Abstract
Significant advances in our understanding of RNA architecture, folding and recognition have emerged from structure-function studies on riboswicthes, non-coding RNAs whose sensing domains bind small ligands and whose adjacent expression platforms contain RNA elements involved in the control of gene regulation. We now report on the ligand-bound structure of the Thermotoga petrophila fluoride riboswitch, which adopts a higher-order RNA architecture stabilized by pseudoknot and long-range reversed Watson-Crick and Hoogsteen A•U pair formation. The bound fluoride ion is encapsulated within the junctional architecture, anchored in place through direct coordination to three Mg2+ ions, which in turn are octahedrally coordinated to waters and five inwardly-pointing backbone phosphates. Our structure of the fluoride riboswitch in the bound state defines how RNA can form a binding pocket selective for fluoride, while discriminating against larger halide ions. The T. petrophila fluoride riboswitch most likely functions in gene regulation through a transcription termination mechanism.
The field of RNA structure, folding and recognition has been propelled forward by the discovery of metabolite-sensing bacterial non-coding RNA elements, termed riboswitches, that have been shown to impact on RNA-mediated gene regulation1,2. Most riboswitches are positioned within 5′-untranslated regions of genes associated with transport and metabolism of their cognate metabolites. Riboswitches interconvert between metabolite-free and -bound conformations depending on metabolite concentration, with the sensing domain involved in metabolite recognition, while the adjacent expression platform contains RNA elements that control translational initiation, transcription termination or ribozyme-mediated cleavage. To date, structure-function studies have been undertaken on riboswitches that target purines and their analogs, amino acids, coenzymes and phosphoamino sugars, as well as Mg2+ ions3,4. Structural studies on the compact ligand-bound sensing domains of the above riboswitches have elucidated the structural principles underlying RNA folding topology and recognition, whereby endogenous RNA molecules can fold and form pockets that recognize small metabolites and discriminate against closely related analogs5,6.
Of particular interest is how RNA as a negatively charged polyphosphate can bind ligands that are also negatively charged. The earliest studies to address this question focused on the cofactor thiamine pyrophosphate (TPP) riboswitch2, where structural insights established that a pair of hydrated Mg2+ ions mediated interactions between the diphosphates of TPP and guanine base edges (rather than backbone phosphates) of the RNA7,8. This structural information was critical for guiding subsequent studies on ligand-induced folding of the TPP riboswitch9,10. Similar structural principles involving a bridging hydrated Mg2+ ion were also utilized for recognition of the monophosphate of flavin mononucleotide (FMN) by its riboswitch11,12.
Recently, a riboswitch associated with crcB motif non-coding RNAs from Pseudomonas syringae has been identified that targets fluoride ion with a Kd of approx. 60 μM and discriminates against other halogen ions13. This riboswitch is common to bacterial and archaeal species and found to activate the expression of genes that encode putative fluoride transporters. Given the small size and negative charge of the fluoride ion, it seems remarkable that RNA can form a small enough pocket to target it and discriminate against larger halide ions.
Structure of fluoride-bound riboswitch
The conserved secondary fold of crcB RNA motif fluoride-sensing riboswitches was deduced following sequence conservation and covariational analysis amongst bacterial and archaeal species, as well as in-line probing and mutational studies13. The resulting analysis identified two helical stems connected by a large asymmetric internal loop, with overhangs at both ends, with that at the 5′-end capable of adopting pseudoknot-like higher-order interactions.
We used isothermal titration calorimetry (ITC) to establish that the 52-mer RNA sequence corresponding to the sensing domain of the T. petrophila fluoride riboswitch (Fig. 1a) bound fluoride ion (on addition of KF) with a Kd of 134 ± 9 μM under 5 mM Mg2+ conditions (Fig. 1b). The stoichiometry of binding approaches 1:1 (N = 0.87), with estimated thermodynamic parameters of ΔH = −2.5 ± 0.1 Kcal/mol and ΔS = 9.3 cal/mol/deg. Similar binding parameters were observed under 1 mM Mg2+ conditions, but no binding was observed in the absence of Mg2+ ion (Supplementary Fig. 1a, b, respectively).
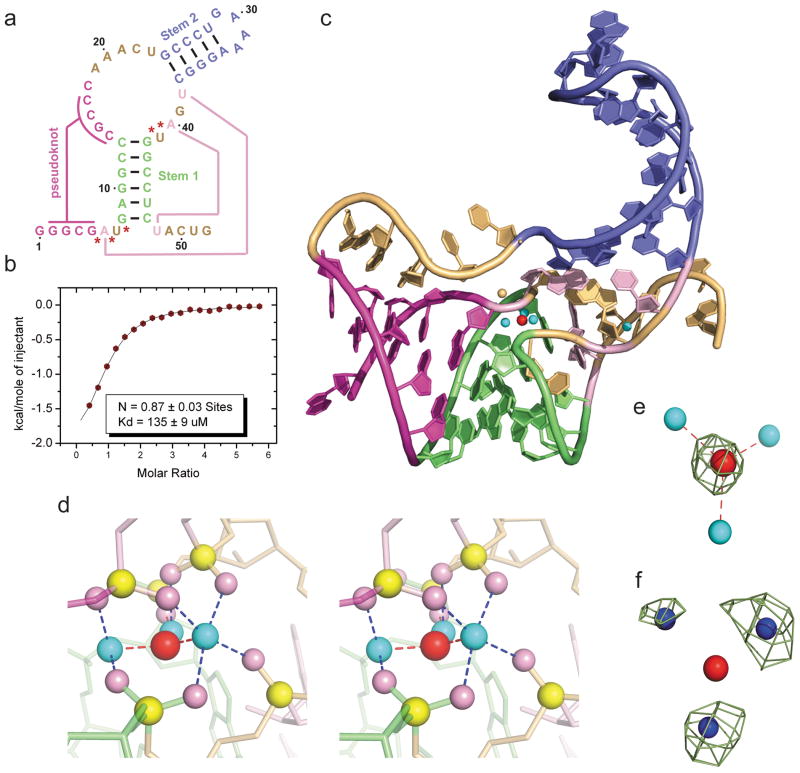
Figure 1. Sequence, binding affinity and structure of the sensing domain of the T. petrophila fluoride riboswitch in the ligand bound state.
a, Secondary structure schematic of the 52-nt sensing domain of the T. petrophila fluoride riboswitch used in this study. The color-coding highlights regular stem segments (blue and green) and long-range pseudoknot pairing interactions (magenta and pink; connected by lines), and bases not involved in pairing interactions (gold). Five phosphates, whose non-bridging oxygens form a shell around the three hydrated metal ions (in cyan), with the metal ions in turn coordinated to the fluoride ion (in red), are labeled by red stars. b, ITC-binding curve for binding of KF to the T. petrophila fluoride riboswitch in 5 mM Mg2+-containing buffer (for experimental conditions, see Methods section). c, 2.3 Å crystal structure of the fluoride riboswitch in the ligand bound state. The color-coding of RNA segments follows that shown in panel a, with the fluoride ion shown by a red ball and directly-coordinated metal ions by cyan balls. d, A close-up stereo view of the ligand-binding pocket in the same perspective as in panel c, with the emphasis on the fluoride ion, three coordinating metal ions and five inwardly-pointing backbone phosphates. e, Fo-Fc omit electron density map contoured a 4σ level calculated following deletion of the fluoride ion. f, Anomalous density at 9σ level at positions of metal ions 1, 2 and 3 for crystals of the complex soaked in 50 mM Mn2+-containing solution.
The sensing domain of the T. petrophila fluoride riboswitch (Fig. 1a) yielded crystals in the presence of fluoride ion that diffracted to 2.3 Å resolution. We solved the structure of this fluoride riboswitch by cocrystallizing it with Ir(NH3)63+, and capitalizing on the anomalous properties of iridium to solve the phase problem. (Supplementary Fig. 2). The three dimensional structure in the bound state is shown in Fig. 1c with different elements color-coded as shown in Fig. 1a. The most striking feature is that the bound fluoride ion (red ball, Fig. 1c), positioned within the center of the riboswitch fold, is surrounded and directly coordinated by three metal ions (cyan balls, Fig. 1c; metal-fluoride distances of 1.8–2.0 Å). A close-up stereo view of the ligand-binding pocket in the same perspective as in Fig. 1c, with the emphasis on the fluoride ion (in red), three coordinating metal ions (in cyan) and five inwardly-pointing backbone phosphates (non-bridging oxygens in pink and phosphorus atoms in yellow) is shown in stereo in Fig. 1d.
Fluoride coordinated to three Mg2+ ions
We have identified the fluoride ion binding site in the Fo-Fc omit map for fluoride ion (Fig. 1e) and assign all three metal ions coordinated to the fluoride ion, to Mg2+ ions, based on observation of anomalous signals following soaking of the crystals in 50 mM Mn2+ ion-containing solution (Fig. 1f and Supplementary Fig. 3). The additional metal ion (in orange, Fig. 1c) positioned close by but not coordinated to the fluoride ion (metal-fluoride distance of 4.0 Å), is assigned to a K+ cation (buffer contained K-acetate), based on the observed anomalous signal at its position following soaking of crystals in Cs+ ion-containing (Supplementary Fig. 4a) and Tl+ ion-containing (Supplementary Fig. 4b) solutions.
Riboswitch adopts pseudoknot scaffold
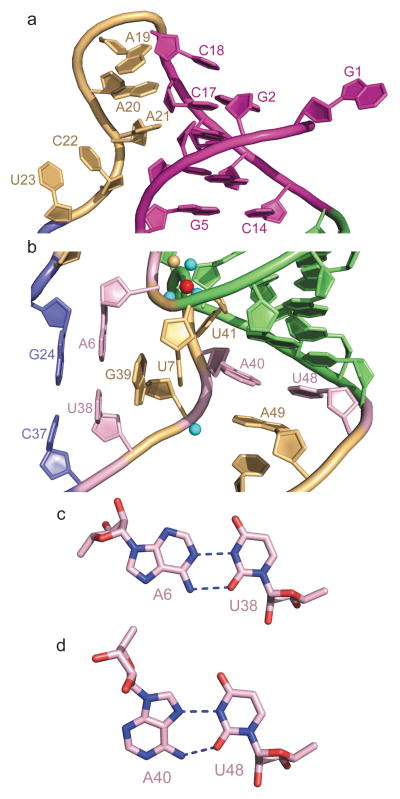
The higher order fold of the fluoride-bound riboswitch is stabilized by pseudoknot formation involving residues G2-G3-C4-G5 of the 5′-overhang segment and residues C14-G15-C16-C17 of the internal loop, thereby forming a regular duplex composed of four stacked Watson-Crick G-C base pairs (in magenta, Fig. 2a). This validates the prediction of pseudoknot formation amongst the same residues13, though our crystal structure shows stabilization by four rather than the postulated five base pairs anticipated in solution. In this regard, 5′-terminal G1 and 3′-terminal U51 and G52 are involved in crystal packing contacts in our structure of the fluoride riboswitch (Supplementary Fig. 5). None of the residues in the C18 to U23 segment of the internal loop are involved in pairing interactions, but rather C18 stacks on terminal Watson-Crick G2-C17 pair, with a sharp turn at the C18-A19 step, followed by continuous stacking within the A19-A20-A21 and C22-U23 steps (Fig. 2a). The junctional architecture in the vicinity of the fluoride-binding site is additionally stabilized by formation of long-range single-base pseudo-knot like pairing between A6 and U38 (predicted previously13) and between A40 and U48 (Fig. 2b). Both form non-canonical pairs, with A6•U38 pairing through a reversed Watson-Crick alignment (Fig. 2c) and A40•U48 pairing through a reversed Hoogsteen alignment (Fig. 2d). Note that unpaired U7 and G39 are mutually interdigitated (Fig. 2b) and contribute to formation of the junctional architecture. The tracing of these RNA segments in the 2Fo-Fc electron density maps of the fluoride riboswitch are shown in Supplementary Fig. 6a, b.
Figure 2. Details of long-range interactions within the structure of the T. petrophila fluoride riboswitch in the ligand bound state.
a, An expanded region of the structure highlighting pseudoknot formation involving residues G2-G3-C4-G5 of the 5′-overhang segment and residues C14-G15-C16-C17 of the large internal loop (in magenta), as well as continuous stacking within the A19-A20-A21 and C22-U23 steps. b, An expanded region of the structure highlighting long-range single-base pseudo-knot like pairing between A6 and U38, and between A40 and U48. Note the mutual interdigitation between unpaired U7 and G39 that contribute to formation of the junctional architecture. c, Long-range reversed Watson-Crick A6•U38 pair formation. d, Long-range reversed Hoogsteen A40•U48 pair formation.
Coordination of Mg2+ ions by phosphates
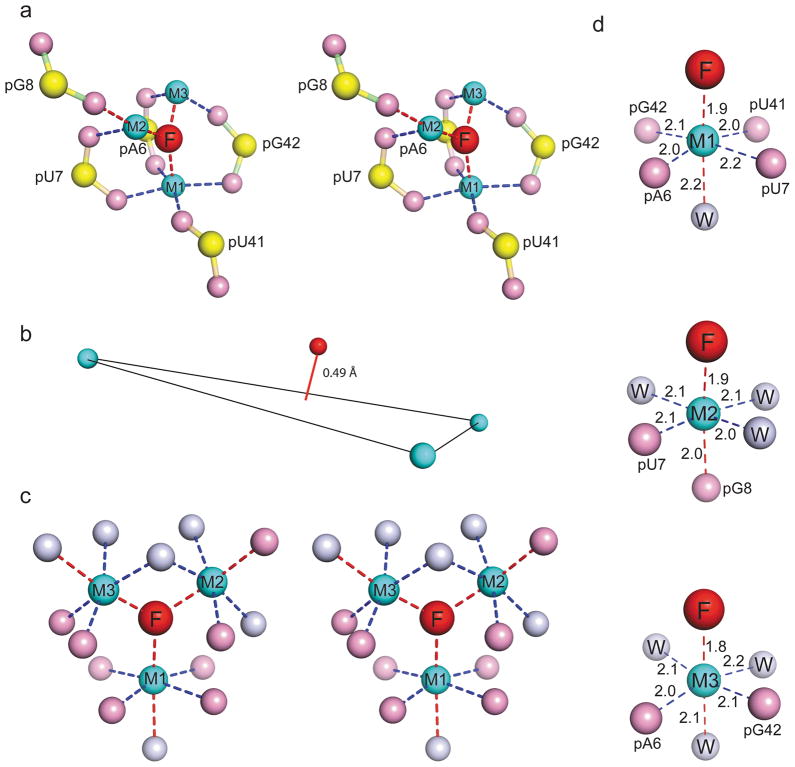
The negatively charged fluoride ion (in red) is directly coordinated to three Mg2+ ions (in cyan) labeled M1, M2 and M3 (stereo view in Fig. 3a), with the fluoride ion positioned somewhat out of the plane (0.49 Å) formed by the three Mg2+ ions (Fig. 3b). A stereo-view of the corresponding Fo-Fc omit electron density maps for fluoride ion, three Mg2+ ions and bound waters are shown in Supplementary Fig. 7a (contoured at 3σ) and 7b (contoured at 7σ). The three Mg2+ ions in turn are coordinated by waters and five inwardly-pointing non-bridging phosphate oxygens (in pink; phosphorus in yellow), with three of these phosphates involved in bidentate coordination (pA6, pU7 and pG42), while two others involved in monodentate coordination (pU8 and pU41) (Fig. 3a). A stereo-view of the corresponding Fo-Fc omit electron density maps for the five nucleotides with inwardly-pointing phosphates are shown in Supplementary Fig. 7c (contoured at 3σ) and 7d (contoured at 5σ).
Figure 3. Details of fluoride ion binding site in the T. petrophila fluoride riboswitch in the ligand bound state.
a, An expanded stereo view highlighting the non-bridging phosphate oxygens directly coordinated to the three Mg2+ ions labeled M1, M2 and M3, that are in turn directly coordinated to the fluoride ion. In terms of nomenclature, the phosphate labeled pA6 corresponds to the G5pA6 step. Three of the phosphates are involved in bidentate coordination with the metals, while two are involved in mondentate coordination. b, The fluoride ion is positioned 0.49 Å above the plane formed by the three metal ions. c, A stereo view of the coordination geometries of fluoride ion with surrounding Mg2+ ions, and in turn of Mg2+ with surrounding non-bridging phosphate oxygens and waters. Metal coordination within square-planar positions are shown by dashed blue lines, while coordination in apical positions are shown by dashed red lines in panels a, c and d. d, These panels show the octahedral coordination geometries around Mg2+ ions M1, M2 and M3, together with coordination distances in Å.
In essence, our structure identifies a unique solution for how a negatively-charged RNA scaffold can target a negatively-charged fluoride ligand. The fluoride ion is surrounded by and coordinated to an inner shell of three Mg2+ ions, which in turn are surrounded and coordinated to an outer shell of five backbone phosphates and waters. Notably, the five participating phosphates are located within two distinct segments of the sequence, with three of them residing within the 5′-overhang G5pA6pU7pG8 segment, while two other reside within the A40pU41pG42 internal loop segment, as labeled in Fig. 1a by red asterisks. Because of the unique orientation of these five inwardly-directing backbone phosphates, 8 of the 14 internal loop residues are stacked but not involved in hydrogen-bond pairing, and differ from large internal loops of other structurally characterized riboswitches, where ligand-binding results in a compaction mediated by maximal hydrogen-bond pairing and stacking of loop residues14–19.
Metal ions and their coordinated waters were identified based on 2Fo-Fc and Fo-Fc maps guided by the coordination geometries. The positioning of fluoride ion, non-bridging phosphate oxygens and water molecules that constitute the three Mg2+ ion cluster that is coordinated to the fluoride ion is shown in stereo in Fig. 3c, with individual Mg2+ ions adopting the anticipated octahedral-like alignments as shown in Fig. 3d. Mg2+ coordination within the square-planar arrangements are shown by dashed blue lines, while coordination in apical positions are shown by dashed red lines, together with distances listed in Å in Fig. 3d. Notably, the fluoride riboswitch does not bind chloride ion (added KCl) in the presence of 20 mM Mg2+ as monitored by ITC (Supplementary Fig. 1c). In addition, it does not bind fluoride ion (added KF) in the presence of 20 mM Li+ ions (Supplementary Fig. 1d), in contrast to the observed binding under Mg2+ ion conditions (Fig. 1b).
Fluoride-Mg2+ coordination in proteins
Of additional note, the fluoride ion adopts an apical position in the octahedral coordination geometries for all three Mg2+ ions (Fig. 3d). Mg2+ ion M1 is coordinated by four non-bridging phosphate oxygens aligned in a square planar arrangement, while Mg2+ ions M2 and M3 are coordinated by two non-bridging phosphate oxygens (Fig. 3d). Additional support for our model of the coordination geometry of three Mg2+ ions around fluoride in the fluoride riboswitch comes from the highly similar coordination geometry observed in the 1.9 Å structure of fluoride-inhibited pyrophosphotase (crystals grown from 1 mM MnCl2 and 5 mM NaF-containing buffer)20. Here, a fluoride ion is coordinated by a similar three-metal ion arrangement in the pyrophosphase system (Supplementary Fig. 8). In the fluoride-inhibited pyrophosphotase structure, two Mn2+ ions and one Na+ ion are coordinated by oxygens atoms from one POP molecule and from four aspartate carboxylate groups, and water molecules (Supplementary Fig. 8a).
We are aware that fluoride anion is a potent hydrogen bond acceptor13,21, but currently have no evidence in support of direct F--H hydrogen bonding in the structure of the fluoride riboswitch, nor was such hydrogen bonding reported for the fluoride-inhibited pyrophosphatase system20.
Previous studies of metals in RNA folding, stability and catalysis have focused on monovalent and divalent cations22, including Mg2+ clusters bridging closely-positioned phosphates in 5S RNA23, P4-P5-P6 fragment of group I introns24 and Mg2+-sensing riboswitches25,26. Our current contribution based on the discovery of a fluoride riboswitch13, has defined how a RNA scaffold combines an inner shell of metal ions and an outer shell of phosphates to completely encapsulate a fluoride ion.
Folding and mechanism of action
We have recorded 900 MHz imino proton NMR spectra of the fluoride riboswitch in the free and fluoride bound states, which establish a conformational transition (chemical shift changes) and compaction (additional peaks) through higher order structure generation on complex formation, with slow exchange between the free and bound forms (Supplementary Fig. 9). We have not been able to crystallize the ligand-free form of the fluoride riboswitch in the presence of Mg2+ ions. In the absence of this structure, we can only speculate on the folding energy landscape of the fluoride riboswitch as to how it dynamically assembles to recognize its ligand. This could occur through either hierarchical or simultaneous positioning of Mg2+ ions, RNA functional groups and ligand for recognition. This issue of folding energy landscape, as well as energetic costs associated with transfer of the fluoride ion from free to the riboswitch-bound state, could perhaps be addressed in the future by comprehensive single molecule studies.
Previous functional studies implied that the B. cereus and P. syringae fluoride-responsive riboswitches control gene expression by regulating transcription termination and translational initiation, respectively13. To deduce mechanistic insights into gene regulation by the T. petrophila fluoride riboswitch from a structural perspective would require crystallographic characterization of the complete riboswitch containing both the sensing domain and expression platform in the ligand-free and bound states, which is beyond our current capabilities. Nevertheless, we note that that the sensing domain and adjacent expression platform of the T. petrophila fluoride riboswitch could interconvert between two conformations, one in which stem 1 forms in the presence of bound fluoride (ON state) and an alternate conformation at low fluoride concentrations where stem 1 is disrupted as a result of forming a rho-independent transcription terminator (OFF state) involving a stable stem loop ending in uracils at the 3′-end (Supplementary Fig. 10). Thus, it appears that both the B. cereus (supported by functional data)13 and T. petrophila fluoride riboswitches are likely to adopt a transcription termination mechanism for gene regulation.
METHODS SUMMARY
Details of RNA preparation, purification and complex formation, as well as crystallization, x-ray data collection and refinement are described in detail in Full Methods.
Full Methods and any associated reference are available in the on line version of the paper at www.nature.com/nature
METHODS
RNA preparation, purification and complex formation
The crcB motif of the T. petrophila fluoride riboswitch followed by the HDV ribozyme was transcribed in vitro using T7 RNA polymerase27. The transcribed RNA was purified by denaturing polyacrylamide gel electrophoresis (PAGE), followed by anion-exchange chromatography and ethanol precipitation. To generate the complex of the sensing domain of the T. petrophila fluoride riboswitch with fluoride ion, 10 mM KF was added to the buffer consisting of 100 mM potassium-acetate, pH 6.8 and 5 mM MgCl2. After annealing at 60 °C for 10 min, the complex was purified by gel-filtration chromatography, prior to setting up crystallization trials.
Crystallization
Crystals of the fluoride anion-bound T. petrophila fluoride riboswitch grew at 20 °C over a period of 1 week using sitting-drop vapor diffusion approach after mixing the complex at an equimolar ratio with the reservoir solution containing 0.1 M MES-sodium, pH 6.5, 35–40% (w/v) MPD. To generate heavy-atom-bound crystals, 5 mM Ir(NH3)63+ was mixed with 0.5 mM complex before setting up crystallization trials. The Ir(NH3)63+ bound crystals grew from a solution containing 0.1 M sodium-cacodylate, pH 7.0, 20 mM spermine, 0.2 M strontium chloride and 20% MPD over a period of 4 days.
For heavy atom and cation soaking, crystals grown from 0.1 M sodium-cacodylate, pH 7.0, 20 mM spermine, 160 mM KCl, 50 mM MgCl2 and 20% MPD were transferred into the mother solution of the crystals with MgCl2 replaced with 50 mM MnCl2, or with KCl replaced with 50 mM Tl-acetate or 100 mM CsCl for 24 hr.
For cryoprotection, crystals were passed through several 5 μL drops of the stabilizing solution, which was the reservoir solution with MPD replaced by 30% MPD and 5% glycerol.
X-ray data collection and refinement
X-ray diffraction data were collected on flash frozen crystals of the fluoride anion-bound T. petrophila fluoride riboswitch at NE-CAT beamlines at the Advanced Photon Source, Argonne National Laboratory and processed using the HKL2000 program (HKL Research). The structure (space group: P21212) was determined using SAD technique employing anomalous signal from four iridium atoms using HKL2MAP program28 and PHENIX29 suite. The RNA model was built in COOT30 and refined in PHENIX29 and REFMAC31 using 2.3 Å native data set of the ligand-bound fluoride riboswitch. Metal ions and their coordinated waters were identified based on 2Fo-Fc and Fo-Fc maps guided by the coordination geometries. Fluoride ion was added to the model at the last stage based on the experimental and refined maps, coupled with electrostatic analysis. The x-ray statistics of the native and iridium-containing crystals are listed in Supplementary Table 1. Anomalous data sets of Mn2+, Cs+, and Tl+ were collected at the wavelength of 1.7712 Å. The x-ray statistics of the complex crystals soaked in Mn2+-, Cs+-, and Tl+-containing solutions are listed in Supplementary Table 1. Mn2+, Cs+, and Tl+ cations were positioned based on the anomalous electron density maps (Supplementary Figs. 3, 4a and 4b). Cations were interpreted as Mg2+ or K+ on the basis of the anomalous maps of their mimics, coordination geometry and distances32.
NMR Spectra
Imino proton NMR spectra (10 to 15 ppm) of the fluoride riboswitch in the free and bound state were recorded on a 900 MHz Bruker NMR spectrometer with cryoprobe using a jump-and-return pulse for H2O solvent suppression. NMR spectra were recorded in buffer containing 50 mM K-acetate-d3, 5 mM Mg-sulfate, 90%H2O/10%D2O, pH 6.8 at 25 °C.
Isothermal Titration Calorimetry
All experiments were performed on a Microcal ITC200 calorimeter at 20 °C. Prior to titration, 0.2–0.3 mM RNA samples of the fluoride-bound riboswitch were dialyzed overnight at 4 °C against experimental buffer containing 50 mM potassium acetate, pH 6.8, and 0 to 20 mM MgSO4 or other cations to remove bound fluoride. RNAs were refolded by heating at 60 °C for 10 min and followed by cooling on ice. For measurements, KF dissolved in the dialysis buffer at 5 mM concentration, was typically titrated into the RNA in the sample cell (V = 207 μL) by 20 serial injections of 2 μl each, with a 0.5 μl/s rate, 180 s intervals between injections, and a reference power of 6 μcal/s. The thermograms were integrated and analyzed by using Origin 7.0 software (Microcal, Inc.).
Supplementary Material
Acknowledgments
We thank Jonathan Goldberg and Christopher Lima for discussion and helpful insights, Yizhou Liu for recording imino proton NMR spectra on a 900 MHz Bruker NMR spectrometer at the New York Structure Center, and the personnel of the synchrotron beamlines 24-ID-C/E at the Advanced Photon Source, Argonne National Laboratory for their assistance. This research was funded by NIH grant GM34504 to D.J.P.
Footnotes
Author Contributions A.R. generated and purified RNA constructs, grew diffraction quality crystals, collected synchrotron data sets and solved the structure of the fluoride riboswitch in the bound state under the supervision of D.J.P. K.R. assisted in crystallographic aspects of the structure determination, including finding unique solutions to the positioning of the Mg and fluoride ions. D.J.P. wrote the manuscript with the assistance of the other authors, all of whom discussed the results and commented on the manuscript.
Author Information Atomic coordinates of the structure of the fluoride riboswitch in the bound state have been deposited in the RCSB Protein Data Bank under the accession code 4ENC for the native structure and 4ENB for the Ir(NH3)63+-containing structure of the fluoride-bound riboswitch, as well as 3VRS, 4ENA and 4EN5 for crystals of the complex soaked in Mn2+-, Cs+-, and Tl+-containing solutions. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Mironov AS, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 2.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 3.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Ann Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 5.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montange RK, Batey RT. Riboswitches: Emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 7.Serganov A, et al. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thore S, Leinungdut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 9.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anthony PC, Perez CF, Garcia-Garcia C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc Natl Acad Sci USA. 2012;109:1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler WC, Cohen-Chalamish S, Breaker RR. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 12.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker JL, et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandal M, et al. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 15.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxantine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 16.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 18.Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heikinheimo P, et al. Toward a quantum-mechanical description of metal-assisted phosphoryl transfer in pyrophosphatase. Proc Natl Acad Sci USA. 2001;98:3121–3126. doi: 10.1073/pnas.061612498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Auffinger P, Hays FH, Westhof E, Ho PS. Halogen bonds in biological molecules. Proc Natl Acad Scis USA. 2004;101:16789–16794. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanna R, Doudna JA. Metal ions in ribozyme folding and catalysis. Curr Opin Chem Biol. 2000;4:166–1170. doi: 10.1016/s1367-5931(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 23.Correll CC, Freeborn B, Moore PB, Steitz TA. Metals, motifs and recognition in the crystal structure of a 5S RNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 24.Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;7:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- 25.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg2+ Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 26.Dann CE, III, et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 27.Pikovskaya O, et al. Preparation and crystallization of riboswitch-ligand complexes. Methods Mol Biol. 2009;540:115–128. doi: 10.1007/978-1-59745-558-9_9. [DOI] [PubMed] [Google Scholar]
- 28.Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 29.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:212–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 32.Feig AL, Uhlenbeck OC. In: The RNA World. 2. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor Laboratory Press; 1999. pp. 287–319. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.