Abstract
There is increasing evidence to support the hypothesis of adaptive response, a phenomenon in which protection arises from a low-dose radiation (<0.1 Gy) against damage induced by subsequent exposure to high-dose radiation. The molecular mechanisms underlying such protection are poorly understood. The goal of this study was to fill this knowledge gap. Mass spectrometry-based proteomics was used to characterize global protein expression profiles in the medium collected from human lymphocyte cultures given sham irradiation (0 Gy) or a priming low dose of 0.03 Gy 137Cs γ rays 4 h prior to a challenging dose of 1 Gy 137Cs γ rays. Adaptive response was determined by decreased micronucleus frequencies in lymphocytes receiving low dose irradiation prior to high dose irradiation compared to those receiving only high dose irradiation. Adaptive response was found in these experiments. Proteomic analysis of media revealed: (a) 55 proteins with similar abundance in both groups; (b) 23 proteins in both groups, but 7 of them were high abundance in medium with adaptive environment, while 16 high abundance proteins were in medium without adaptive environment; (c) 17 proteins in medium with adaptive environment only; and (d) 8 proteins in medium without adaptive environment only. The results provide a foundation for improving understanding of the molecular mechanisms associated with the beneficial effects of low dose radiation that, in turn, will have an important impact on radiation risk estimation. Hence, these studies are highly relevant to radiation protection due to an increased use of low dose radiation in daily life (e.g., medical diagnosis or airport safety) or an unavoidable exposure to low level background radiation.
Keywords: biological indicators, health effects, hormesis, radiation, biology
Introduction
There is no doubt that high doses of radiation are harmful to cells or tissues. However, the results from many studies using a variety of biological endpoints (i.e., metaphase chromosome aberration, micronucleus, DNA damage, mutation, neoplastic transformation, and cancer) have shown that exposure to low doses (<0.1 Gy) of low linear energy transfer (LET) radiation (Olivieri et al. 1984; Bond et al. 1991; Azzam et al. 1996; Wolff 1996; Redpath et al. 2001; Feinendegen 2005; Scott and Di Palma 2006; Elmore et al. 2008; Mitchel 2010) or high LET radiation (Iyer and Lehnert 2002; Varés et al. 2011) can protect against damage induced by a subsequent exposure to a relatively high dose of radiation. This protection phenomenon by low dose radiation (initially demonstrated many years ago in human lymphocytes irradiated in vitro) is normally known as the “adaptive response (AR),” the term originally coined by Sheldon Wolff and his colleagues (Olivieri et al. 1984; Wiencke et al. 1986). It has also been found that the protective effects of low dose radiation against the induction of cytogenetic damage by high dose radiation varied among blood samples from different subjects (Sankaranarayanan et al. 1989) or among different lymphoblastoid cell lines (Sorensen et al. 2002), suggesting inter-individual variation in response to priming low dose radiation. This AR phenomenon has also been detected in in vitro studies using other cell types such as human skin fibroblasts (Pinto et al. 2010) and human hybrid (Hela X skin fibroblast) cells (Elmore et al. 2008).
The capability of low doses of low LET radiation, without a challenging dose, to reduce cytogenetic damage to below the spontaneous rate has been detected in both in vivo* (Hooker et al. 2004) and in vitro (Rithidech and Scott 2008) studies. Likewise, increases in proliferation and survival of bone marrow cells have been detected after exposure of mice to a single dose of 0.05 Gy of x-rays (Wang and Cai 2000). There is also evidence of a radiation-induced AR in animal studies when the “priming” low dose is given before or after a high “challenging” dose, with varying intervals (from 2 h to 2 wk) between the two radiation treatments (Farooqi and Kesavan 1993; Mitchel et al. 2003; Day et al. 2006; Ito et al. 2007). The efficiency of protection is inversely correlated to the level of the priming dose and the duration of the interval between the priming and the challenging doses. Further, the results from these studies showed that both acute or multiple exposures to a priming low dose are effective in reducing the damage from subsequent exposure to high dose radiation. However, normally priming low doses given at a low dose rate are more efficient in protecting cells than those given at a higher dose rate (Broome et al. 2002; Mitchel 2010).
Although the manifestation of the AR has been established in varying biological systems, the exact molecular mechanisms underlying protection against injury induced by subsequent acute exposure to high dose radiation are poorly understood. A variety of biological processes have been implicated in radiation-induced AR, depending on the cell type. These include the modulation of the cell cycle (Miura 2004; Cramers et al. 2005; Feinendegen 2005), stimulation of DNA repair (Ikushima et al. 1996; Coleman et al. 2005; Hafer et al. 2007), and activation of antioxidant defense systems (de Toledo et al. 2006; Otsuka et al. 2006; Fan et al. 2007). However, no definite molecular events have been determined. Hence, the identification of proteins potentially involved in the induction of adaptive response would greatly enhance understanding of the molecular mechanisms associated with the protection of low dose radiation against harmful effects of succeeding exposure to high dose radiation. The resulting data will have a significant impact on the assessment of health risk from exposure to radiation, which is a key component of radiation protection.
As an initial step in identifying proteins potentially involved in radiation-induced AR by means of mass spectrometry (MS)-based proteomics, the authors characterized global protein-expression profiles in the medium collected from lymphocyte cultures given sham-irradiation (0 Gy) or a priming low dose of 0.03 Gy of 137Cs γ rays prior to a challenging dose of 1 Gy of 137Cs γ rays. The protection (or the induction of the hypothesized AR) by a priming low dose irradiation was determined by a reduction in the frequencies of micronuclei (MN) in human blood lymphocyte cultures receiving a priming low dose radiation prior to a challenging high-dose radiation (an adaptive environment) related to those receiving only challenging high-dose radiation (a non-adaptive environment).
The focus of this study was on proteomics analyses of secreted proteins in the media from human lymphocyte cultures exposed to high dose radiation with or without protection by priming low dose radiation; i.e., the adaptive and non-adaptive environment. The authors hypothesized that the priming low dose radiation induced the synthesis of a specific subset of proteins capable of cell protection in exposed human lymphocytes and that those proteins were secreted into the culture medium, resulting in attenuation of the detrimental effects induced by succeeding exposure to challenging high dose radiation. In the past, the role of secreted soluble factors (proteins) in radiation-induced bystander effects (Mothersill and Seymour 1997) and radiation-induced genomic instability (Sowa Resat and Morgan 2004) has been suggested. However, information on the contribution of secreted proteins in radiation-induced AR is lacking. Hence, the resulting data obtained from this study will fill this knowledge gap.
In this study, a dose of 0.03 Gy of 137Cs γ rays was selected as a priming low dose radiation because of its reported beneficial effects (Feinendegen 2005; Mitchel 2010). It also has been established that a single dose of 1.0 Gy of low LET radiation induces a significant increase in the frequency of MN in exposed human-lymphocytes (Fenech and Morley 1985; Balasem and Ali 1991; Silva et al. 1994). Liquid chromatography tandem mass-spectrometry (LC-MS/MS) with the Linear Trap Quadrupole (LTQ) mass spectrometer was used to identify proteins potentially involved in AR in media from cultures with or without adaptive environment. The relative abundance of each identified protein was determined by spectral counting of peptides. Subsequently, the extensive online protein databases were used to search for what is known about the biological function of the identified proteins. Knowing the functions of these proteins will enhance understanding of the network of molecular signaling pathways associated with the potential beneficial effects of the hypothesized adaptive response of exposure to low dose radiation that, in turn, would improve the estimation of health risk from exposure to radiation.
Materials and Methods
Cytokinesis block micronucleus (CBMN) assay
Chemicals
Chemicals for blood lymphocyte cultures (RPMI 1640, penicillin/streptomycin, L-glutamine, phytohaemagglutinin M) were purchased from Invitrogen Corporation (Carlsbad, CA). Heat-inactivated fetal bovine serum was purchased from Gemini Bio-Products (Woodland, CA). Gurr-Giemsa stain was purchased from BDH (Santa Monica, CA).
Whole blood-lymphocyte cultures
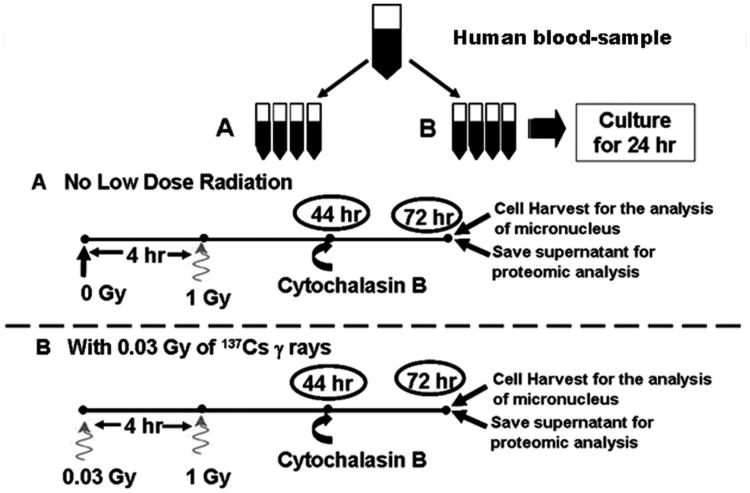
Fig. 1 shows the experimental design of the study. Briefly, peripheral blood samples (approximately 5 mL) were collected by venipuncture into heparinized syringes, using established bloodborne pathogen/biohazard safety protocols from two nonsmoking healthy female volunteers who were of similar ages. These two individuals had no known history of previous exposure to other clastogenic agents. Blood sample collections were performed under the approved guidelines by the Institutional Committees on Research Involving Human Subjects (CORIHS) at Stony Brook University. The informed consents were documented. The method of culturing whole blood lymphocyte cultures was similar to that routinely used in the authors' laboratory (Rithidech and Scott 2008). All experiments were done in duplicate for each subject. In each experiment for each donor, eight lymphocyte culture tubes were prepared. Cells were incubated at 37°C in humidified 5% CO2 atmosphere for 24 h. Thereafter, blood-lymphocyte culture tubes were divided into two groups (four culture tubes in each). These included: Group A (without adaptive environment), no priming low dose (0 Gy, sham-control irradiation) followed by a single dose of 1 Gy of 137Cs γ rays 4 h later (at the dose rate of 0.70 Gy min−1) and Group B (with adaptive environment), a priming low dose of 0.03 Gy of 137Cs γ rays given at 4 h before a single dose of 1 Gy of 137Cs γ rays. A gamma-irradiator (Gamma Cell40, Atomic Energy of Canada, Ltd, Ontario, Canada) located at Stony Brook University was used for irradiation. The exposure time for the 0-Gy sham-control irradiation (prior to a challenging high dose) was the same as that for the priming low dose of 0.03 Gy.
Fig. 1.
Diagram of the experimental design.
The protocol for the CBMN assay routinely used in this laboratory (Rithidech et al. 2005; Rithidech and Scott 2008) was followed. Briefly, at 44 h after culture-initiation, 3 μg/mL of Cytochalasin-B (Cyt-B) was added to each culture tube to block cytokinesis (which normally occurs in the telophase stage of the cell cycle). Cells were harvested 28 h after the addition of Cyt-B. The total culture time was 72 h, which resulted in the formation of many first division binucleated (BN) cells that were scored for the induction of MN (Fenech 2000). At harvest, the medium (supernatant) from each treatment of each subject was collected, concentrated using Agilent spin concentrators with 5 kDa cut-off (Agilent Technologies, Inc, Wilmington, DE), lyophilized, and stored at −80°C until shipment in dry ice by overnight carrier to Indiana University School of Medicine for proteomic analysis.
Micronucleus analysis
The slides were coded before scoring (under a light microscope with a 40 × 10 magnification). The criteria for selection of BN cells and identification of MN given in the HUMN project website [http://HUMN.org] and routinely used in this laboratory (Rithidech et al. 2005; Rithidech and Scott 2008) were applied. The numbers of BN cells with one, two, three, or more MN were then tabulated.
Statistical analysis
Generally, cytogenetic data, particularly in control animals and in those induced by low doses of toxic agents, contain a large number of cells (i.e., BN lymphocytes in this study) with zero or very few aberrations (i.e., MN in this study). Hence, the frequencies of aberrations are not normally distributed, and the variances are not homogeneous. Therefore, it is important to transform the data prior to statistical analysis to achieve reasonable normality and reasonably homogeneous inter-individual variability within experiment groups. In this study and before the statistical analysis, the average square root transformation [ASQRT, √X + √(X+1) where X is the observed MN frequency] (Whorton 1985; Albertini et al. 2000) was applied to each replication of each subject's measured MN frequency. The ASQRT has been routinely used in this laboratory for conducting statistical analyses of cytogenetic data using parametric statistical analysis methods (Rithidech et al. 1988, 2007). The frequencies of MN per 1,000 BN cells in human lymphocyte cultures exposed to 1.0 Gy of 137Cs γ rays with or without receiving preexposure to priming low dose radiation were evaluated statistically using Student's t test with a significance value of p < 0.05.
Proteomic analysis—Materials
Iodoethanol (>99% purity), triethylphosphine (TEP, >99% purity), and ammonium bicarbonate (ReagentPlus® grade) were obtained from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN) and MS grade water were purchased from EMD Chemicals (Gibbstown, NJ, USA). Modified sequencing grade porcine trypsin was obtained from Princeton Separations (Freehold, NJ).
Protein reduction, alkylation, and digestion
Proteins (from concentrated samples mentioned above) in 200 μL of 4 M urea were reduced and alkylated using TEP and iodoethanol as described previously (Lai et al. 2008). Briefly, 200 μL of the reduction/alkylation cocktail were added to the protein solution. The sample was incubated at 37°C for 90 min, dried by SpeedVac, and reconstituted with 100 μL of 100 mM NH4HCO3 at pH 8.0. A 150-μL aliquot of a 20 μg/mL trypsin solution was added to the sample and incubated at 37°C for 3 h, after which another 150 μL of trypsin were added, and the solution was incubated at 37°C overnight.
LC-MS/MS
The digested samples were analyzed using a Thermo-Finnigan linear ion-trap (LTQ) mass spectrometer coupled with a Surveyor autosampler and MS HPLC system (Thermo-Finnigan). Tryptic peptides were injected onto the C18 microbore RP column (Zorbax SB-C18,1.0 mm × 150 mm) at a flow rate of 50 μL/min. The mobile phases A, B, and C were 0.1% formic acid in water, 50% ACN with 0.1% formic acid in water, and 80% ACN with 0.1% formic acid in water, respectively. The gradient elution profile was as follows: 10% B (90% A) for 5 min, 10–95% B (90–5% A) for 120 min, 100% C for 5 min, and 10% B (90% A) for 12 min. The data were collected in the “Triple-Play” (MS scan, Zoom scan, and MS/MS scan) mode with the ESI interface using normalized collision energy of 35%. Dynamic exclusion settings were set to repeat count 1, repeat duration 30 s, exclusion duration 120 s, and exclusion mass width 0.75 m/z (low) and 2.0 m/z (high). Each sample was injected twice.
Protein identification and classification
The acquired data were searched against the International Protein Index (IPI) human database (ipi.HUMAN.v3.37) using SEQUEST (v. 28 rev. 12) algorithms in Bioworks (v. 3.3). General parameters were set as follows: peptide tolerance 2.0 amu, fragment ion tolerance 1.0 amu, enzyme limits set as “fully enzymatic—cleaves at both ends,” and missed cleavage sites set at 2. The searched peptides and proteins were validated by PeptideProphet (Keller et al. 2002) and ProteinProphet (Nesvizhskii et al. 2003) in the Trans-Proteomic Pipeline (TPP, v. 3.3.0, http://tools.proteomecenter.org/software.php). Quantitative analysis of proteins' relative abundances was performed using spectral counting of peptides (Liu et al. 2004), whose data were from TPP. Significant difference analysis was completed using the t-test in Microsoft Excel.
Following identification by LC-MS/MS, proteins were classified into different categories based upon their distribution in cellular compartments and their biological function by searching the gene ontology (GO) database (http://www.geneontology.org/), the public Swiss-Prot-TrEMBL (http://www.expasy.org), and NCBI (protein, http://.ncbi.nlm.nih.gov) servers.
Results
Frequencies of MN in human-lymphocyte cultures with or without treatment of 0.03 Gy of 137Cs γ rays prior to 1 Gy of 137Cs γ rays
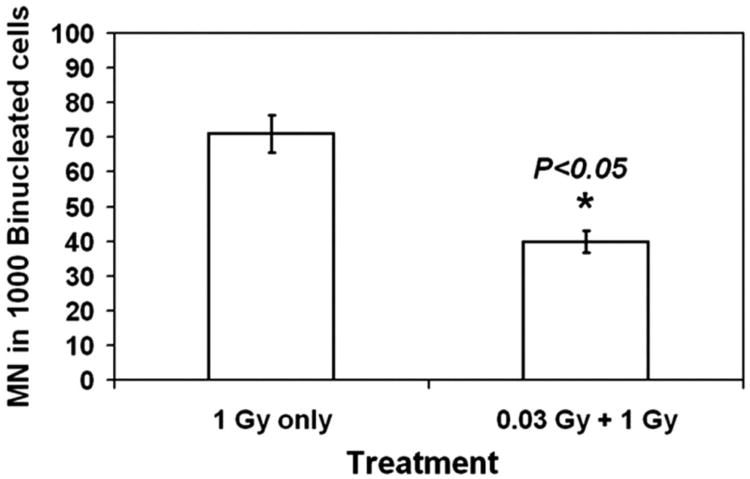
Fig. 2 shows the frequencies of MN in BN-lymphocytes induced by a single dose of 1 Gy of 137Cs γ rays alone or in combination with pre-exposure to a priming low dose of 0.03 Gy of 137Cs γ rays. Table 1 shows the information on the total number of BN-cells scored in each treatment from each subject and the distribution of MN. While the resulting data were presented as original unit rates, statistical significance was assessed using the ASQRT-transformation numbers. The current data showed a significant reduction in MN frequencies when a priming low dose was given to human lymphocyte cultures at 4 hr before a high dose of 1 Gy of 137Cs γ rays (p < 0.05), illustrating the induction of AR by a priming low dose radiation. Further, such AR was detected in lymphocytes from both subjects.
Fig. 2.
Frequency of MN in BN lymphocytes treated with 1 Gy γ-irradiation with or without pretreatment of 0.03 Gy γ-irradiation. Each bar represents mean of MN in 1,000 BN-lymphocytes scored ± S.E. from four measurements (two subjects with two replications).
Table 1.
Frequency and distribution of micronuclei (MN) in binucleated human-lymphocytes.
| Distribution of MN | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Treatment | Total cells scored | Total MN in 1000 binucleated cells (±S.E.) | 0 | 1 | 2 | 3 |
| 1 Gy | ||||||
| Subject 1 | 3467 | 72.7 ± 5.37 | 3243 | 198 | 24 | 2 |
| Subject 2 | 1180 | 66.1 ± 5.28 | 1110 | 62 | 8 | 0 |
| 0.03 Gy + 1 Gy | ||||||
| Subject 1 | 2957 | 40.92 ± 2.66 | 2846 | 101 | 10 | 0 |
| Subject 2 | 1500 | 35.45 ± 2.17 | 1465 | 47 | 3 | 0 |
Protein identification and classification
In total, 103 proteins with ≥90.00% confidence were identified by peptides with ≥90.00% confidence via TPP validation. The lists of these proteins are presented in Tables 2 to 4 with the following information for individual proteins: the IPI number, the common name of the protein, the biological function, the cellular component, the TPP confidence, the percent sequence coverage, and the number of peptides detected. Table 2 shows a list of 55 proteins with similar abundance in media with and without adaptive environments. Table 3a and b provide the lists of proteins found in media from both groups showing high abundance proteins in media with adaptive environment (seven proteins, Table 3a) and those with high abundance in media without adaptive environment (16 proteins, Table 3b). Table 4a and b present secreted proteins found only in media with an adaptive environment (17 proteins, Table 4a) and only in media without an adaptive environment (eight proteins, Table 4b).
Table 2.
List of secreted proteins detected in media from both groups with similar abundance.
| Protein number | International Protein Index (IPI) number | Common name of protein | Biological function | Cellular component | Trans-proteomic Pipeline (TPP) confidence | % Sequence coverage | Number of peptides |
|---|---|---|---|---|---|---|---|
| 1 | IPI00744503 | 17 kDa protein (Hemoglobin, gamma A) | Oxygen transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 2 | IPI00830113 | 19 kDa protein | Oxygen transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 3 | IPI00807522 | Actin, Beta (Fragment) | Cytoskeleton | Cytoplasm | 0.9999 | 29.1 | 1 |
| 4 | IPI00794523 | Actin Gamma 1 (ACTG1 protein) | Cytoskeleton | Cytoplasm | 0.9999 | 29.1 | 1 |
| 5 | IPI00848058 | Actin | Cytoskeleton | Cytoplasm | 0.9999 | 29.1 | 1 |
| IPI00021440 | 0.9999 | 29.1 | 1 | ||||
| IPI00021439 | 0.9999 | 29.1 | 1 | ||||
| 6 | IPI00853068 | Alpha 2 globin variant (Fragment) | Oxygen transporter | Extracellular Region | 1 | 74.6 | 12 |
| 7 | IPI00022429 | Alpha-1-acid glycoprotein 1 precursor | Stress and inflammatory Protein | Extracellular Region | 1 | 14.4 | 3 |
| IPI00020091 | Alpha-1-acid glycoprotein 2 precursor | 1 | 11.9 | 2 | |||
| 8 | IPI00029863 | Alpha-2-antiplasmin precursor | Stress and inflammatory Protein | Extracellular Region | 0.9999 | 5.5 | 1 |
| 9 | IPI00853045 | Anti-RhD monoclonal T125 kappa light chain precursor | Immune Response | Extracellular region | 1 | 8.7 | 5 |
| 10 | IPI00021841 | Apolipoprotein A-I precursor | Activation of anti-oxidant defense system | Extracellular Region | 1 | 67 | 31 |
| 11 | IPI00304273 | Apolipoprotein A-IV precursor | Activation of anti-oxidant defense system | Extracellular Region | 0.9677 | 3.5 | 1 |
| IPI00847179 | 0.9677 | 3.5 | 1 | ||||
| 12 | IPI00021855 | Apolipoprotein C-I precursor | Activation of anti-oxidant defense system | Extracellular Region | 0.9999 | 26.5 | 2 |
| 13 | IPI00021727 | Complement component 4 binding protein, alpha chain precursor | Immune Response | Extracellular Region | 0.9743 | 3.2 | 1 |
| 14 | IPI00386879 | cDNA FLJ14473 fis or immunoglobulin heavy constant alpha 1 | Immune response | Extracellular Region | 1 | 17 | 9 |
| 15 | IPI00017601 | Ceruloplasmin precursor | Ion Transport | Extracellular Region | 1 | 11.9 | 7 |
| 16 | IPI00783987 | Complement C3 precursor (Fragment) | Immune Response | Extracellular Region | 1 | 17 | 23 |
| 17 | IPI00006902 | Fascin-3 | Cytoskeleton | Cytoplasm | 0.9842 | 4.4 | 1 |
| 18 | IPI00219713 | Fibrinogen gamma chain precursor, Isoform Gamma-A | Stress and inflammatory Protein | Extracellular Matrix | 1 | 44.4 | 15 |
| IPI00021891 | Fibrinogen gamma chain precursor, Isoform Gamma-B | 1 | 44.4 | 15 | |||
| 19 | IPI00749035 | Gamma-G globin | Oxygen transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 20 | IPI00657911 | Gamma-globin | Oxygen transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 21 | IPI00641737 | Haptoglobin precursor | Immune Response | Extracellular region | 1 | 30.5 | 11 |
| 22 | IPI00796636 | Hemoglobin (Fragment) | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 23 | IPI00657660 | Hemoglobin delta-beta fusion protein | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 24 | IPI00816618 | Hemoglobin gamma-G (Fragment) | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 25 | IPI00829896 | Hemoglobin Lepore-Baltimore (Fragment) | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 26 | IPI00410714 | Hemoglobin subunit alpha | Oxygen Transporter | Extracellular Region | 1 | 74.6 | 12 |
| 27 | IPI00473011 | Hemoglobin subunit delta | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| IPI00791558 | 0.9101 | 6.8 | 1 | ||||
| 28 | IPI00217471 | Hemoglobin subunit epsilon | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 29 | IPI00220706 | Hemoglobin subunit gamma-1 | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 30 | IPI00554676 | Hemoglobin subunit gamma-2 | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 31 | IPI00022371 | Histidine-rich glycoprotein precursor | Protease inhibitor | Extracellular Region | 0.995 | 4.8 | 2 |
| 32 | IPI00876869 | Immunoglobulin C1-set Domain | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| 33 | IPI00472961 | Immunoglobulin kappa constant (IGKC) | Immune response | Extracellular Region | 1 | 8.7 | 5 |
| IPI00430847 | |||||||
| IPI00746963 | |||||||
| IPI00761125 | |||||||
| IPI00784070 | |||||||
| IPI00845354 | |||||||
| IPI00816118 | |||||||
| IPI00827488 | |||||||
| 34 | IPI00430820 | Immunoglobulin kappa | Immune response | Extracellular | 1 | 8.7 | 5 |
| IPI00854806 | variable 1-5 | Region | |||||
| IPI00478600 | (IGKV1-5) | ||||||
| IPI0049424 | |||||||
| 35 | IPI00440577 | Immunoglobulin kappa variable 2-24 (IGKV2-24) | Immune response | Extracellular Region | 1 | 8.7 | 5 |
| 36 | IPI00852577 | Immunoglobulin lambda light chain C region (IGLC1) | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| 37 | IPI00719373 | Immunoglobulin lambda locus (IGL@) | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| IPI00450309 | 0.9985 | 8.6 | 1 | ||||
| IPI00658130 | 0.9985 | 8.6 | 1 | ||||
| IPI00555945 | 0.9985 | 8.6 | 1 | ||||
| IPI00796167 | 0.9985 | 8.6 | 1 | ||||
| IPI00154742 | 0.9985 | 8.6 | 1 | ||||
| IPI00829626 | 0.9985 | 8.6 | 1 | ||||
| IPI00829640 | 0.9985 | 8.6 | 1 | ||||
| IPI00829877 | 0.9985 | 8.6 | 1 | ||||
| IPI00744476 | 0.9985 | 8.6 | 1 | ||||
| IPI00745660 | 0.9985 | 8.6 | 1 | ||||
| IPI00718819 | 0.9985 | 8.6 | 1 | ||||
| IPI00815938 | 0.9985 | 8.6 | 1 | ||||
| 38 | IPI00718819 | Immunoglobulin lambda variable 2-14 (IGLV2-14) | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| IPI00877071 | 0.9985 | 8.6 | 1 | ||||
| 39 | IPI00815938 | Immunoglobulin lambda variable 3-21 (IGLV3-21) | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| 40 | IPI00550162 | Immunoglobulin lambda variable 3-25 (IGLV3-25) | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| 41 | IPI00382938 | Immunoglobulin lambda variable 4-3 (IGLV4-3) | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| 42 | IPI00430808 | Immunoglobulin light chain (Fragment) | Immune response | Extracellular Region | 1 | 8.7 | 5 |
| 43 | IPI00645038 | Inter-alpha (Globulin) inhibitor H2 | Protease Inhibitor | Extracellular Region | 1 | 4.2 | 3 |
| 44 | IPI00305461 | Inter-alpha-trypsin inhibitor heavy chain H2 precursor | Protease Inhibitor | Extracellular Region | 1 | 4.2 | 3 |
| 45 | IPI00827875 | Lambda-chain precursor | Immune response | Extracellular Region | 0.9985 | 8.6 | 1 |
| 46 | IPI00019580 | Plasminogen precursor | Protease Inhibitor | Extracellular Region | 0.9822 | 1.4 | 1 |
| 47 | IPI00807459 | Putative uncharacterized protein | Immune response | Extracellular Region | 1 | 8.7 | 5 |
| 48 | IPI00784661 | Serotransferrin precursor | Stress and inflammatory Protein | Extracellular Region | 1 | 8.7 | 5 |
| 49 | IPI00784773 | Serpin peptidase inhibitor, clade C (antithrombin), member 1 (SERPINC1) | Protease Inhibitor | Extracellular Region | 1 | 8.7 | 5 |
| IPI00784519 | 0.9985 | 8.6 | 1 | ||||
| IPI00784711 | 0.9985 | 8.6 | 1 | ||||
| IPI00022463 | 1 | 46.7 | 35 | ||||
| IPI00844156 | 0.9974 | 6.9 | 1 | ||||
| 50 | IPI00745872 | Serum albumin precursor, isoform 1 | Activation of anti-oxidant defense system | Extracellular region | 1 | 84.1 | 157 |
| 51 | IPI00300117 | Sodium channel protein type 7 subunit alpha | Sodium Transport | Membrane | 0.9891 | 1.3 | 3 |
| 52 | IPI00798430 | Transferrin variant (Fragment) | Stress and inflammatory Protein | Extracellular Region | 1 | 46.7 | 35 |
| 53 | IPI00815947 | Truncated beta-globin (Fragment) | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
| 54 | IPI00022434 | Uncharacterized protein ALB | Activation of anti-oxidant defense system | Extracellular region | 1 | 14.6 | 11 |
| 55 | IPI00853641 | Uncharacterized protein HBE1 | Oxygen Transporter | Extracellular Region | 0.9101 | 6.8 | 1 |
Table 4.
List of 17 secreted proteins found only in the medium obtained from human-lymphocyte cultures with an adaptive environment (4a) and 8 secreted proteins found only in the medium from human-lymphocyte cultures without an adaptive environment (4b).
| 4a | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Protein number | International Protein Index (IPI) number | Common name of protein | Biological function | Cellular component | Trans-proteomic Pipeline (TPP) confidence | % Sequence coverage | Number of peptides |
| 1 | IPI00019943 | Afamin precursor | Activation of anti-oxidant defense system | Extracellular Matrix | 0.9999 | 3.7 | 1 |
| 2 | IPI00021842 | Apolipoprotein E precursor | Activation of anti-oxidant defense system | Extracellular region | 0.9966 | 6.3 | 2 |
| 3 | IPI00028095 | Bis(5′-adenosyl)-triphosphatase | DNA repair | Cytoplasm | 0.9492 | 11.6 | 1 |
| 4 | IPI00001707 | Bromodomain-containing protein 7 | Cell Cycle Control | Cytoplasm | 0.9864 | 4.1 | 1 |
| IPI00647008 | 0.9864 | 4.1 | 1 | ||||
| 5 | IPI00479116 | Carboxypeptidase N subunit 2 precursor | Immune Response | Extracellular region | 0.9999 | 5 | 1 |
| IPI00738433 | 0.9999 | 5 | 1 | ||||
| 6 | IPI00795633 | Clusterin | Activation of anti-oxidant defense system | Extracellular matrix | 0.9999 | 5 | 1 |
| IPI00400826 | Clusterin, isoform 1 | 0.9976 | 8.7 | 2 | |||
| 7 | IPI00477618 | Collagen alpha-1(XXV) chain | Activation of anti-oxidant defense system | Extracellular Matrix | 0.9286 | 3 | 1 |
| IPI00797164 | 0.9286 | 3 | 1 | ||||
| 8 | IPI00019591 | Complement factor B precursor, isoform 1 (Fragment) | Immune Response | Extracellular region | 1 | 3.1 | 2 |
| 9 | IPI00296534 | Fibulin-1 precursor | Cell Cycle Control | Cytoplasm | 0.9002 | 1.6 | 1 |
| 10 | IPI00001755 | Glypican-6 precursor | Cell Cycle Control | Extracellular matrix | 0.9172 | 4.5 | 1 |
| 11 | IPI00021109 | Hydroxyacid oxidase 2 | Oxidation-Reduction | Cytoplasm | 0.9336 | 4.8 | 1 |
| 12 | IPI00385264 | Ig mu heavy chain disease protein | Immune Response | Extracellular region | 1 | 14.1 | 2 |
| 13 | IPI00032328 | Kininogen-1 precursor, isoform HMW | Activation of anti-oxidant defense system | Extracellular region | 0.9662 | 5.1 | 1 |
| IPI00215894 | Kininogen-1 precursor, isoform LMW | 0.9662 | 5.1 | 1 | |||
| 14 | IPI00297208 | Myosin-10 | Cytoskeleton | Extracellular region | 0.9819 | 1.8 | 1 |
| 15 | IPI00022420 | Retinol-binding protein 4 | Activation of anti-oxidant defense system | Extracellular Matrix | 0.9613 | 5 | 1 |
| 16 | IPI00852806 | Exocyst complex component 6B; SEC15-like 2 (ras like binding protein) | Cell Cycle Control | Organelle | 0.9075 | 6 | 1 |
| 17 | IPI00847746 | Small G protein signaling modulator | Cell Cycle Control | Organelle | 0.9144 | 5.3 | 1 |
| 4b | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Protein number | International Protein Index (IPI) number | Common name of protein | Biological function | Cellular component | Trans-proteomic Pipeline (TPP) confidence | % Sequence coverage | Number of peptides |
| 1 | IPI00790784 | Alpha-1-antitrypsin precursor, isoform 2 | Stress and inflammatory Protein | Extracellular Matrix | 0.9899 | 2.6 | 1 |
| IPI00869004 | Alpha-1-antitrypsin precursor, Isoform 3 | 0.9899 | 2.6 | 1 | |||
| 2 | IPI00032258 | Complement Component 4A precursor | Stress and inflammatory Response | Extracellular Matrix | 1 | 7.5 | 7 |
| 3 | IPI00654875 | Complement Component 4B precursor | Stress and inflammatory Response | Extracellular Matrix | 1 | 7.5 | 7 |
| 4 | IPI00853174 | DNAJ homolog subfamily A member 4 | Molecular Chaperone | Cytoplasm | 0.9769 | 7.1 | 2 |
| 5 | IPI00852643 | Hypothetical protein LOC84162 | Fragile Site Association | Cytoplasm | 0.9936 | 1.5 | 1 |
| 6 | IPI00294193 | Inter-alpha-trypsin inhibitor heavy chain H4 precursor, Isoform 1 | Protease Inhibitor | Extracellular Region | 0.9999 | 5.8 | 2 |
| IPI00218192 | Inter-alpha-trypsin inhibitor heavy chain H4 precursor, Isoform 2 | 0.9999 | 5.8 | 2 | |||
| 7 | IPI00251161 | Putative uncharacterized protein DKFZp434M0126 | Fragile Site Association | Cytoplasm | 0.9936 | 1.5 | 1 |
| 8 | IPI00658128 | Stromal interaction molecule 2 protein | Inhibition of Ca++ influx | Organelle (Golgi apparatus) | 0.9422 | 1.1 | 2 |
Table 3.
List of secreted proteins found in media from both groups, but 7 proteins with high abundance in the medium obtained from human-lymphocyte cultures with an adaptive environment (3a); while 16 proteins with high abundance in the medium from human-lymphocyte cultures without an adaptive environment (3b).
| 3a | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Protein number | International Protein Index (IPI) number | Common name of protein | Biological function | Cellular component | Trans-proteomic Pipeline (TPP) confidence | % Sequence coverage | Number of peptides |
| 1 | IPI00550991 | Alpha-1-antichymotrypsin precursor | Cell Survival | Extracellular Region | 1.000 | 15.10 | 4 |
| IPI00847635 | Alpha-1-antichymotrypsin precursor, isoform 1 | 1.000 | 15.10 | 4 | |||
| 2 | IPI00022895 | Alpha-1B-glycoprotein precursor | Immune Response | Extracellular Region | 1.000 | 16.20 | 4 |
| 3 | IPI00021857 | Apolipoprotein C-III precursor | Activation of anti-oxidant defense system | Extracellular Region | 1.000 | 16.2 | 2 |
| IPI00657670 | Apolipoprotein C-III precursor variant 1 | 1.000 | 16.2 | 2 | |||
| 4 | IPI00449920 | cDNA FLJ90170 fis | Immune Response | Extracellular Region | 1.000 | 17.00 | 9 |
| 5 | IPI00647556 | Gelsolin | Cell Survival | Extracellular Region | 0.998 | 10.50 | 1 |
| IPI00641047 | 0.998 | 10.50 | 1 | ||||
| 6 | IPI00719233 | Immunoglobulin heavy constant alpha 1 (IGHA1) protein | Immune Response | Extracellular Region | 1.000 | 17 | 9 |
| IPI00430842 | 1.000 | 17 | 9 | ||||
| 7 | IPI00742696 | Vitamin D-binding protein precursor | Cell Survival | Extracellular Matrix | 1.000 | 25.3 | 10 |
| IPI00555812 | 1.000 | 25.3 | 10 | ||||
| 3b | |||||||
|
| |||||||
| 1 | IPI00745089 | Alpha-1B-glycoprotein | Stress and inflammatory Protein | Extracellular Region | 1 | 16.2 | 4 |
| 2 | IPI00553177 | Alpha-1-antitrypsin precursor, Isoform 1 | Protease Inhibitor | Extracellular Matrix | 1 | 54.1 | 29 |
| 3 | IPI00478003 | Alpha-2-macroglobulin precursor | Stress and inflammatory Protein | Extracellular Matrix | 1 | 19.8 | 23 |
| 4 | IPI00032179 | Antithrombin III variant | Protease Inhibitor | Extracellular Region | 1 | 15.9 | 4 |
| 5 | IPI00021854 | Apolipoprotein A-II precursor | Stress and inflammatory Protein | Extracellular Region | 1 | 58 | 6 |
| 6 | IPI00298828 | Beta-2-glycoprotein 1 precursor | Stress and inflammatory Protein | Cytoplasm | 1 | 9.6 | 2 |
| 7 | IPI00418163 | Complement component 4B1 | Stress and inflammatory Protein | Extracellular Matrix | 0.9998 | 1.8 | 2 |
| 8 | IPI00022431 | Fetuin-A (Alpha-2-HS-glycoprotein precursor) | Stress and inflammatory Protein | Extracellular Matrix | 1 | 11.7 | 5 |
| 9 | IPI00021885 | Fibrinogen alpha chain precursor | Stress and inflammatory Protein | Extracellular Matrix | 1 | 14 | 8 |
| 10 | IPI00298497 | Fibrinogen beta chain precursor | Stress and inflammatory Protein | Extracellular Matrix | 1 | 30.1 | 10 |
| 11 | IPI00298860 | Growth-inhibiting protein 12 | Stress and inflammatory Protein | Extracellular Region | 1 | 7.8 | 2 |
| 12 | IPI00654755 | Hemoglobin subunit beta | Oxygen Transporter | Extracellular Region | 1 | 74.1 | 11 |
| 13 | IPI00022488 | Hemopexin precursor | Stress and inflammatory Protein | Extracellular Matrix | 1 | 40.5 | 12 |
| 14 | IPI00292530 | Inter-alpha-trypsin inhibitor heavy chain H1 precursor | Protease inhibitor | Extracellular Region | 0.9999 | 2.1 | 1 |
| 15 | IPI00848342 | Lactotransferrin precursor | Stress and inflammatory Protein | Extracellular Region | 1 | 7.8 | 2 |
| 16 | IPI00855916 | Transthyretin | Stress and inflammatory Protein | Extracellular Region | 1 | 32.7 | 3 |
| IPI00022432 | Transthyretin precursor | 1 | 32.7 | 3 | |||
Discussion
This data indicated differential expression, both qualitatively and quantitatively, of human proteins in media from lymphocyte cultures exposed to high dose radiation with or without adaptive environment; i.e., pre-exposure to 0 Gy (sham control, without an adaptive environment) or 0.03 Gy (with an adaptive environment). However, there were several secreted proteins with similar abundance (such as immunoglobulin and actin) in the media from both groups. These proteins are presumably responsible for normal regulatory processes of cells. The authors reported only secreted human proteins since the medium for growing human lymphocytes in cultures contained fetal bovine serum. The data indicated that human lymphocytes exposed to low dose radiation secreted a subset of proteins capable of altering the consequences of subsequent high dose irradiation. Hence, these proteins may act as a molecular switch that regulates the protection against damage induced by succeeding high-dose irradiation. This study is novel in that the samples used for proteomics and determining, as asserted, the biological evidence for AR (a decrease in MN frequency in cultures given priming low dose radiation) were obtained from the same culture of primary human cells, not that of human cell-lines. The results also demonstrated that the MS-based proteomic approach used in this study is highly sensitive not only in the identification of secreted proteins potentially involved in the radiation-induced AR but also in the determination of their abundances in the human lymphocyte system. Previously, a similar proteomic approach was used to characterize secreted proteins after an in vitro γ-irradiation (0.1 Gy) of human mammary epithelial cell lines, in which significant alterations (related to a sham-control exposed group) in the abundance of proteins were undetected (Springer et al. 2005). Differences in cell types (human mammary epithelial cells vs. human lymphocytes), radiation dose (0.1 Gy vs. 0.05 Gy γ-irradiation), and experimental design (single dose of low dose γ-irradiation vs. priming low dose prior to high dose γ-irradiation) used in that study may contribute to obtaining dissimilar results from this study. Of note, although the protective effects of low dose radiation against the damage induced by subsequent exposure to high dose radiation have been shown in human lymphocytes or human skin fibroblasts, such a phenomenon in human mammary epithelial cells has yet to be reported. Further, proteomics is a relatively new technique and still under development. Different instruments and software applied often generate dissimilar results. It has previously been observed that the LTQ (Linear Trap Quadrupole, linear ion trap) mass spectrometer (used in this study) has several advantages in protein identification over the LCQ (Liquid Chromatography Quadrupole, 3D ion trap) mass spectrometer used in a study conducted by another group of investigators (Springer et al. 2005). The advantages of LTQ over LCQ include: (a) increased ion-trapping efficiency, and (b) quicker ion-ejection rate, resulting in greater than five-fold more protein identifications, better identification of low-abundance proteins, and higher confidence protein identifications (Blackler et al. 2006).
Based upon biological function, the unique 17 proteins found only in the media with adaptive environment (Table 4a) were classified into six groups. These include proteins involved in: (1) activation of anti-oxidant defense system (35%), (2) cell-cycle control (29%), (3) immune response (18%), (4) DNA repair (6%), (5) oxidation reduction (6%), and (6) cytoskeleton (6%). The potential involvement of the first five groups of proteins in AR has been suggested (Luckey 1982; James et al. 1990; Ikushima et al. 1996; Matsubara et al. 2000; Wang and Cai 2000; Miura 2004; Scott 2004; Cramers et al. 2005; Feinendegen 2005; de Toledo et al. 2006; Otsuka et al. 2006; Bauer 2007; Fan et al. 2007; Hafer et al. 2007; Liu et al. 2007; Portess et al. 2007). This study is the first to identify the contribution of cytoskeleton protein, namely myosin-10, to what the authors argue is radiation-induced AR. It is known that cellular myosin is a microtubule binding protein and that it has a crucial role not only in spindle-fiber assembly during cell division for proper cytokinesis but also in maintaining cell shape and movement for homeostasis of the cell/tissue (Wu et al. 1998; Weber et al. 2004). Currently, the exact molecular function of myosin-10 contributing to what is believed to be radiation-induced AR remains unclear. However, it has been suggested that microtubules may mediate the AR to the low phosphate of Na/Pi (Hansch et al. 1993; Lottscher et al. 1997). Hence, this finding of myosin abundance in media from the adaptive environment warrants further investigation on the involvement of cytoskeleton in this phenomenon.
It is clear that the majority of proteins observed in this phenomenon (which may be radiation-induced AR) found in the cell culture system used in this study were those capable of activating the antioxidant defense system. Among the unique 17 proteins found only in media with adaptive environment (Table 4a), secreted clusterin has previously been suggested to play an important role in radiation-induced AR both in human skin cells in culture and in mouse bone marrow cells in vivo (Klokov et al. 2004). Consequently, these findings presented information on a new subset of secreted proteins that may be associated with radiation-induced AR. Protective effects of these proteins have been reported in other cell systems. For examples, neuroprotective effects of afamin (Vitamin E-binding protein) have previously been found (Heise et al. 2002), antioxidative activity of ApoE has been reported in the brain (Ramassamy et al. 2001), and enhanced kininogen synthesis has a protective role for the cardiovascular system (Chao et al. 1996). The results also showed that a specific subset of secreted proteins (involved in activation of the antioxidant defense system, or immune response, or anti-apoptosis) was higher in abundance in the medium with an AR environment than that without (Table 3a). Of note, protective effects of gelsolin and Vitamin D-binding protein after irradiation have previously been suggested in a study using a mouse model (Rithidech et al. 2009). In that study, a striking depletion of these two proteins was found in plasma samples collected at 3 and 7 d after exposure of mice to a single dose of 3 Gy of 137Cs γ rays, as compared to those in plasma samples of the corresponding sham controls. Gelsolin and Vitamin D-binding protein are known to be responsible for removal of actin (i.e., actin scavenging system) that is released from dying cells to prevent cell death (Dahl 2005; Bucki et al. 2008). Hence, prolonged depletion of gelsolin and Vitamin D-binding protein may ultimately lead to cell death (Osborn et al. 2008). Taken together, the abundance of gelsolin and Vitamin D-binding protein appears to be associated with cell survival.
In contrast, the majority of proteins found in medium without AR was a specific subset of stress and inflammatory proteins (Table 4b). The finding of a protein involved in the inhibition of Ca++-influx (stromal interaction molecule-2 protein) only in the medium without an AR pretreatment suggests that a balance in Ca++-concentration is important in cell protection. A transient increase in Ca++-influx has been linked to what others have speculated to be AR induction (Mattson 2008). However, prolonged accumulation of Ca+ + can cause cell damage (Lyng et al. 2006). The abundance of another subset of stress and inflammatory proteins was higher in medium without AR (Table 3b), although they were detected in media from both groups. High levels of some of these proteins (e.g., Apolipoprotein A-II precursor, Beta-2-glycoprotein, and Fetuin A) were previously detected in plasma of 3-Gy γ-irradiated mice (Rithidech 2009). Although further validation is required, these findings suggest that high expression levels of a specific subset of stress and inflammatory proteins may be indicative of exposure to high dose radiation.
It is, however, important to emphasize that proteomics is a relatively new and challenging technique that engenders as many questions as it answers. Fundamentally, researchers may be identifying the presence of proteins in different environments, but it is not certain what the function of those proteins actually is at this point in time. Moreover, the identification of the proteins really depends on the accuracy of the software involved. For these reasons, the authors validated identification using PeptideProphet (Keller et al. 2002) and ProteinProphet (Nesvizhskii et al. 2003) in the Trans-Proteomic Pipeline (TPP, v. 3.3.0, http://tools.proteomecenter.org/software.php), the two methods widely used worldwide in proteomics. It is recognized that the results from this study were derived from two human subjects (with two replications per subject for each experimental group) and that a false negative result may occur with this small sample size. However, the true positive results are still reliable (Eng 2003). As indicated in the results section, the protective effects of a priming low dose radiation against cytogenetic damage induced by a subsequent high dose irradiation were detected in lymphocytes from both subjects included in this study. Hence, the true-positive result was used to obtain the conclusions, making the outcome of this study trustworthy. In the future, it is likely that more new subsets of proteins potentially associated with the protection effects of low dose radiation will be discovered when a larger sample size and newly improved software for protein identification are applied.
Conclusion
The data reported here demonstrated that when using a proteomic approach with the LC-MS/MS system, the global expression profiles of secreted proteins strongly supported the induction of AR by low dose radiation given to human lymphocytes (obtained from freshly drawn blood) before subsequent exposure to high dose radiation. The data also suggested that a specific subset of proteins (with defense mechanisms) was secreted by 0.03-Gy irradiated lymphocytes within 4 h and that such proteins altered cell response to injury induced by succeeding high dose irradiation. These data indicated that the majority of secreted proteins found in the medium from cultures without an adaptive response are those frequently associated with stress and inflammatory response. It is recognized that the level of background radiation is much lower than the priming low dose used in this study and that exposure to low dose background radiation is chronic. The authors intend to use the findings obtained from this study as the starting point for future investigation on the potential mechanisms associated with the beneficial effects of low dose background radiation or those encountered in daily life such as medical diagnosis or airport safety. Hence, to better mimic human exposure to low dose radiation in daily life and to improve the understanding of mechanisms for protection mediated by the possible radiation-induced AR, further investigation should be conducted to determine the effects of dose series of the priming dose (as a single or a repeated application, or chronic or fractionated exposure), dose rate series of the priming dose, and time series (the interval between the priming and the challenging doses) both in vitro and in vivo.
Acknowledgments
Research funded by the Pathology Department, Stony Brook University, NY; the Department of Cellular & Integrative Physiology, Indiana University, School of Medicine, IN; and NASA grant #NN07AP88G.
Footnotes
Rithidech K, Udomthanakunchai C, Honikle L, Whorton EB. No evidence for the in vivo induction of genomic instability by low doses of 137Cs gamma rays in bone marrow cells of BALB/cJ and C57BL/6J Mice, In press Dose Response. DOI: 10.2203/dose-response.11-002.Rithidech, 2011
References
- Albertini RJ, Anderson D, Douglas GR, Hagmar L, Hemminki K, Merlo F, Natarajan AT, Norppa H, Shuker DEG, Tice R, Waters MD, Aitio A. IPCS guidelines for the monitoring of genotoxic effects of carcinogens in humans. Mutation Res/Rev Mutation Res. 2000;463:111–172. doi: 10.1016/s1383-5742(00)00049-1. [DOI] [PubMed] [Google Scholar]
- Azzam EI, de Toledo SM, Raaphorst GP, Mitchel RE. Low-dose ionizing radiation decreases the frequency of neoplastic transformation to a level below the spontaneous rate in C3H 10T1/2 cells. Radiat Res. 1996;146:369–373. [PubMed] [Google Scholar]
- Balasem AN, Ali AS. Establishment of dose-response relationships between doses of Cs-137 gamma-rays and frequencies of micronuclei in human peripheral blood lymphocytes. Mutat Res. 1991;259:133–135. doi: 10.1016/0165-1218(91)90047-p. [DOI] [PubMed] [Google Scholar]
- Bauer G. Low dose radiation and intercellular induction of apoptosis: potential implications for the control of oncogenesis. Int J Radiat Biol. 2007;83:873–888. doi: 10.1080/09553000701727523. [DOI] [PubMed] [Google Scholar]
- Blackler AR, Klammer AA, MacCoss MJ, Wu CC. Quantitative comparison of proteomic data quality between a 2D and 3D quadrupole ion trap. Analytical Chem. 2006;78:1337–1344. doi: 10.1021/ac051486a. [DOI] [PubMed] [Google Scholar]
- Bond VP, Benary V, Sondhaus CA. A different perception of the linear, nonthreshold hypothesis for low-dose irradiation. Proc Natl Acad Sci USA. 1991;88:8666–8670. doi: 10.1073/pnas.88.19.8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome EJ, Brown DL, Mitchel REJ. Dose responses for adaption to low doses of 60Co γ rays and 3H β-particles in normal human fibroblasts. Radiat Res. 2002;158:181–186. doi: 10.1667/0033-7587(2002)158[0181:drfatl]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bucki R, Levental I, Kulakowska A, Janmey PA. Plasma gelsolin: function, prognostic value, and potential therapeutic use. Curr Protein Pept Sci. 2008;9:541–551. doi: 10.2174/138920308786733912. [DOI] [PubMed] [Google Scholar]
- Chao C, Madeddu P, Wang C, Liang Y, Chao L, Chao J. Differential regulation of kallikrein, kininogen, and kallikrein-binding protein in arterial hypertensive rats. Am J Physiol. 1996;271:78–88. doi: 10.1152/ajprenal.1996.271.1.F78. [DOI] [PubMed] [Google Scholar]
- Coleman MA, Yin E, Peterson LE, Nelson D, Sorensen K, Tucker JD, Wyrobek AJ. Low-dose irradiation alters the transcript profiles of human lymphoblastoid cells including genes associated with cytogenetic radioadaptive response. Radiat Res. 2005;164:369–382. doi: 10.1667/rr3356.1. [DOI] [PubMed] [Google Scholar]
- Cramers P, Atanasova P, Vrolijk H, Darroudi F, van Zeeland AA, Huiskamp R, Mullenders LH, Kleinjans JC. Preexposure to low doses: modulation of x-ray-induced DNA damage and repair? Radiat Res. 2005;164:383–390. doi: 10.1667/rr3430.1. [DOI] [PubMed] [Google Scholar]
- Dahl B. The extracellular actin scavenger system in trauma and major surgery. Clinical and experimental studies. Acta Orthopaedica (Suppl) 2005;76:2–24. [PubMed] [Google Scholar]
- Day TK, Zeng G, Hooker AM, Bhat M, Scott BR, Turner DR, Sykes PJ. Extremely low priming doses of X radiation induce an adaptive response for chromosomal inversions in pKZ1 mouse prostate. Radiat Res. 2006;166:757–766. doi: 10.1667/RR0689.1. [DOI] [PubMed] [Google Scholar]
- de Toledo SM, Asaad N, Venkatachalam P, Li L, Howell RW, Spitz DR, Azzam EI. Adaptive responses to low-dose/low-dose-rate gamma rays in normal human fibroblasts: the role of growth architecture and oxidative metabolism. Radiat Res. 2006;166:849–857. doi: 10.1667/RR0640.1. [DOI] [PubMed] [Google Scholar]
- Elmore E, Lao XY, Kapadia R, Giedzinski E, Limoli C, Redpath JL. Low doses of very low-dose-rate low-LET radiation suppress radiation-induced neoplastic transformation in vitro and induce an adaptive response. Radiat Res. 2008;169:311–318. doi: 10.1667/RR1199.1. [DOI] [PubMed] [Google Scholar]
- Eng J. Sample size estimation: how many individuals should be studied? Radiol. 2003;227:309–313. doi: 10.1148/radiol.2272012051. [DOI] [PubMed] [Google Scholar]
- Fan M, Ahmed KM, Coleman MC, Spitz DR, Li JJ. Nuclear Factor-kappaB and manganese superoxide dismutase mediate adaptive radioresistance in low-dose irradiated mouse skin epithelial cells. Cancer Res. 2007;67:3220–3228. doi: 10.1158/0008-5472.CAN-06-2728. [DOI] [PubMed] [Google Scholar]
- Farooqi Z, Kesavan PC. Low-dose radiation-induced adaptive response in bone marrow cells of mice. Mutat Res. 1993;302:83–89. doi: 10.1016/0165-7992(93)90008-j. [DOI] [PubMed] [Google Scholar]
- Feinendegen LE. Evidence for beneficial low level radiation effects and radiation hormesis. Br J Radiol. 2005;78:3–7. doi: 10.1259/bjr/63353075. [DOI] [PubMed] [Google Scholar]
- Fenech M. The in vitro micronucleus technique. Mutat Res. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- Fenech M, Morley AA. Measurement of micronuclei in lymphocytes. Mutat Res. 1985;147:29–36. doi: 10.1016/0165-1161(85)90015-9. [DOI] [PubMed] [Google Scholar]
- Hafer K, Iwamoto KS, Scuric Z, Schiestl RH. Adaptive response to gamma radiation in mammalian cells proficient and deficient in components of nucleotide excision repair. Radiat Res. 2007;168:168–174. doi: 10.1667/RR0717.1. [DOI] [PubMed] [Google Scholar]
- Hansch E, Forgo J, Murer H, Biber J. Role of microtubules in the adaptive response to low phosphate of Na/P; cotransport in opossum kidney cells. Pflügers Archiv European J Physiol. 1993;422:516–522. doi: 10.1007/BF00375080. [DOI] [PubMed] [Google Scholar]
- Heise M, Hutter-Paier B, Jerkovic L, Pfragner R, Windisch M, Becker-André M, Dieplinge H. Vitamin E binding protein afamin protects neuronal cells in vitro. J Neural Transm Suppl. 2002;62:337–345. doi: 10.1007/978-3-7091-6139-5_32. [DOI] [PubMed] [Google Scholar]
- Hooker AM, Bhat M, Day TK, Lane JM, Swinburne SJ, Morley AA, Sykes PJ. The linear no-threshold model does not hold for low-dose ionizing radiation. Radiat Res. 2004;162:447–452. doi: 10.1667/rr3228. [DOI] [PubMed] [Google Scholar]
- Ikushima T, Aritomi H, Morisita J. Radioadaptive response: efficient repair of radiation-induced DNA damage in adapted cells. Mutat Res. 1996;358:193–198. doi: 10.1016/s0027-5107(96)00120-0. [DOI] [PubMed] [Google Scholar]
- Ito M, Shibamoto Y, Ayakawa S, Tomita N, Sugie C, Ogino H. Low-dose whole-body irradiation induced radioadaptive response in C57BL/6 mice. J Radiat Res (Tokyo) 2007;48:455–460. doi: 10.1269/jrr.07022. [DOI] [PubMed] [Google Scholar]
- Iyer R, Lehnert BE. Alpha-particle-induced increases in the radioresistance of normal human bystander cells. Radiat Res. 2002;157:3–7. doi: 10.1667/0033-7587(2002)157[0003:apiiit]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- James SJ, Enger SM, Peterson WJ, Makinodan T. Immune potentiation after fractionated exposure to very low doses of ionizing radiation and/or caloric restriction in autoimmune-prone and normal C57Bl/6 mice. Clin Immunol Immunopathol. 1990;55:427–437. doi: 10.1016/0090-1229(90)90129-e. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search Analytical Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Klokov D, Criswell T, Leskov KS, Araki S, Mayo L, Booth-man DA. IR-inducible clusterin gene expression: a protein with potential roles in ionizing radiation-induced adaptive responses, genomic instability, and bystander effects. Mutat Res. 2004;568:97–110. doi: 10.1016/j.mrfmmm.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Lai XY, Bacalla RL, Blazer-Yost BL, Hong D, Mason SB, Witzmann FA. Characterization of the renal cyst fluid proteome in autosomal dominant polycystic kidney disease (ADPKD) patients. Proteomics Clin Appl. 2008;2:1140–1152. doi: 10.1002/prca.200780140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Gong P, Bernstein LR, Bi Y, Gong S, Cai L. Apoptotic cell death induced by low-dose radiation in male germ cells: hormesis and adaptation. Crit Rev Toxicol. 2007;37:587–605. doi: 10.1080/10408440701493061. [DOI] [PubMed] [Google Scholar]
- Liu HB, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lottscher M, Kaissling B, Biber J, Murer H, Levi M. Role of microtubules in the rapid regulation of renal phosphate transport in response to acute alterations in dietary phosphate content. J Clin Invest. 1997;99:1302–1312. doi: 10.1172/JCI119289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey TD. Physiological benefits from low levels of ionizing radiation. Health Phys. 1982;43:771–789. doi: 10.1097/00004032-198212000-00001. [DOI] [PubMed] [Google Scholar]
- Lyng FM, Maguire P, McClean B, Seymour C, Mothersill C. The involvement of calcium and MAP kinase signaling pathways in the production of radiation-induced bystander effects. Radiat Res. 2006;165:400–409. doi: 10.1667/rr3527.1. [DOI] [PubMed] [Google Scholar]
- Matsubara J, Turcanu V, Poindron P, Ina Y. Immune effects of low-dose radiation: short-term induction of thymocyte apoptosis and long-term augmentation of T-cell-dependent immune responses. Radiat Res. 2000;153:332–338. doi: 10.1667/0033-7587(2000)153[0332:ieoldr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Hormesis and disease resistance: activation of cellular stress response pathways. Human Experimental Toxicol. 2008;27:155–162. doi: 10.1177/0960327107083417. [DOI] [PubMed] [Google Scholar]
- Mitchel RE, Jackson JS, Morrison DP, Carlisle SM. Low doses of radiation increase the latency of spontaneous lymphomas and spinal osteosarcomas in cancer-prone, radiation-sensitive Trp53 heterozygous mice. Radiat Res. 2003;159:320–327. doi: 10.1667/0033-7587(2003)159[0320:ldorit]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Mitchel REJ. The dose window for radiation-induced protective adaptive responses. Dose Response. 2010;8:192–208. doi: 10.2203/dose-response.09-039.Mitchel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y. Oxidative stress, radiation-adaptive responses, and aging. J Radiat Res (Tokyo) 2004;45:357–372. doi: 10.1269/jrr.45.357. [DOI] [PubMed] [Google Scholar]
- Mothersill C, Seymour C. Survival of human epithelial cells irradiated with cobalt 60 as microcolonies or single cells. International J Radiat Biol. 1997;72:597–606. doi: 10.1080/095530097143095. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Kelle rA, Kolke E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Olivieri G, Bodycote J, Wolff S. Adaptive response of human lymphocytes to low concentrations of radioactive thymidine. Science. 1984;223:594–597. doi: 10.1126/science.6695170. [DOI] [PubMed] [Google Scholar]
- Osborn T, Verdrengh M, Stossel T, Tarkowski A, Bokarewa M. Decreased levels of the gelsolin plasma isoform in patients with rheumatoid arthritis. Arthritis Res Therapy. 2008;10:1–9. doi: 10.1186/ar2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka K, Koana T, Tauchi H, Sakai K. Activation of antioxidative enzymes induced by low-dose-rate whole-body gamma irradiation: adaptive response in terms of initial DNA damage. Radiat Res. 2006;166:474–478. doi: 10.1667/RR0561.1. [DOI] [PubMed] [Google Scholar]
- Pinto M, Azzam EI, Howell RW. Investigation of adaptive responses in bystander cells in 3D cultures containing tritium-labeled and unlabeled normal human fibroblasts. Radiat Res. 2010;174:216–227. doi: 10.1667/RR1866.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portess DI, Bauer G, Hill MA, O'Neill P. Low-dose irradiation of nontransformed cells stimulates the selective removal of precancerous cells via intercellular induction of apoptosis. Cancer Res. 2007;67:1246–1253. doi: 10.1158/0008-5472.CAN-06-2985. [DOI] [PubMed] [Google Scholar]
- Ramassamy C, Krzywkowski P, Averill D, Lussier-Cacan S, Theroux L, Christen Y, Davignon J, Poirier J. Impact of apoE deficiency on oxidative insults and antioxidant levels in the brain. Molec Brain Res. 2001;86:76–83. doi: 10.1016/s0169-328x(00)00268-0. [DOI] [PubMed] [Google Scholar]
- Redpath JL, Liang D, Taylor TH, Christie C, Elmore E. The shape of the dose-response curve for radiation-induced neoplastic transformation in vitro: evidence for an adaptive response against neoplastic transformation at low doses of low-LET radiation. Radiat Res. 2001;156:700–707. doi: 10.1667/0033-7587(2001)156[0700:tsotdr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Rithidech K, Au WW, Ramanujam VM, Whorton EB, Jr, Legator MS. Persistence of micronuclei in peripheral blood normochromatic erythrocytes of subchronically benzene-treated male mice. Environ Mol Mutagen. 1988;12:319–329. doi: 10.1002/em.2860120306. [DOI] [PubMed] [Google Scholar]
- Rithidech K, Honikel L, Rieger R, Xie WP, Fischer T, Simon SR. Protein expression profiles in mouse blood plasma following acute whole body exposure to 137Cs gamma rays. Int J Radiat Biol. 2009;85:432–447. doi: 10.1080/09553000902820390. [DOI] [PubMed] [Google Scholar]
- Rithidech KN, Honikel L, Whorton EB. mFISH analysis of chromosomal damage in bone marrow cells collected from CBA/CaJ mice following whole body exposure to heavy ions (56Fe ions) Radiat Environ Biophys. 2007;46:137–145. doi: 10.1007/s00411-006-0092-x. [DOI] [PubMed] [Google Scholar]
- Rithidech KN, Scott BR. Evidence for radiation hormesis after in vitro exposure of human lymphocytes to low doses of ionizing radiation. Dose-Response. 2008;6:252–271. doi: 10.2203/dose-response.07-024.Rithidech. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rithidech KN, Tungjai M, Whorton EB. Protective effect of apigenin on radiation-induced chromosomal damage in human lymphocytes. Mutat Res. 2005;585:96–104. doi: 10.1016/j.mrgentox.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan K, von Duyn A, Loos MJ, Natarajan AT. Adaptive response of human lymphocytes to low-level radiation from radioisotopes or X-rays. Mutat Res. 1989;211:7–12. doi: 10.1016/0027-5107(89)90101-2. [DOI] [PubMed] [Google Scholar]
- Scott BR. A biological-based model that links genomic instability, bystander effects, and adaptive response. Mutat Res. 2004;568:129–143. doi: 10.1016/j.mrfmmm.2004.06.051. [DOI] [PubMed] [Google Scholar]
- Scott BR, Di Palma J. Sparsely ionizing diagnostic and natural background radiation are likely preventing cancer and other genomic-instability associated diseases. Dose Response. 2006;5:230–255. doi: 10.2203/dose-response.06-002.Scott. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MJ, Carothers A, Dias A, Luis JH, Piper J, Boavida MG. Dose dependence of radiation-induced micronuclei in cytokinesis-blocked human lymphocytes. Mutat Res. 1994;322:117–128. doi: 10.1016/0165-1218(94)00019-0. [DOI] [PubMed] [Google Scholar]
- Sorensen KJ, Attix CM, Christian AT, Wyrobek AJ, Tucker JD. Adaptive response induction and variation in human lymphoblastoid cell lines. Mutation Res/Genetic Toxicol Environmental Mutagenesis. 2002;519:15–24. doi: 10.1016/s1383-5718(02)00110-9. [DOI] [PubMed] [Google Scholar]
- Sowa Resat MB, Morgan WF. Radiation-induced genomic instability: a role for secreted soluble factors in communicating the radiation response to non-irradiated cells. J Cellular Biochem. 2004;92:1013–1019. doi: 10.1002/jcb.20149. [DOI] [PubMed] [Google Scholar]
- Springer DL, Ahram M, Adkins JN, Kathmann LE, Miller JH. Characterization of medium conditioned by irradiated cells using proteome-wide, high-throughput mass spectrometry. Radiat Res. 2005;164:651–654. doi: 10.1667/rr3457.1. [DOI] [PubMed] [Google Scholar]
- Varés G, Wang B, Tanaka K, Kakimoto A, Eguchi-Kasai K, Nenoi M. Mutagenic adaptive response to high-LET radiation in human lymphoblastoid cells exposed to X-rays. Mutation Res/Fundamental Molecular Mechanisms Mutagenesis. 2011;706:46–52. doi: 10.1016/j.mrfmmm.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Wang G-J, Cai L. Induction of cell-proliferation hormesis and cell-survival adaptive response in mouse hematopoietic cells by whole-body low-dose radiation. Toxicol Sci. 2000;53:369–376. doi: 10.1093/toxsci/53.2.369. [DOI] [PubMed] [Google Scholar]
- Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- Whorton EB. Some experimental design and analysis considerations for cytogenetics studies. Environ Mutagen. 1985;7(Suppl 4):9–15. doi: 10.1002/em.2860070804. [DOI] [PubMed] [Google Scholar]
- Wiencke JK, Afzal V, Olivieri G, Wolff S. Evidence that the [3H]thymidine-induced adaptive response of human lymphocytes to subsequent doses of X-rays involves the induction of a chromosomal repair mechanism. Mutagenesis. 1986;1:375–380. doi: 10.1093/mutage/1.5.375. [DOI] [PubMed] [Google Scholar]
- Wolff S. Aspects of the adaptive response to very low doses of radiation and other agents. Mutat Res. 1996;358:135–142. doi: 10.1016/s0027-5107(96)00114-5. [DOI] [PubMed] [Google Scholar]
- Wu X, Kocher B, Wei Q, Hammer JA. Myosin Va associates with microtubule-rich domains in both interphase and dividing cells. Cell Motility Cytoskeleton. 1998;40:286–303. doi: 10.1002/(SICI)1097-0169(1998)40:3<286::AID-CM7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]