Abstract
An increasing number of studies involve integrative analysis of gene and protein expression data taking advantage of new technologies such as next-generation transcriptome sequencing (RNA-Seq) and highly sensitive mass spectrometry (MS) instrumentation. Thus, it becomes interesting to revisit the correlative analysis of gene and protein expression data using more recently generated datasets. Furthermore, within the proteomics community there is a substantial interest in comparing the performance of different label-free quantitative proteomic strategies. Gene expression data can be used as an indirect benchmark for such protein-level comparisons. In this work we use publicly available mouse data to perform a joint analysis of genomic and proteomic data obtained on the same organism. First, we perform a comparative analysis of different label-free protein quantification methods (intensity-based and spectral count based, and using various associated data normalization steps) using several software tools on proteomic side. Similarly, we perform correlative analysis of gene expression data derived using microarray and RNA-Seq methods on genomic side. We also investigate the correlation between gene and protein expression data, and various factors affecting the accuracy of quantitation at both levels. It is observed that spectral count-based protein abundance metrics, which are easy to extract from any published data, are comparable to intensity-base measures with respect to correlation with gene expression data. The results of this work should be useful for designing robust computational pipelines for extraction and joint analysis of gene and protein expression data in the context of integrative studies.
INTRODUCTION
There is a significant interest in high throughput quantitative methods for analyzing gene and protein expression in complex biological systems. In recent years, both genomic and proteomic technologies have improved owing to such new developments as next-generation transcriptome sequencing (RNA-Seq) and highly sensitive mass spectrometry (MS) instrumentation. Thus, it becomes interesting to revisit the correlative analysis of gene and protein expression data using more recently generated datasets. Furthermore, within the proteomics community there is a substantial interest in comparing the performance of different label-free quantitative proteomic strategies. Gene expression data can be used as an indirect benchmark for such protein-level comparisons.
On the proteomic side, liquid chromatography- tandem mass spectrometry (so called LC-MS/MS or shotgun proteomics) remains the method of choice for large-scale protein identification. With respect to protein quantification, label-free MS-based quantification strategies have grown in popularity as alternatives to label-based approaches [1]. There are two major approaches for label-free protein quantification: using integrated peptide ion intensities extracted from the first stage (MS1) spectra [2–5], or using spectral counts (i.e. counting the number of MS/MS spectra identifying peptides from a particular protein) [6–8]. There is a great deal of interest in performing a comparative analysis of intensity-based and spectral count based measures, as well as various normalization steps associated with each method [9–14].
On the gene expression side, next-generation sequencing has recently emerged as a promising alternative to established microarray-based methods [15]. In RNA-Seq, millions of short nucleotide fragments (referred to as “reads”) are aligned to the genome. Gene expression levels are then established by counting the number of reads for each gene. The method can detect more exons and alternative splicing events than microarray technology and generally has a low error rate [15]. The development of improved statistical and computational methods for performing count-based gene expression analysis is an active are of research.
In this work, we use publicly available mouse data to perform a joint analysis of genomic and proteomic data obtained on the same organism. The focus of this analysis is twofold. First, we perform a comparative analysis of different label-free protein quantification methods using several software tools on proteomic side; and perform correlation analysis of gene expression data derived using microarray and RNA-Seq methods on genomic side. Second, we seek to gain a better understanding of the degree of correlation between gene and protein expression data. Early studies, based on data generated using gene expression microarrays and low sensitivity proteomic platforms, generally showed a low correlation [16–18]. More recent studies, however, suggested that the correlation may be significantly higher than previously thought, at least for a certain cases of proteins [19],[20]. This was further investigated in a series of recent studies showing that protein and transcript levels are linked but regulated by a series of dynamic and complex processes, including protein physico-chemical and structural properties and mRNA and protein degradation rates [21–24]. Here, by means of comparative analysis of two data types, we supplement previous efforts by investigating various factors affecting the accuracy of quantitation both at gene and protein levels. In doing so, we attempt to minimize the number of biological factors affecting the correlation by focusing on genes and proteins from a single cellular compartment, mouse mitochondria.
MATERIALS AND METHODS
Experimental data
The proteomic dataset was taken from Ref. [25] which comprehensively analyzed mouse mitochondrial proteins in various mouse tissues. In the original study, MS data were combined with other genome-scale datasets, including an extensive GFP tagging study, to define a set of 1098 mitochondrial genes [25] (MitoCarta database). Here we have selected MS data from two tissues only, brainstem and liver. For each tissue, the protein sample (mitochondrial fraction) was first separated using a 1D SDS-PAGE gel. Each gel was then cut into 20 bands, proteins extracted from each band were trypsin digested, and the digests analyzed using an LTQ-Orbitrap mass spectrometer.
Microarray gene expression profiles were obtained from mRNA expression data (gcRMA) in the GNF1M tissue atlas [26] (http://wombat.gnf.org/). For mRNA-Seq, raw short reads generated from Illumina/Solexa sequencer (San Diego, CA) as well as processed data RPKM (reads per kilobase of exon model per million mapped reads) gene expression values [15]) for brainstem and liver mouse tissues were obtained from (http://woldlab.caltech.edu/rnaseq/).
Protein identification
MS/MS spectra for brainstem and liver tissue were searched using X! TANDEM [27] with the k-score plugin [28]. Searches were done using the IPI mouse protein sequence database v3.50 with reverse protein sequences appended as decoys [29]. Each search was performed with the following parameters: parent mass error of +/− 100 ppm, fragment ion monoisotopic error of 0.8 Da; tryptic digest allowing for 1 missed cleavage; allowing for potential modification of +16 Da on methionine and +42 Da on the N-terminus of the peptide. The search results were then processed using TPP tools [30] PeptideProphet [31] and ProteinProphet [32]. ProteinProphet collapses proteins that share common peptide evidence into protein groups [33]. From this point on, references to proteins refer to these protein groups. At the protein level, proteins identified with ProteinProphet probability > 0.9 (decoy estimated [34] estimated FDR< 1% in both brainstem and liver tissues) were selected for protein abundance calculation. For proteins passing this threshold, their associated peptides with PeptideProphet probability >0.5 (to achieve a peptide-level probability FDR threshold < 5% for spectra in both brainstem and liver tissues) were considered for protein abundance estimation. Allowing lower probability peptide threshold (for proteins identified with high confidence) increases the number of peptide features that could be retrieved, and leverages the under-sampling problem for both intensity-based and spectral count-based methods. Note that raw MS data was reanalyzed to extract quantitative information using alternative methods, but otherwise all analysis was limited to proteins identified with high confidence and classified as mitochondrial proteins in the original study [25].
Intensity-based label-free quantification
Peptide features were extracted using label-free quantification software tools msInspect [3] and msBID [35]. msInspect first uses peptide identifications from MS/MS spectra to generate an accurate mass and time tag (AMT) database. Then it identifies peptide locations from MS1 data, and generates LC-MS peptide features. These LC-MS peptide features are then matched against the AMT database to extract peak features for each peptide. msBID program extracts peptide features in a similar way as msInspect, albeit using a different peak detection and intensity normalizing approach.
Each peptide feature was extracted from the MS1data in two ways: ‘apex’ (the intensity at the height of the peak representing the peptide signal in the MS1 spectrum) and ‘area’ (area under the curve). Peptide intensities (both apex and area) were also recorded after normalization. For normalization, peptide intensities were divided by the sum of all the peptide intensities (peptides identified with probability above 0.5) in the entire LC-MS run. Two different approaches were used to combine peptide-level intensity features to compute protein abundance. The first method is simply to sum the three largest peptide intensities (similar to [36]) per protein. The second method is to sum over all peptides then dividing by the protein's length. Thus, there were 8 configurations in total for intensity-based protein quantification (apex/area; with or without normalization; top3/all peptides) for both msInspect and msBID tools. In addition, we used the protein abundance estimations as provided in the MitoCarta database [25]. These estimates were generated by SpectrumMill software (Agilent Technologies, Santa Clara, California) and were based on the total peptide intensity per protein.
Spectral count-based label-free quantification
Spectral counting [6] is based on the assumption that the MS/MS sampling rate of a particular peptide is related to the abundance of the peptide represented by its precursor ion in the sample mixture. The abundance of a peptide is measured as the number of peptide-to-spectrum matching (PSM) events. A protein's abundance is measured by the total number of MS/MS spectra associated with it from all of its supporting peptides. Spectral counts were further normalized to derive the normalized spectral abundance factor (NSAF) [7]. The NSAF method involves normalizing for protein length and the total number of identified MS/MS spectra in an experiment. This length normalization is executed since longer proteins are expected to produce more peptides.
Gene expression analysis
Gene expression by mRNA-Seq was taken from Ref. [15]. It was computed based on aligning short reads (each of 25 bases) to mRNA sequences, recreating the gene models based on the compilation of aligned short reads, and then computing the expression profile of these genes. For each of the genes in brainstem and liver tissues, gene expressions based on mRNA-Seq have been measured by RPKM values [15] for the gene model. Three different sets of RPKM values were reported in [15]: first, expanded, and final. Except where noted, all analysis was performed using final values. Gene expression by microarray was retrieved directly from GNF1M tissue atlas [26].
Linking gene and protein data
The lists of identified proteins and the quantification data were parsed into a relational database. The resulting database table mapped protein groups to all of their associated gene symbols. Due to protein isoforms, a single gene symbol could be associated with multiple protein identifiers. In such cases the protein identifiers with the largest protein abundance values were chosen as the representative records.
RESULTS AND DISCUSSION
Overview of the analysis procedure
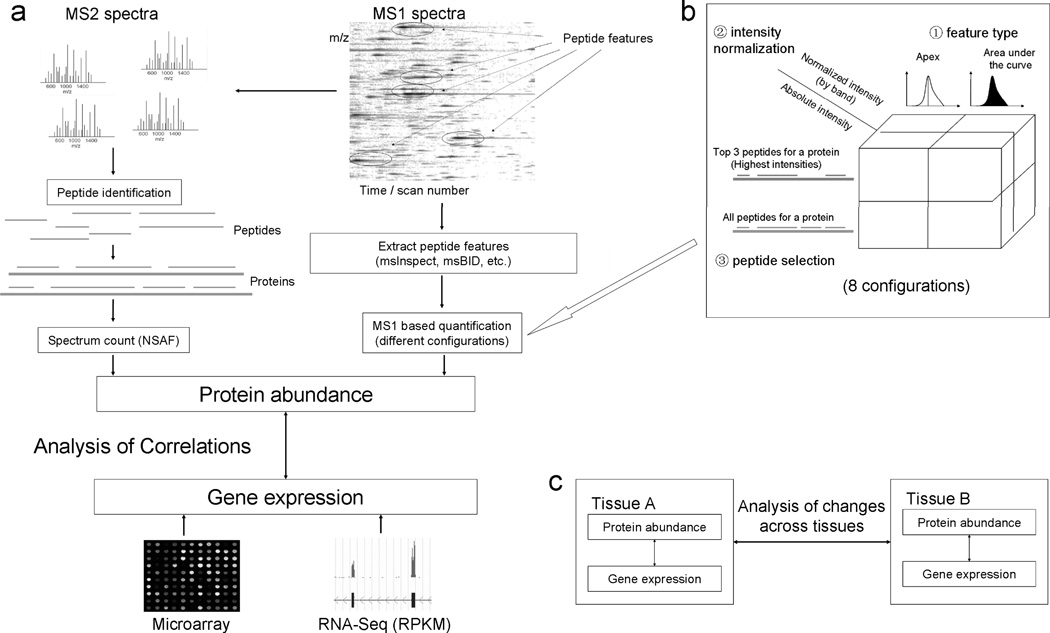
In this study, we have conducted a comprehensive comparison of gene expression with protein abundance for mouse mitochondrial genes (Figure 1). Mass spectrometry data for protein abundance analysis were retrieved from the MitoCarta database [25]. All MS/MS spectral searches were performed using X! TANDEM and post processed through the TPP tools [30] (see Materials and Methods for details). Two representative MS1 (i.e. intensity-based) based quantification tools, msInspect and msBID, were used to extract peptide features from MS1 spectra, and protein abundances were computed from these peptide features based on different configurations (Figure 1a). There were a total of 8 configurations for each of the two tools used, generated by extracting peptide features by apex or area, calculating peptide abundance with or without total peptide intensity normalization step, and measuring protein abundance based on either top three most intense peptides or using all peptides with length normalization (Figure 1b). Gene expression data for mouse mitochondrial genes were retrieved from both microarray and RNA-Seq data. Protein abundances computed by different methods were then correlated with gene expression data generated by both microarray and mRNA-Seq for mitochondrial genes (Figure 1a). Additionally, changes in gene expression and protein abundance across two different tissues (brainstem and liver) were analyzed in tandem (Figure 1c).
Figure 1. Framework for the comparison between gene expression and protein abundance based on different methods.
(A) Generation of protein abundance and analysis of the correlation between protein abundance and gene expression. (B) 8 configurations for MS1 based label-free protein quantification. (C) Analysis of the changes of gene expression and protein abundance across different tissues.
Comparison of intensity-based and spectral count-based protein quantification measures
For the brainstem tissue data, a total of 1,693 proteins were identified and spectral counts were obtained for all of these proteins, of which 650 were mitochondrial proteins as annotated in the MitoCarta database. Intensity-based abundance measurements (using msInspect and msBID) could be obtained for approximately 90% of the identified proteins (Supplementary Table 1). Similar results were obtained for the liver tissue data (Supplementary Table 2). The correlations between protein abundance estimates by different methods (and later with gene expression data) were analyzed only on mouse mitochondrial genes (Table 1). Of the total of 1098 mouse genes in MitoCarta, 611, 566, 650 were quantified by msInspect, msBID, spectral counts in the brainstem tissue samples, respectively. On the same dataset SpectrumMill (data from the original publication [25]) quantified 679 proteins. Likewise, the numbers of genes that could be compared between the gene-expression and proteomics data of the liver tissue were 596, 586, 641, and 700 proteins, respectively. The number of genes/proteins for which quantitative information was available at both gene and at protein level in all methods was 443 and 515 in brainstem and liver tissues, respectively.
Table 1. The number of mouse mitochondrial genes that have protein abundance estimates by different methods.
1098 genes identified in MitoCarta database were considered as the mouse mitochondrial genes. Shown in parentheses is the total number of proteins quantified by each method (not only mitochondrial genes) in the proteomic data. SpectrumMill data was taken from [25] which contained quantitative information for mitochondrial genes only.
| Method | Brainstem | Liver |
|---|---|---|
| msInspect | 611 (1457) | 596 (1102) |
| msBID | 566 (1197) | 586 (996) |
| Spectral count | 650 (1693) | 641 (1267) |
| SpectrumMill | 679 | 700 |
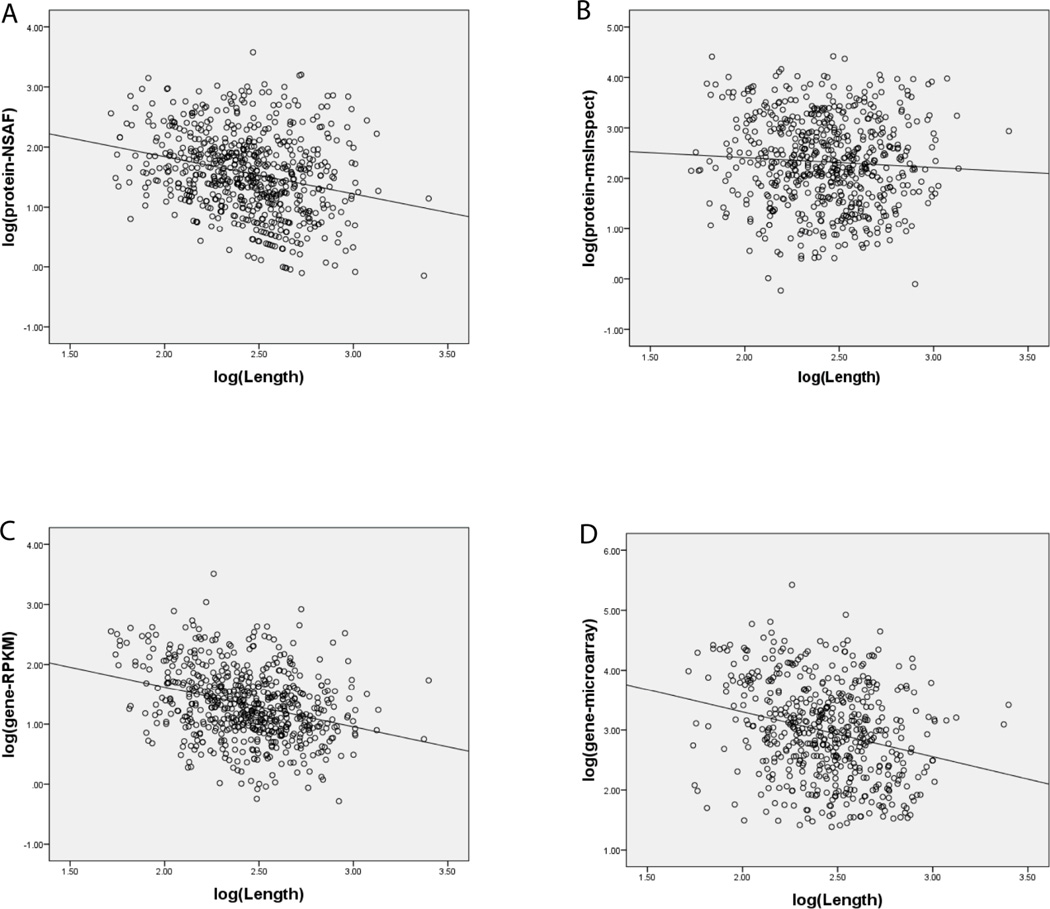
For msInspect and msBID data, 8 different quantitative measures were computed (see Methods). Despite normalizing for protein length, a non-negligible correlation (e.g. r = −0.26, i.e. a negative correlation, in brainstem tissue) between NSAF factors and protein length, log transformed, was observed (Figure 2A). For comparison, protein abundance estimates obtained using top 3 normalized peptide area intensities had a statistically insignificant correlation with protein length (e.g., r = −0.06 for msInspect using top 3 normalized area intensity, brainstem tissue; see Figure 2B). It has been suggested that protein length (and other protein sequence features) can explain a substantial fraction of protein abundance variation in human cells [37]. However, such a significant difference observed here between NSAF and top 3 normalized peptide area intensities with respect to protein length suggests that length normalization of raw spectral counts in NSAF overcorrects for protein length and introduces a bias. It should be noted that in [37] spectral count based abundance measures (APEX scores) were normalized not to protein length but to the number of predicted ‘identifiable’ peptides. However, these two normalization factors - protein length and the number of predicted peptides - in our data had a similar effect. Thus, we elected to use partial Pearson correlation controlling for protein length (i.e., use protein length as the partial correlation’s controlling variable) when comparing different protein abundance measures (and later when comparing protein and gene expression data). Using partial Spearman correlation produced similar results (data not shown).
Figure 2. Correlation between protein length and both genes expression and protein abundance.
(A) Correlation between protein length and spectral counts (NSAF). (B) Same for protein abundance by msInspect - top 3 normalized area intensities. (C) Same for gene expression by RPKM, log transformed. (D) Same for gene expression by microarrays, log transformed.
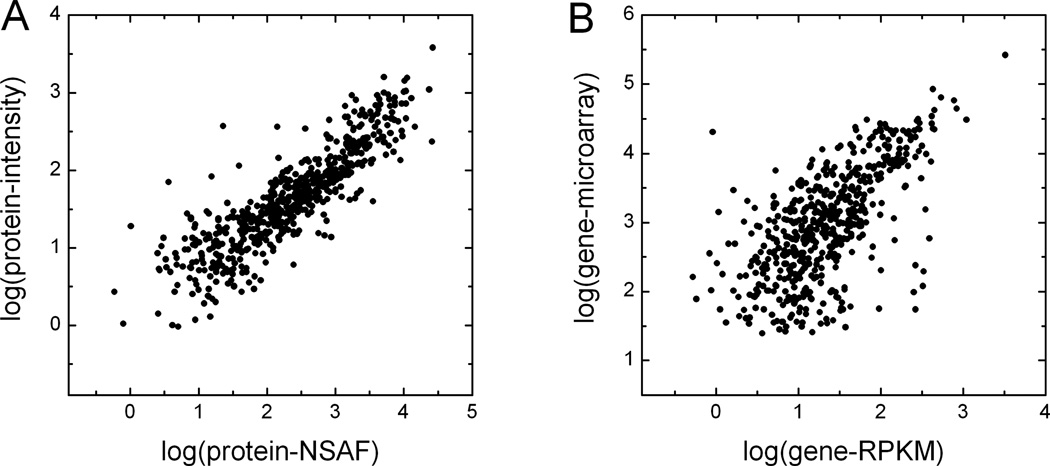
Different protein abundance estimates were compared to one another (see Supplementary Tables 3 and 4 for all pair-wise comparisons in brainstem and liver tissues). High partial correlation coefficients were observed among all pair-wise comparisons between msInspect, msBID, and SpectrumMill. The correlation coefficients for these comparisons were in 0.9 to 0.93 range, with the best correlation observed when using top 3 normalized area intensities as a measure of protein abundance (Table 2). The correlation between intensity-based and spectral count-based measures was lower but still high (see Table 2), as illustrated in Figure 3A for the comparison between msInspect and NSAF.
Table 2. Correlation between gene expression and protein abundances computed by different methods.
For each pair of methods, the partial correlation is computed based on all mitochondrial genes for which comparison could be made. Data for mouse brainstem tissue. Each cell shows partial correlation based on all genes for which quantitative information is available by both methods that are compared, and based on genes quantified by all methods (shown in brackets). Both msInspect and msBID’s results are based on top 3 peptides with largest normalized area intensity. All correlation coefficients are computed using log-transformed gene and protein quantification measures.
| SpectrumMill | msInspect | msBID | NSAF | RPKM | Microarray | |
|---|---|---|---|---|---|---|
| SpectrumMill | - | 0.91 (0.92) | 0.91 (0.91) | 0.90 (0.90) | 0.49 (0.51) | 0.36 (0.40) |
| msInspect | 0.91 (0.92) | - | 0.89 (0.91) | 0.87 (0.88) | 0.51 (0.53) | 0.40 (0.44) |
| msBID | 0.91 (0.91) | 0.89 (0.91) | - | 0.84 (0.89) | 0.54 (0.54) | 0.41 (0.42) |
| NSAF | 0.90 (0.90) | 0.87 (0.88) | 0.84 (0.89) | - | 0.51 (0.53) | 0.42 (0.44) |
| RPKM | 0.49 (0.51) | 0.51 (0.53) | 0.54 (0.54) | 0.51 (0.53) | - | 0.62 (0.61) |
| Microarray | 0.36 (0.40) | 0.40 (0.44) | 0.41 (0.42) | 0.42 (0.44) | 0.62 (0.61) | - |
Figure 3. Comparison between different quantitative measures at gene and protein levels.
(A) Protein abundance estimates by MS2 spectral counts (NSAF) plotted against protein abundance estimates from MS1 intensity data by msInspect, log transformed. Data for mitochondrial genes in brainstem tissue. Protein abundance by msInspect is based on top 3 normalized peptide area intensities. (B) Correlation of gene expression based on RPKM by mRNA-Seq and based on microarrays, log transformed.
Each of the MS1 quantitation methods was able to quantify a different number of proteins. As a result, the correlations discussed above were computed with differing numbers of value pairs for different comparisons. We repeated the analysis focusing on those proteins that had been quantified by all 3 intensity-based tools (msInspect, msBID, and SpectrumMill). This slightly increased the correlation and further diminished the differences between different approaches for estimating protein abundance (see Table 2). Overall, the results demonstrated a good agreement between the two quantitative approaches for estimating protein abundance (intensity based and spectral counts) in these data.
Correlation between gene expression and proteins abundance measures
Prior to considering correlation between gene and protein data, we compared different gene expression measurements with each other. Three different sets of RPKM values were computed for each gene in [15], referred to as first, expanded, and final RPKM values. As previously reported [15], final RPKM values (which take into account both unique short reads and multi-reads and also count reads in extended exon regions) had the best correlation with gene expression by microarrays. Based on the 528 mitochondrial genes for which both RPKM and microarray values were available in the brainstem tissue, the correlation coefficients between microarray and first, expanded, and final RPKM values were 0.54, 0.45, and 0.64, respectively (see Figure 3B for microarray – final RPKM comparison). A significant correlation between gene expression and protein length was observed for both microarray and RPKM measures (e.g. r = −0.26 and −0.31 (negative correlation) for microarray and RPKM in brainstem tissue, respectively; see Figure 2C and 2D). Similar results were observed for data from the liver tissue.
Each of the various protein abundance measurements (msInspect, msBID, SpecturmMill, and NSAF) were then correlated with the gene expression data. As a first step, different gene expression data (microarray and three RPKM measurements) were compared to each of the protein abundance values computed by the different methods mentioned above. The analysis was again done using partial Pearson correlation controlling for protein length to eliminate potential length bias in both gene and protein expression data. Among the three RPKM measures, the correlation between protein expression and final RPKM values was consistently higher than with first or expanded RPKM values. For example, protein abundance computed using msBID and top 3 normalized area intensities had a 0.54 correlation with final RPKM values, compared with 0.48 and 0.49 for first and expanded RPKM values, respectively. Thus, unless where noted, the analysis presented below was restricted to final RPKM values.
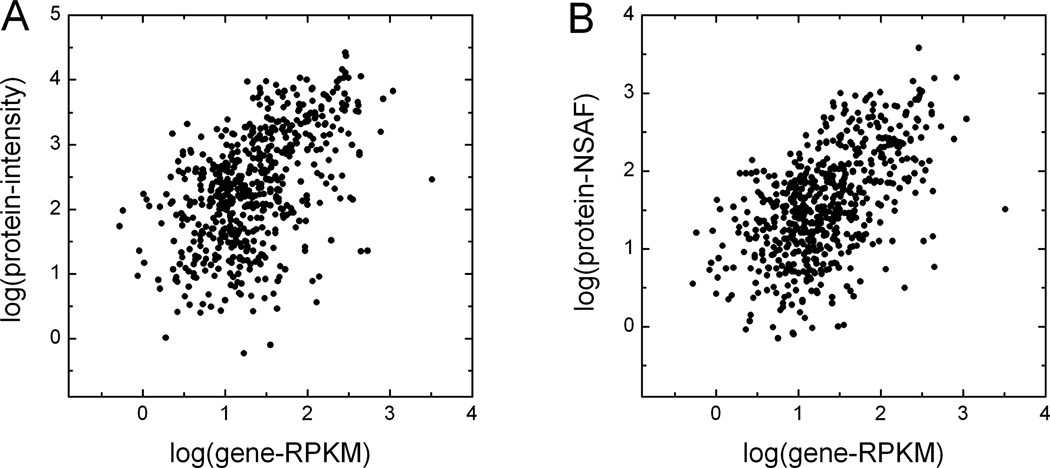
Among all of the intensity-based protein quantification methods, using the top 3 normalized area intensities computed by either msInspect or msBID gave the best correlation coefficients between gene expression and protein abundance. Moderate correlations (0.5 to 0.55 range) were observed for brainstem tissue comparisons of protein abundance to gene expression (Table 2 and Figure 4). For the brainstem tissue, the highest correlation was observed between the protein abundance as computed by msBID using the top 3 normalized area intensities and the RPKM values. For the liver tissue, NSAF showed best correlation with gene expression by RNA-Seq. However, it is important to note that the correlation coefficients based on other protein quantification methods were not significantly lower for either tissue type (see Table 2). We have also repeated the analysis focusing on those proteins that had been quantified by all 3 intensity-based tools (msInspect, msBID, and SpectrumMill). This again resulted in an improvement in the protein to gene expression correlations (Table 2 and Supplementary Table 3). The best correlation was still observed for msBID top 3 intense peptides versus the RNA-Seq gene expression data.
Figure 4. Correlation of gene expression based on RPKM by RNA-Seq and protein abundance by label-free method.
(A) MS1 based quantification by msInspect plotted against RPKM, log transformed. (B) Normalized MS2 spectral counts (NSAF)) plotted against RPKM, log transformed. Data for mouse mitochondrial genes in brainstem tissue. Protein abundance by msInspect is based on top 3 normalized peptide area intensities.
In all gene expression to protein abundance comparisons, the RNA-Seq data correlated better with the proteomics data than did the microarray results (Table 2). The differences between microarray and RNA-Seq gene expression estimates are expected, with several recent studies providing an in-depth comparison between these two platforms, see e.g. [38]. The comparison between RNA-seq and microarray data from the same samples performed in [39] showed that Pearson correlations ranged between 0.76 and 0.8. Furthermore, RNA-seq measurements provided more accurate gene expression estimates than microarrays based on the analysis of 34 selected genes by real-time polymerase chain reaction (RT-PCR). The correlation between RT-PCR and RNA-seq and microarray data for these 34 genes was 0.66 and 0.46, respectively. Our study provides an indirect, protein level confirmation of higher accuracy of gene expression estimates by RNA-seq data.
Analysis of technical factors affecting the correlation between gene and protein abundances
Before considering biological reasons affecting the correlation between gene and protein abundances, described in the next section, we performed additional analysis investigating several confounding factors more technical in nature. In addition to the non-negligible bias introduced by protein length normalization discussed above, protein to gene correlation is affected by the overall abundance of genes and proteins that are compared, and by the number of gene/protein pairs for which the comparison is made.
The overall gene/protein abundance could be a contributing factor because lower abundance proteins (transcripts) are quantified less accurately due their low spectral counts (low short read counts in RNA-seq). Intensity-based approaches (msBID, msInspect) provide a wider range of quantitative values for proteins identified by a small number of spectra, but the accuracy of these abundance estimates is still dependent on the number of quantifiable peptides [40]. Thus, we investigated how the observed correlation between gene and protein abundances changes when the analysis is restricted to genes and proteins with abundance measures passing a certain minimum threshold (at both gene and protein levels). Unless otherwise noted, all correlations reported below were computed for brainstem tissue, using partial Pearson correlation coefficient controlling for protein length, using log transformed NSAF, RPKM, and msBID abundance measures (msBID options: top 3 most intense peptides, peak area, normalized to the total intensity in the run). Using all genes in our dataset annotated as mitochondrial according to MitoCarta, the correlation coefficients between RPKM gene expression measures and both NSAF and msBID protein abundance values were 0.54 (based on 605 and 527 gene/protein pairs for which RPKM values could be compared with NSAF and msBID values, respectively). After removing gene/protein pairs with abundance measures below −1 on the standardized scale (after z-score transformation of log transformed RPKM, NSAF, and msBID measures; see Supplementary Table 5) at either gene or protein level, the degree of correlation remained essentially unchanged: 0.54 for RPKM-NSAF (based on 442 gene/protein pairs) and 0.57 for RPKM-msBID (389 pairs). This result indicates that, in this dataset, the inclusion of lower intensity genes and proteins does not significantly affect the overall correlation. It should be noted, however, that here the analysis was restricted to mitochondrial proteins which were enriched in proteomic samples at the sample preparation stage. Thus, this conclusion may not apply to data from unbiased shotgun proteomic datasets exhibiting a higher dynamic range of protein abundances and containing a larger proportion of proteins that are of low abundance.
Second, we investigated how the correlation coefficients are affected by the overall number of gene/protein pairs. Starting with 527 genes/proteins for which all three (RPKM, NSAF, and msBID) measurement were available, we performed a simulation analysis randomly selecting subsets containing 200, 100, 50, and 20 gene/protein pairs (10 instances in each case). We then performed the correlation analysis independently on each subset, and averaged the correlation coefficients obtained for multiple instances of the same size. Reducing the set size to 200, 100, and even 50 gene/protein pairs did not significantly affect the average correlation coefficients (see Table 3), however the standard deviation of correlation coefficients gradually increased. When using 20 gene/protein pairs (Table 3) or less (data not shown), we observed a noticeable shift in the correlation coefficients toward lower values. This analysis suggests that a special caution should be taken when interpreting correlation coefficients computed for selected subsets of genes and proteins (e.g. when performing the analysis separately for different Gene Ontology categories), especially when the number of gene/proteins in a particular category is small.
Table 3. Correlation between gene and protein abundances as a function of sample size.
Subsets of size 200, 100, 50, and 20 were randomly generated (10 instances for subset of each size) from the entire set of 527 genes/protein pairs for which RPKM, NSAF, and msBID values were available. Average partial Pearson correlation coefficients (controlling for protein length) in brainstem tissue. In parenthesis: standard deviation of the correlation coefficient.
| Set size | RPKM-NSAF | RPKM-msBID | NSAF-msBID |
|---|---|---|---|
| all (527) | 0.54 | 0.54 | 0.89 |
| 200 | 0.53 (0.03) | 0.54 (0.03) | 0.89 (0.01) |
| 100 | 0.55 (0.06) | 0.56 (0.06) | 0.88 (0.02) |
| 50 | 0.57 (0.10) | 0.58 (0.11) | 0.89 (0.03) |
| 20 | 0.50 (0.12) | 0.50 (0.11) | 0.88 (0.05) |
We then examined to see if curating the RNA-Seq based gene expression data could further improve the correlation with the proteomics results. With RNA-Seq technology, reads are not uniformly distributed across an alignment. As such, a gene can be reported to have a very high RPKM value that does not reflect true expression of the whole gene but only a small fragment of its coding region. This can cause inaccurate quantification data to be reported for a gene. Indeed, we observed that for some gene loci, a majority of the reads were aligned to untranslated regions. The RPKM values for these genes were substantially higher than the corresponding protein abundance levels. Thus, we computed a new measure, ‘normalized number of reads in coding regions’, using only reads aligned to known coding regions of the corresponding gene model (based on mouse gene model from mm9 of UCSC [41]).
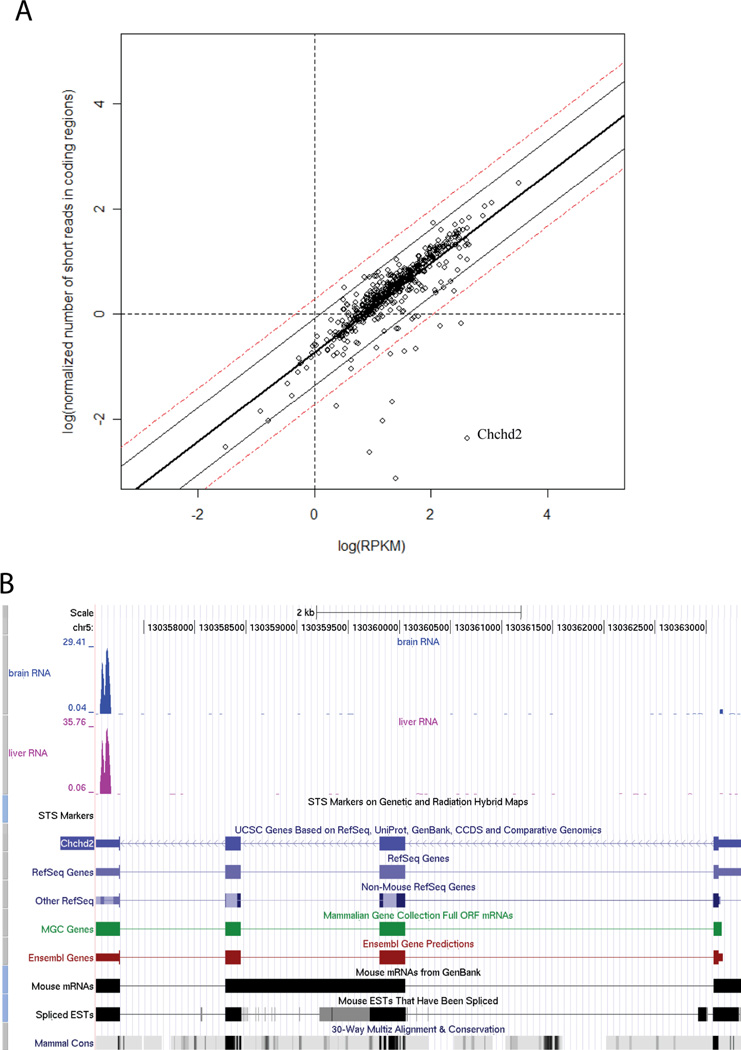
For the majority of genes the normalized number of short reads in coding regions did not differ significantly (within+/− 2 standard deviation of the mean) from the corresponding normalized total number of reads (Figure 5A). Restricting the analysis to these genes only (termed “coding region dominant” genes) improved the correlation slightly. We further analyzed 11 outlier genes which had poor agreement between gene and protein abundances. It was observed that for many of these genes, the number of reads mapping to the coding regions were a small fraction of the total number of reads aligning to the gene. For example, in the case of Chchd2 (marked in Figure 5A), more than 90% of reads aligned to untranslated regions (Figure 5B). Repeated reads [42] and reads that do not match exactly within the gene loci could also impact the accuracy of gene expression measurements. To examine this, we recomputed the gene expression values by removing redundant or mismatched reads. However, this did not result in a significant increase in the correlation coefficients in these data.
Figure 5. The analysis of the “coding region dominant” genes and selected example.
(a) Comparison of normalized number of short reads in coding regions (log transformed) and log(RPKM). Data for mitochondrial genes in brainstem tissue. The number of short reads in coding regions is normalized and it is always smaller than RPKM for the same gene. The boundaries of 90% and 99% confidence intervals are shown in solid and dashed lines. (b) The screen shots of UCSC genome browser of the outlier gene Chchd2. Most of the short reads for this gene are aligned to untranslated regions.
Another way to minimize technical issues affecting the accuracy of quantification is to compare changes in gene expression and protein abundance levels across different tissues. Both the gene expression and protein abundance levels were normalized (by RPKM for gene expression; and by top 3 normalized peptide area intensities for protein abundance) before carrying out this comparison. We observed that approximately 74% of genes exhibited same direction of change based on gene expression by mRNA-Seq and protein abundance by msInspect for brainstem against liver (Figure 6). We performed the same cross-tissue comparison using other gene expression and protein abundance measurements. In doing so, it was found that gene expression by mRNA-Seq and microarray showed consistent changes in direction with each other for 83% of the genes (Supplementary Figure 1A). Protein abundance based on NSAF and msInspect were also consistent in their direction of changes between brainstem and liver for the majority (85%) of genes (Supplementary Figure 1B).
Figure 6. The change of protein abundance by msInspect and gene expression by mRNA-Seq (RPKM) across brainstem and liver tissues.
Consistency between fold changes in protein and gene abundances across brainstem and liver tissues, log(protein_abundancebrainstem / protein_abundanceliver) vs. log(gene_expressionbrainstem / gene_expressionliver). Protein estimates are by msInspect using top 3 normalized peptide area intensities, and gene expression by mRNA-Seq (RPKM).
Analysis of biological factors affecting the correlation between gene and protein expression
Next, we investigated biological factors affecting the correlation between gene and protein expression data. First, we investigated the correlation between genes and proteins separately for subsets of genes and proteins co-annotated to the same biological process (BP), molecular function (MF), or cellular component (CC) according to GO (http://www.geneontology.org). To do this, we first performed gene enrichment analysis using DAVID [43] with 650 genes taken as input. Note that of all MitoCarta genes, approximately 75% were annotated in GO as mitochondrial. Computed separately for this core subset of mitochondrial genes, the correlation was further improved, especially for the spectral count-based measure (e.g. RPKM-NSAF values increased to 0.58). For each cluster of enriched GO categories containing at least one term with more than 20 members in our dataset, we selected one representative entry having the largest count (see Supplementary Table 6).
The correlation coefficients, as well as the average abundance of genes/proteins from each category (using z-transformed abundance measures), are shown in Table 4. Overall, there were noticeable differences in the degree of correlation among genes and proteins from different categories. For example, with respect to various cellular compartments (within the mitochondria), the RPKM-NSAF correlation coefficients ranged from 0.67 for inner membrane mitochondrial genes, to 0.52 for mitochondrial lumen, to 0.32 for mitochondrial other membrane (RPKM-msBID results were very similar, see Table 4). Furthermore, genes/proteins annotated as ribosomal demonstrated no significant correlation between gene expression and protein abundance (however, this may partially be explained by the bias in RNA-seq toward ribosomal RNA). Overall, given the size of the corresponding GO categories, and referring to the analysis performed above, the observed differences in the correlation coefficients are unlikely to be attributed solely to random fluctuation or to differences in the average abundance of genes and proteins in each category. More detailed biological interpretation of these observations, however, goes beyond the scope of this work.
Table 4. Correlation between gene and protein abundances for selected GO categories.
Brainstem tissue, partial Pearson correlation coefficient (controlling for protein length). In parentheses: number of gene/protein pairs for which quantitative abundance values were available at both gene and protein level. Mean RPKM, NSAF, and msBID values for genes/proteins from a particular GO category were computed on the standardised scale (after z-transformation of original values).
| Term | Count | RPKM-NSAF | RPKM-msBID | Mean RPKM |
Mean NSAF |
Mean msBID |
|---|---|---|---|---|---|---|
| CC:organelle inner membrane | 189 | 0.67 (183) | 0.64 (165) | 0.44 | 0.52 | 0.49 |
| CC:mitochondrial lumen | 120 | 0.51 (113) | 0.52 (102) | −0.21 | 0.01 | −0.05 |
| BP:generation of precursor metabolites and energy | 94 | 0.60 (90) | 0.62 (84) | 0.92 | 1.06 | 1.02 |
| BP:cofactor metabolic process | 52 | 0.67 (48) | 0.70 (43) | 0.07 | 0.12 | 0.25 |
| CC:ribosome | 68 | 0.00 (65) | 0.22 (51) | −0.07 | −0.4 | −0.61 |
| CC:mitochondrial outer membrane | 33 | 0.32 (27) | 0.35 (25) | 0.16 | 0.15 | 0.17 |
| CC:respiratory chain | 45 | 0.13 (42) | 0.27 (42) | 1.22 | 1.21 | 1.14 |
| MF:hydrogen ion transmembrane transporter activity | 29 | 0.57 (26) | 0.61 (21) | 1.41 | 1.36 | 0.96 |
| BP:organic acid catabolic process | 22 | 0.48 (18) | 0.64 (15) | −0.41 | −0.2 | 0.1 |
| MF:iron ion binding | 34 | 0.58 (26) | 0.45 (23) | 0.53 | 0.39 | 0.51 |
| MF:nucleotide binding | 136 | 0.57 (125) | 0.57 (108) | −0.1 | −0.08 | 0 |
| BP:nitrogen compound biosynthetic process | 38 | 0.74 (35) | 0.73 (23) | 0.43 | 0.33 | 0.48 |
We also investigated the degree of correlation between gene and protein abundances depending on the degree of mRNA and protein stability. To perform this analysis, we used data from a recent large-scale analysis of mRNA and protein half-lives in mouse. After mapping data from this study to our dataset, we obtained mRNA and protein half-life measurements for 443 and 466 mRNA and proteins, respectively (Supplementary Table 5). We then subdivided the data into two subsets: more stable proteins (protein half-life above average) and less stable (below average). When analyzed separately, the correlation coefficients between RPKM and NSAF values were substantially different for these two subsets, 0.57 (based on 212 gene/protein pairs) and 0.5 (based on 225 gene/protein pairs) for more stable and less stable proteins, respectively. Similarly, when dividing into two subsets based on mRNA half-life, the correlation coefficients showed similar differences. This is in agreement with previous observations that protein and mRNA stability are among the most significant factors governing the correlation between gene and protein abundances. We have also observed that there is a substantial correlation between the abundance of the protein and its half-life (and similarly for mRNA). Thus, the difference in the correlation between gene and protein abundances between more stable and less stable mRNA and proteins may in part be due to the overall differences in their abundance. However, when we repeated the analysis described earlier in the manuscript in which we removed genes and proteins in the low abundance range, the trend remained the same.
CONCLUDING REMARKS
In this work, we computed label-free mass spectrometry based protein abundance measures for mouse mitochondrial proteins in two different tissue types using several different methods. Although a number of recent studies compared gene and protein abundance data using larger datasets, these studies generally utilized a single protein quantification strategy (and often labeling-based such as SILAC [44, 45]). The work presented here is one of the first attempts to compare different gene expression technologies (RNA-Seq and microarray) to both MS1 and MS2 label-free proteomic data, and to compare different label-free proteomic strategies and software tools to each other.
The results show that using the top 3 normalized peptide area intensities from MS1 data as a measure of protein abundance correlated best with gene expression data collected through RNA-Seq. However, spectral count based measures did not perform significantly worse as measured by their correlation coefficients. This suggests that spectral counts, which can be easily extracted from published proteomic datasets, can be used as a basis for a more comprehensive strategy of evaluating protein abundance trends and the correlation between protein and gene expression data in a wide range of different cell types and tissues.
Additional analysis, however, may be required to find a more accurate way to normalize spectral counts to account for protein length (this also applies to intensity-based approaches that use sum over all peptides followed by division by protein length [22]). Using conventional approaches such as division by length introduces a bias in protein abundance estimates that complicates the analysis of the relationship between steady state protein abundance and underlying biological processes such as transcription, translation, and mRNA decay and protein degradation [46]. We also investigated other confounding factors such as the effect of the absolute protein or mRNA abundance, and the dataset size. We found that such analysis is useful for separating meaningful biological factors from random effects, and believe it should be performed as a part of every study involving joint analysis of gene expression and protein abundance data. Ultimately, better understand of these biases and selection of most appropriate data normalization schemes for label-free quantification will require generation of credible benchmark datasets with a help of more accurate, targeted quantification methods [47].
By focusing on mitochondrial proteins in this study we sought to minimize the contribution from biological sources to variability in correlation between gene and protein expression. This allowed us to focus on the technical sources of variability such as differences between different modes of quantitation and software tools used. We found that some of the outliers, i.e. gene/protein pairs with significantly inconsistent gene and protein abundance, had an unbalanced distribution of short reads on gene models. This warrants further investigation of this and other technical issues in RNA-Seq data for improved gene quantification. Even within the set of mitochondrial proteins, abundances of proteins in different GO categories showed varying degree of correlation with gene expression data, in agreement with previous observations [46] and recent studies showing that average protein and mRNA stability vary significantly between different GO functional categories [22, 48]. We were limited in our ability to investigate these effects in more detail given the nature of the dataset that was available to us. However, the continuing growth of data repositories (e.g. PeptideAtlas [49], Tranche [50]) containing publicly available proteomic data sets for which one can identify matching gene expression data will undoubtedly lead to many additional and more informative integrative analyses. In addition to the analysis of global signatures of gene and protein expression, it could include systematic confirmation (or lack thereof [51]) of RNA-Seq derived alternative splice forms at the protein level, cross-species analysis [52, 53], and many other applications. The results of this work should thus be useful for designing robust computational pipelines for extracting gene and label-free protein abundance information from these data.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants R01-CA-126239 and R01-GM-094231 and by National Natural Science Foundation of China (NSFC Grant No. 61103167).
REFERENCES
- 1.Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 2.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 3.May D, Fitzgibbon M, Liu Y, Holzman T, et al. A platform for accurate mass and time analyses of mass spectrometry data. J Proteome Res. 2007;6:2685–2694. doi: 10.1021/pr070146y. [DOI] [PubMed] [Google Scholar]
- 4.Mueller LN, Rinner O, Schmidt A, Letarte S, et al. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 5.Sturm M, Bertsch A, Gropl C, Hildebrandt A, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 7.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3:354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 8.Lundgren DH, Hwang SI, Wu LF, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics. 2010;7:39–53. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 9.Trudgian DC, Ridlova G, Fischer R, Mackeen MM, et al. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics. 2011;11:2790–2797. doi: 10.1002/pmic.201000800. [DOI] [PubMed] [Google Scholar]
- 10.Sandin M, Krogh M, Hansson K, Levander F. Generic workflow for quality assessment of quantitative label-free LC-MS analysis. Proteomics. 2011;11:1114–1124. doi: 10.1002/pmic.201000493. [DOI] [PubMed] [Google Scholar]
- 11.Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. Journal of Proteome Research. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 12.Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. doi: 10.1002/pmic.200700426. [DOI] [PubMed] [Google Scholar]
- 13.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of Relative Abundance Ratios Derived from Peptide Ion Chromatograms and Spectral counting for Quantitative Proteomic Analysis Using Stable Isotope Labeling. Anal. Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 14.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, et al. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum D, Jansen R, Gerstein M. Analysis of mRNA expression and protein abundance data: an approach for the comparison of the enrichment of features in the cellular population of proteins and transcripts. Bioinformatics. 2002;18:585–596. doi: 10.1093/bioinformatics/18.4.585. [DOI] [PubMed] [Google Scholar]
- 17.Washburn MP, Koller A, Oshiro G, Ulaszek RR, et al. Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3107–3112. doi: 10.1073/pnas.0634629100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie L, Wu G, Zhang W. Correlation of mRNA expression and protein abundance affected by multiple sequence features related to translational efficiency in Desulfovibrio vulgaris: a quantitative analysis. Genetics. 2006;174:2229–2243. doi: 10.1534/genetics.106.065862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 20.Kislinger T, Cox B, Kannan A, Chung C, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 21.Vogel C. Translation's coming of age. Mol Syst Biol. 2011;7:498. doi: 10.1038/msb.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwanhausser B, Busse D, Li N, Dittmar G, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 23.Fournier ML, Paulson A, Pavelka N, Mosley AL, et al. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol Cell Proteomics. 2010;9:271–284. doi: 10.1074/mcp.M900415-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MV, Topper SE, Hubler SL, Hose J, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol. 2011;7:514. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su AI, Wiltshire T, Batalov S, Lapp H, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 28.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 29.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 30.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 32.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 33.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data - The protein inference problem. Molecular & Cellular Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang D, Zhang N, Lee H, Yi E, et al. MS-BID: a Java package for label-free LC-MS-based comparative proteomic analysis. Bioinformatics. 2008;24:2641–2642. doi: 10.1093/bioinformatics/btn491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva JC, Gorenstein MV, Li GZ, Vissers JPC, Geromanos SJ. Absolute Quantification of Proteins by LCMSE: A Virtue of Parallel ms Acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Vogel C, Abreu Rde S, Ko D, Le SY, et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Molecular systems biology. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su Z, Li Z, Chen T, Li QZ, et al. Comparing Next-Generation Sequencing and Microarray Technologies in a Toxicological Study of the Effects of Aristolochic Acid on Rat Kidneys. Chemical Research in Toxicology. 2011;24:1486–1493. doi: 10.1021/tx200103b. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Liu Z, Gong S, Wu X, et al. Genome-wide gene expression profiling of nucleus accumbens neurons projecting to ventral pallidum using both microarray and transcriptome sequencing. Frontiers in Neuroscience. 2011;5 doi: 10.3389/fnins.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsou CC, Tsai CF, Tsui YH, Sudhir PR, et al. IDEAL-Q, an Automated Tool for Label-free Quantitation Analysis Using an Efficient Peptide Alignment Approach and Spectral Data Validation. Molecular & Cellular Proteomics. 2010;9:131–144. doi: 10.1074/mcp.M900177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karolchik D, Baertsch R, Diekhans M, Furey TS, et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2008 doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:U1251–U1260. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 45.Lundberg E, Fagerberg L, Klevebring D, Matic I, et al. Defining the transcriptome and proteome in three functionally different human cell lines. Molecular systems biology. 2010;6 doi: 10.1038/msb.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abreu RD, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Molecular Biosystems. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan S, Aebersold R, Chen R, Rush J, et al. Mass Spectrometry Based Targeted Protein Quantification: Methods and Applications. Journal of Proteome Research. 2008;8:787–797. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boisvert FM, Ahmad Y, Gierliński M, Charrière F, et al. A quantitative spatial proteomics analysis of proteome turnover in human cells. Molecular & Cellular Proteomics. 2011 doi: 10.1074/mcp.M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desiere F, Deutsch EW, Nesvizhskii AI, Mallick P, et al. Integration with the human genome of peptide sequences obtained by high-throughput mass spectrometry. Genome Biology. 2005;6 doi: 10.1186/gb-2004-6-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill JA, Smith BE, Papoulias PG, Andrews PC. ProteomeCommons.org Collaborative Annotation and Project Management Resource Integrated With the Tranche Repository. Journal of Proteome Research. 2010;9:2809–2811. doi: 10.1021/pr1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ning K, Nesvizhskii AI. The Utility of Mass Spectrometry-based Proteomic Data for Validation of Novel Alternative Splice Forms Reconstructed from RNA-Seq Data: A Preliminary Assessment. BMC Bioinformatics. 2010;11:S14. doi: 10.1186/1471-2105-11-S11-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss M, Schrimpf S, Hengartner MO, Lercher MJ, von Mering C. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics. 2010;10:1297–1306. doi: 10.1002/pmic.200900414. [DOI] [PubMed] [Google Scholar]
- 53.Laurent JM, Vogel C, Kwon T, Craig SA, et al. Protein abundances are more conserved than mRNA abundances across diverse taxa. Proteomics. 2010;10:4209–4212. doi: 10.1002/pmic.201000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.