (Spheres representation) of Cys mutations for tethering Fe(II): Ser36 (green), Lys48 (magenta), Met105 (blue) and Thr126 (red).
(B) Scheme for directed hydroxyl radical cleavage by Fe(II)-BABE modified forms of eIF5A in 80S complexes.
(C) Directed hydroxyl radical cleavage of Met-[32P]tRNAiMet by Fe(II)-BABE- derivatized eIF5A in 80S complexes. Cleavage products were resolved on 10% (w/v) denatured polyacrylamide gels, and cleavage sites on [32P]tRNAiMet were determined by comparison to samples containing eIF5A-ΔC [WT(CysΔ), lane 8]. The tRNA ladders were prepared by digesting Met-[32P]tRNAiMet with RNase T1 (cleaves 3′ of G residue) or by base cleavage (lane 2). The tRNA residue numbers are shown at the left, and cleavage fragments are boxed.
(D) Primer extension analysis of 25S rRNA cleavage fragments produced by Fe(II)-tethered to the indicated positions in eIF5A. U, C: 25S rRNA sequencing reactions using reverse transcriptase and dideoxynucleotides ddATP and ddGTP, respectively. 25S rRNA helices and the position of the L1 stalk are indicated on the left.
(E) Sites of eIF5A-Fe(II)-BABE cleavages are shown on the secondary (left) and three- dimensional (pdb 1YFG, (Basavappa and Sigler, 1991)) structures of tRNAiMet. Cleavage sites are color-coded according to the site where Fe(II) was tethered on eIF5A (see panel A).
(F) Summary of 25S rRNA cleavages by eIF5A-Fe(II)-BABE derivatives.
Cleavage of Met-[32P]tRNAiMet by hydroxyl radicals generated in the vicinity of the ferrous iron by the Fenton reaction was monitored by 10% denaturing gel electrophoresis. Compared to reactions employing eIF5A-ΔC, which due to the absence of Cys residues is not modified by Fe(II)-BABE, hydroxyl radicals generated by Fe(II)-BABE tethered to eIF5A-ΔC-K48C yielded cleavages in the tRNAiMet acceptor stem bases G70-U72 (Fig. 6C, compare lanes 4 and 8, and Fig. 6E). As this K48C site of Fe(II)-BABE modification is located only three residues from the site of hypusine modification (K51) on eIF5A, these results place the hypusine side chain in the vicinity of the amino acid attached to the 3′-CCA-end of the P-site tRNA. In contrast, hydroxyl radicals generated using Fe(II)-modified eIF5A-ΔC-M105C cleaved bases A54 and U55 in the T stem region of Met-tRNAiMet (Fig. 6C, lane 7, and Fig. 6E), while hydroxyl radicals generated using Fe(II)-modified eIF5A-ΔC-T126C cleaved two different regions in tRNAiMet: bases U16-A20 in the D stem loop region, and bases A38-C41 in anticodon stem region (Fig. 6C, lane 6, and Fig. 6E). No noticeable tRNAiMet cleavages were observed using the iron-modified form of eIF5A-ΔC-S36C (Fig. 6C, lane 5). From these data, we conclude that the eIF5A binds alongside the P-site tRNA on the 80S ribosome, a position similar to the EFP binding site on the bacterial 70S ribosome (Blaha et al., 2009), with the hypusine residue at the top of eIF5A near the aminoacyl end of the tRNA and eIF5A domain II residue Thr126 near the anticodon stem of the tRNA.
In order to further define the eIF5A-binding site on the ribosome, hydroxyl radicals were generated in 80S complexes containing Fe(II)-BABE-modified forms of eIF5A and rRNA cleavages were analyzed by primer extension using [32P]-labeled primers. Whereas Fe(II)-modified eIF5A-ΔC-S36C did not generate cleavages in tRNAiMet, this eIF5A derivative yielded cleavages in helices H68, H76 and H88 of 25S rRNA that were not seen with the eIF5A-ΔC control (Figure 6D, compare lanes 1 and 2, and 8 and 9; see summaries of cleavage sites in Figs. 6F and S4C). These cleavage sites are clustered around the E-site tRNA binding region of the 60S subunit (Fig. 7A, green color). Primer extension analysis of rRNA cleavages generated using Fe(II)-BABE- tethered eIF5A-ΔC-K48C revealed enhanced cleavages in helices H68, H74 and H93 (Fig. 6D, lanes 3 and 10, Fig. 6F, and Fig. S4C). Noticeably, these cleavage sites map near the peptidyl transferase center (PTC) of the 60S subunit (Fig. 7A, magenta color), consistent with the idea that the hypusine residue (modified Lys 51 side chain) is close to the PTC active site of the ribosome.
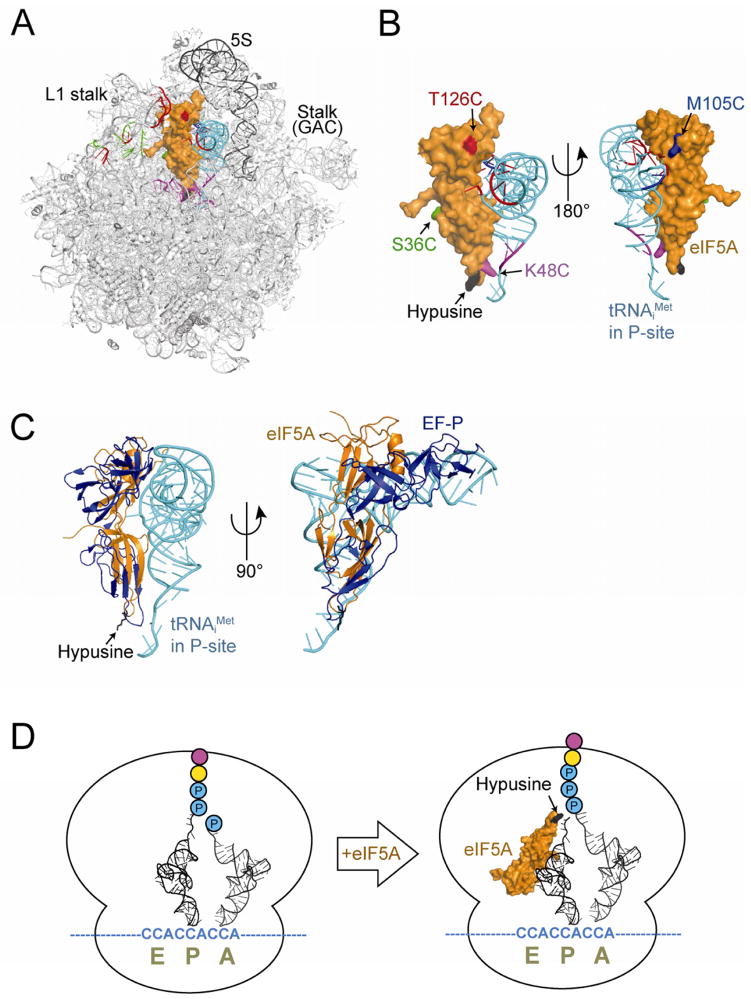
Figure 7. Models of 60S–Met-tRNAiMet–eIF5A Complex and eIF5A Stimulating Polyproline Synthesis.
(A) Docking model of a surface representation of yeast eIF5A (orange, pdb 3ER0) and ribbons representation of tRNAiMet (cyan, pdb 1YFG, (Basavappa and Sigler, 1991)) on the ribbons structure of the yeast 60S ribosome (pdb 3O58, (Ben-Shem et al., 2010)) as viewed from the subunit interface. The position of tRNAiMet was modeled by alignment with P-site tRNA on the bacterial ribosome (pdb 2J00, (Selmer et al., 2006)), and eIF5A was docked on the 60S subunit according to the cleavage data for Met-tRNAiMet and 25S rRNA. Cleavage sites in 25S rRNA and tRNAiMet are color-coded according to the sites of Fe(II) attachment on eIF5A (see Fig. 6A). Positions of L1 stalk, 5S rRNA (black), and GTPase activating center (GAC) Stalk on the 60S subunit are indicated.
(B) Magnified view of docked eIF5A and P-site tRNAiMet structure as shown in panel A (left) and rotated 180° (right). Lys51, the site of hypusine modification, is colored black.
(C) Magnified view of docked eIF5A and P-site tRNAiMet (from A) overlaid on the structure of EFP (blue) from the EFP–70S structure (pdb 3HUW, (Blaha et al., 2009)) oriented as shown in panel A (left) and rotated 90° (right).
(D) Model of ribosome stalled on polyproline sequence with di-proline attached to the P-site tRNA and Pro-tRNAPro in the A site (left). (Right) Binding of eIF5A near the E site places the hypusine side chain (Lys51, black) adjacent to the peptidyl-tRNA in the peptidyl transferase center (PTC) of the ribosome where it can help promote peptide bond formation with the amino acid attached to the A-site tRNA.
The rRNA cleavages generated using iron-modified eIF5A-ΔC-M105C mapped to helices H83 and H88 (Fig. 6D, lanes 4 and 11, Fig. 6F). Interestingly, these helix H88 cleavages partially overlap with the cleavages generated by eIF5A-ΔC-S36C (Figs. 6D, F and S4C), consistent with the presentation of these two residues on the same surface of eIF5A. Finally, the Fe(II)-BABE-modified form of eIF5A-ΔC-T126C generated cleavages near the L1 stalk region of the 60S subunit (Fig. 6D, lanes 5 and 12, Fig. 6F, and Fig. S4C).
It is interesting to note that all of the eIF5A generated cleavages map to the 3′ half of 25S rRNA (Fig. S4C) and there are no detectable cleavage sites in 18S rRNA (data not shown). Thus in contrast to EFP, which contacts both the small and large ribosomal subunits (Blaha et al., 2009), eIF5A appears to principally contact the 60S subunit when binding to 80S ribosomal complexes. To generate a model of eIF5A binding to the ribosome, the hydroxyl radical cleavage data was used to orient yeast eIF5A (pdb 3ER0) on the yeast 60S subunit (pdb 3O58, Ben-Shem et al., 2010) with tRNAiMet (pdb 1YFG, Basavappa and Sigler, 1991) bound in the P site. To initiate the docking process the eIF5A residue K48 was positioned near the PTC and centered among its 25S rRNA and tRNAiMet acceptor stem cleavage sites. Next, the residue T126 was positioned between the ribosomal L1 stalk and the D loop and anticodon stem of the P-site tRNAiMet. After fixing the K48 and T126 locations in the model, the eIF5A structure was rotated so that residues S36 and M105 were oriented toward their respective cleavage sites (Fig. 7A). In the 60S–tRNAiMet–eIF5A complex model, eIF5A binds between the P-site tRNAiMet and the E site such that the N-terminal domain of eIF5A (residues 1–82) and the C-terminal domain (residues 87–157) are close to the acceptor stem and D-loop region of tRNAiMet, respectively (Fig. 7B). This eIF5A binding position, which is based on the hydroxyl radical mapping studies presented here, overlaps with the position of EFP domains I and II as found in the co-crystal structure of EFP bound to the bacterial 70S ribosome (Fig. 7C, Blaha et al., 2009). It is noteworthy that domain III of EFP, which is missing from eIF5A (Fig. 6A), contacts the small ribosomal subunit adjacent to the anticodon stem region of fMet-tRNAifMet, which is bound in the P site (Blaha et al., 2009). Thus, despite their structural and functional similarities, the C-terminal truncation of eIF5A relative to EFP apparently limits eIF5A ribosomal contacts to the large subunit and may confer a functional distinction between the two factors.
DISCUSSION
In addition to the universally conserved translation factors, eEF1A/EFTu and eEF2/EFG, three other factors have been implicated in translation elongation. However, these latter factors are not universally conserved. The factor eEF3 is proposed to coordinate E-site tRNA release with eEF1A–aminoacyl-tRNA binding to the A site of the ribosome in some fungi including S. cerevisiae (Triana-Alonso et al., 1995; Anand et al., 2006). Bacterial EF4 (LepA) is proposed to maintain rapid protein synthesis under stress conditions such as high ionic strength and low temperature (Pech et al., 2011), and SelB/eEFsec is an EFTu ortholog required for the delivery of selenocysteinyl-tRNA to the ribosome (Driscoll and Copeland, 2003). In this paper we demonstrate that eIF5A is required for translation of polyproline sequences. As our results concur with the findings of the recent studies on EFP, the bacterial ortholog of eIF5A (Doerfel et al., 2013; Ude et al., 2013), we conclude that eIF5A/EFP is the third universally conserved translation elongation factor.
Our data demonstrating that eIF5A promotes translation of polyproline sequences are consistent with the recent reports on EFP (Doerfel et al., 2013; Ude et al., 2013). Partial inactivation of eIF5A-S149P in yeast (Figs. 1–3) like deletion of the efp gene in E coli (Ude et al., 2013) impaired expression of reporters or native proteins containing polyproline sequences in vivo. Moreover, peptide synthesis assays demonstrated that eIF5A (Fig. 4) and EFP are critical for the in vitro synthesis of polyproline peptides (Doerfel et al., 2013). Finally, toe-printing analyses revealed that in the absence of eIF5A translating ribosomes stall on polyproline motifs with the second or third Pro codon in the P site (Fig. 5). Thus, di-proline or tri-proline will be attached to the 3′-CCA end of the peptidyl-tRNA (P site) and Pro-tRNA will be bound in the A site. These results suggest that eIF5A and, likewise, EFP are required to promote synthesis of the proline–proline peptide bonds needed to convert di-proline to tri-proline and higher-order polyproline sequences.
In addition to amino acid sequence and structural similarities, both eIF5A and EFP are post-translationally modified. As described earlier, the ε-amino group of a conserved Lys residue in eIF5A from all archaea and eukaryotes is modified by deoxyhypusine synthase and deoxyhypusine hydroxylase to generate hypusine (reviewed in (Park et al., 2010). Similarly, the E. coli gene products YjeA, YjeK and YfcM attach a β-lysine residue to the ε-amino group of Lys34 in E. coli EFP and then hydroxylate the side chain (Navarre et al., 2010; Yanagisawa et al., 2010; Roy et al., 2011; Peil et al., 2012). Both the hypusine modification of eIF5A and the β-lysine modification of EFP are required for these factors to stimulate polyproline synthesis (Fig. 4D, and Doerfel et al., 2013; Ude et al., 2013). Loss of β-lysylation in E. coli impairs translation of a polyproline motif in CadC, the transcriptional activator of the CadBA operon; whereas, loss of β-lysylation in Salmonella enterica alters the expression of a variety of cellular proteins, impairs virulence in mice, and alters resistance to antibiotics (Navarre et al., 2010), presumably due to impaired translation of polyproline sequences. In a similar manner, perhaps the assorted genes identified in suppressor and synthetic enhancement screens with eIF5A mutants in yeast (Valentini et al., 2002; Frigieri et al., 2007), as well as the connections between eIF5A, hypusine, cancer and tumorigenesis in humans and other mammals (Silvera et al., 2010; Scuoppo et al., 2012), reflects altered expression of proteins containing polyproline motifs.
eIF5A and EFP were originally identified based on their ability to stimulate methionyl-puromycin synthesis (Glick and Ganoza, 1975; Kemper et al., 1976; Schreier et al., 1977; Benne and Hershey, 1978). Whereas eIF5A was thought to function as translation initiation factor, it is noteworthy that puromycin, an aminoacyl analog that reacts well with most peptidyl-tRNA substrates, reacts poorly with fMet-tRNAfMet and with peptidyl-tRNA substrates with a C-terminal proline (Wohlgemuth et al., 2008). In contrast, these latter substrates react well with authentic aminoacyl-tRNAs (Wohlgemuth et al., 2008). The poor reactivity with puromycin has been attributed to poor substrate positioning in the active site of the ribosome (Youngman et al., 2004; Wohlgemuth et al., 2008). Thus, the methionyl-puromycin synthesis assay is likely not a good mimic of first peptide bond synthesis. Consistent with this notion, the kobs (data not shown) and the Ymax for MP or MF synthesis were only modestly affected by adding eIF5A (Figs. 4 and S3C); and recent studies, likewise, showed that EFP does not stimulate dipeptide formation (Bullwinkle et al., 2013; Doerfel et al., 2013).
Consistent with a function in translation elongation, inactivation of eIF5A (Fig. S1B, and Saini et al., 2009) or of EFP, or its β-lysine modification, mimics the effects of elongation inhibitors and causes polysome retention (Bullwinkle et al., 2013). It is unclear at present whether the polysome retention upon inactivation of eIF5A observed in Fig. S1B reflects impaired translation elongation on the majority of cellular mRNAs or could be due to impaired translation of just the mRNAs containing polyproline motifs (549 of 5889 ORFs in S. cerevisiae contain a polyproline tract consisting of three or more proline residues). This prevalence of polyproline motifs in yeast (95 proteins with motifs containing 4 or more consecutive prolines) is consistent with eIF5A being essential in yeast; whereas, the efp gene can be deleted in E. coli in which only 9 out of ~4000 proteins contain motifs of four or more prolines (Doerfel et al., 2013). Taken together, the puromycin, dipeptide, and polysome analyses indicate that eIF5A and EFP do not substantially stimulate first peptide bond formation, consistent with the notion that the primary function of these factors is to promote peptide bond formation especially for poor substrates like polyproline.
Previous studies revealed that peptidyl-prolyl-tRNA in the ribosomal P site reacts poorly with puromycin (Muto and Ito, 2008) and that proline is an inefficient A-site substrate for peptide bond formation (Pavlov et al., 2009). Our data, as well as the recent studies with EFP (Doerfel et al., 2013; Ude et al., 2013; Woolstenhulme et al., 2013), demonstrate that combining peptidyl-prolyl-tRNA in the P site with Pro-tRNA in the A site dramatically impairs protein synthesis and establishes a dependency for eIF5A/EFP. At present, it is not clear why polyproline is such a poor substrate for protein synthesis; however, it may reflect the imino acid nature of proline, the geometrical or steric constraints of a cyclic side chain, or the unique ability of proline to readily sample both cis and trans conformations of peptide bonds (Ramachandran and Mitra, 1976). Perhaps insertion of the extended hypusine (or β-lysine) side chain into the PTC (Fig. 7D) stabilizes the proper conformation of the PTC or restricts the conformation of proline in the P site enabling a favorable geometry for peptide bond formation with the A-site amino acid. While the data reported here and the recent studies on EFP establish that translation of homopolyproline motifs requires eIF5A/EFP, additional studies, including genome-wide ribosomal profiling (Ingolia et al., 2009) of wild type and eIF5A mutant cells, will be needed to define the spectrum of amino acids and motifs that rely on eIF5A for their efficient translation.
EXPERIMENTAL PROCEDURES
Details of the construction of plasmids and yeast strains (Supplemental Experimental Procedures) are available upon request.
Dual Luciferase Assay
Dual luciferase reporter constructs were obtained from Elizabeth Grayhack (Letzring et al., 2010). WCEs from yeast transformants were assayed for firefly and Renilla luciferase activity (details are in the Supplemental Experimental Procedures).
Peptide Formation and Toeprinting Assays
Initiation complexes were prepared as described previously (Acker et al., 2007) using [35S]Met-tRNAiMet and purified translation initiation factors. Limiting amounts of initiation complexes were mixed with purified eEF1A, eEF2, eEF3, Phe- (or Pro) tRNA, GTP and ATP in the presence or absence of eIF5A in buffer containing 30 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 1 mM magnesium acetate, 2 mM magnesium chloride, 1 mM spermidine, and 2 mM DTT. All reactions were performed at 26°C. Peptide formation was monitored by electrophoretic TLC as described previously (Eyler and Green, 2011), and the fractional yield of the peptides and free [35S]Met in each reaction at different times were quantified and fit to the single exponential equation: y = Ymax (1−exp (−kobst)), where Ymax is the maximum fraction of peptide formed and kobs is observed rate constant. Toeprinting assays were performed as described (Mitchell et al., 2010) with minor variations. Further information is provided in the Supplemental Experimental Procedures.
Preparation of Initiation and Elongation Factors
Recombinant initiation factors eIF1, eIF1A and eIF5 were purified as described previously (Shin et al., 2011). Initiation factor eIF5B (Shin and Dever, 2007), native elongation factor eEF1A (Eyler and Green, 2011), and poly-histidine tagged versions of elongation factors eEF2 and eEF3 (Andersen et al., 2004; Ortiz et al., 2006) were purified from yeast using published protocols with some modifications. Recombinant, hypusinated eIF5A was prepared by co-expressing His6-eIF5A, Dys1, and Lia1 in E. coli, and the hypusine modification was analyzed by mass spectrometry. Cys mutants of eIF5A used for hydroxyl radical cleavage studies were purified from yeast as previously described (Saini et al., 2009). Details of the expression and purification procedures are provided in the Supplemental Experimental Procedures.
Preparation of mRNA, tRNA, and Ribosomes
Unstructured model mRNAs based on the template: 5′-GGAA(UC)7U-peptide-coding-sequence-(CU)10C-3′ with codons Met(AUG), Pro(CCA), Phe(UUC), and Stop (UAA) were prepared by T7 in vitro transcription or purchased. The UGG isoacceptor of tRNAPro was purified from bulk S. cerevisiae tRNA using a biotinylated oligonucleotide (Yokogawa et al., 2010), and tRNAPhe (Chemical Block) and tRNALys (tRNA Probes, College Station, TX) were purchased. The tRNAs were aminoacylated using S. cerevisiae His6-tagged ProRS or yeast post-ribosomal supernatant (S100). Ribosomal subunits were prepared from the yeast strain YRP840 as described previously (Shin et al., 2007). Further information is provided in the Supplemental Experimental Procedures.
Directed Hydroxyl Radical Cleavage Analysis
Following addition of Fe(II)-BABE-modified eIF5A to 80S initiation complexes, hydroxyl radicals were generated by the Fenton reaction, and primer extension analyses were used to monitor rRNA cleavage sites as described previously (Shin et al., 2011).
Supplementary Material
HIGHLIGHTS.
eIF5A mutant is defective for translation of polyproline sequences in vivo
eIF5A alleviates ribosomal stalling on polyproline sequences in vitro
hypusine modification of eIF5A stimulates polyproline synthesis in vitro
ribosome binding of eIF5A places hypusine adjacent to acceptor stem of P-site tRNA
Acknowledgments
We thank Beth Grayhack for dual-luciferase constructs, Daniel Wilson for discussing results prior to publication, Peter Backlund for mass spectrometry services, and Rachel Green, Hani Zaher, Dan Eyler, Julie Brunelle, Alan Hinnebusch, Jon Lorsch, and members of the Dever, Buskirk, Hinnebusch, Lorsch, and Green labs for helpful discussions. This work was supported in part by the Intramural Research Program of the NIH, NICHD, and by NIH Grant GM77633 (A. B.).
Footnotes
Supplemental Information includes 4 figures, Supplemental Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- Anand M, Balar B, Ulloque R, Gross SR, Kinzy TG. Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J Biol Chem. 2006;281:32318–32326. doi: 10.1074/jbc.M601899200. [DOI] [PubMed] [Google Scholar]
- Andersen CF, Anand M, Boesen T, Van LB, Kinzy TG, Andersen GR. Purification and crystallization of the yeast translation elongation factor eEF3. Acta Crystallogr D Biol Crystallogr. 2004;60:1304–1307. doi: 10.1107/S0907444904010716. [DOI] [PubMed] [Google Scholar]
- Basavappa R, Sigler PB. The 3 Å crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J. 1991;10:3105–3111. doi: 10.1002/j.1460-2075.1991.tb07864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- Benne R, Hershey JWB. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullwinkle TJ, Zou SB, Rajkovic A, Hersch SJ, Elgamal S, Robinson N, Smil D, Bolshan Y, Navarre WW, Ibba M. (R)-β-Lysine-modified Elongation Factor P Functions in Translation Elongation. J Biol Chem. 2013;288:4416–4423. doi: 10.1074/jbc.M112.438879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- Eyler DE, Green R. Distinct response of yeast ribosomes to a miscoding event during translation. RNA. 2011;17:925–932. doi: 10.1261/rna.2623711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigieri MC, Thompson GM, Pandolfi JR, Zanelli CF, Valentini SR. Use of a synthetic lethal screen to identify genes related to TIF51A in Saccharomyces cerevisiae. Genet Mol Res. 2007;6:152–165. [PubMed] [Google Scholar]
- Ganoza MC, Aoki H. Peptide bond synthesis: function of the efp gene product. Biol Chem. 2000;381:553–559. doi: 10.1515/BC.2000.071. [DOI] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Chladek S, Ganoza MC. Peptide bond formation stimulated by protein synthesis factor EF-P depends on the aminoacyl moiety of the acceptor. Eur J Biochem. 1979;97:23–28. doi: 10.1111/j.1432-1033.1979.tb13081.x. [DOI] [PubMed] [Google Scholar]
- Glick BR, Ganoza MC. Identification of a soluble protein that stimulates peptide bond synthesis. Proc Natl Acad Sci U S A. 1975;72:4257–4260. doi: 10.1073/pnas.72.11.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregio AP, Cano VP, Avaca JS, Valentini SR, Zanelli CF. eIF5A has a function in the elongation step of translation in yeast. Biochem Biophys Res Commun. 2009;380:785–790. doi: 10.1016/j.bbrc.2009.01.148. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper WM, Berry KW, Merrick WC. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J Biol Chem. 1976;251:5551–5557. [PubMed] [Google Scholar]
- Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. RNA. 2010;16:2516–2528. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and block an alternative pathway. Mol Cell. 2010;39:950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem Biophys Res Commun. 2008;366:1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz PA, Ulloque R, Kihara GK, Zheng H, Kinzy TG. Translation elongation factor 2 anticodon mimicry domain mutants affect fidelity and diphtheria toxin resistance. J Biol Chem. 2006;281:32639–32648. doi: 10.1074/jbc.M607076200. [DOI] [PubMed] [Google Scholar]
- Park JH, Johansson HE, Aoki H, Huang BX, Kim HY, Ganoza MC, Park MH. Post-translational modification by beta-lysylation is required for activity of Escherichia coli elongation factor P (EF-P) J Biol Chem. 2012;287:2579–2590. doi: 10.1074/jbc.M111.309633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH. The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J Biol Chem. 1989;264:18531–18535. [PubMed] [Google Scholar]
- Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2010;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Wolff EC, Smit-McBride Z, Hershey JW, Folk JE. Comparison of the activities of variant forms of eIF-4D. The requirement for hypusine or deoxyhypusine. J Biol Chem. 1991;266:7988–7994. [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci U S A. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M, Karim Z, Yamamoto H, Kitakawa M, Qin Y, Nierhaus KH. Elongation factor 4 (EF4/LepA) accelerates protein synthesis at increased Mg2+ concentrations. Proc Natl Acad Sci U S A. 2011;108:3199–3203. doi: 10.1073/pnas.1012994108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Virumae K, Atkinson GC, Tenson T, Remme J, Wilson DN. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat Chem Biol. 2012;8:695–697. doi: 10.1038/nchembio.1001. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Mitra AK. An explanation for the rare occurrence of cis peptide units in proteins and polypeptides. J Mol Biol. 1976;107:85–92. doi: 10.1016/s0022-2836(76)80019-8. [DOI] [PubMed] [Google Scholar]
- Roy H, Zou SB, Bullwinkle TJ, Wolfe BS, Gilreath MS, Forsyth CJ, Navarre WW, Ibba M. The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine. Nat Chem Biol. 2011;7:667–669. doi: 10.1038/nchembio.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis: purification and characterization of seven initiation factors. J Mol Biol. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- Scuoppo C, Miething C, Lindqvist L, Reyes J, Ruse C, Appelmann I, Yoon S, Krasnitz A, Teruya-Feldstein J, Pappin D, et al. A tumour suppressor network relying on the polyamine-hypusine axis. Nature. 2012;487:244–248. doi: 10.1038/nature11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shin BS, Acker MG, Maag D, Kim JR, Lorsch JR, Dever TE. Intragenic suppressor mutations restore GTPase and translation functions of a eukaryotic initiation factor 5B switch II mutant. Mol Cell Biol. 2007;27:1677–1685. doi: 10.1128/MCB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Dever TE. Molecular genetic structure-function analysis of translation initiation factor eIF5B. Methods Enzymol. 2007;429:185–201. doi: 10.1016/S0076-6879(07)29009-3. [DOI] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE. Initiation factor eIF2γ promotes eIF2–GTP–Met-tRNAiMet ternary complex binding to the 40S ribosome. Nat Struct Mol Biol. 2011;18:1227–1234. doi: 10.1038/nsmb.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer. 2010;10:254–266. doi: 10.1038/nrc2824. [DOI] [PubMed] [Google Scholar]
- Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem. 2009;284:34809–34818. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Valentini SR, Casolari JM, Oliveira CC, Silver PA, McBride AE. Genetic interactions of yeast eukaryotic translation initiation factor 5A (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics. 2002;160:393–405. doi: 10.1093/genetics/160.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valvarde DP, Ptersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci U S A. 2013;110:E878–887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol. 2010;17:1136–1143. doi: 10.1038/nsmb.1889. [DOI] [PubMed] [Google Scholar]
- Yokogawa T, Kitamura Y, Nakamura D, Ohno S, Nishikawa K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010;38:e89. doi: 10.1093/nar/gkp1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Zuk D, Jacobson A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998;17:2914–2925. doi: 10.1093/emboj/17.10.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.