Abstract
To identify C-MYC targets rate-limiting for proliferation of malignant melanoma, we stably inhibited C-MYC in several human metastatic melanoma lines via lentivirus-based shRNAs approximately to the levels detected in normal melanocytes. C-MYC depletion did not significantly affect levels of E2F1 protein reported to regulate expression of many S-phase specific genes, but resulted in the repression of several genes encoding enzymes rate-limiting for dNTP metabolism. These included thymidylate synthase (TS), inosine monophosphate dehydrogenase 2 (IMPDH2) and phosphoribosyl pyrophosphate synthetase 2 (PRPS2). C-MYC depletion also resulted in reduction in the amounts of deoxyribonucleoside triphosphates (dNTPs) and inhibition of proliferation. shRNA-mediated suppression of TS, IMPDH2 or PRPS2 resulted in the decrease of dNTP pools and retardation of the cell cycle progression of melanoma cells in a manner similar to that of C-MYC-depletion in those cells. Reciprocally, concurrent overexpression of cDNAs for TS, IMPDH2 and PRPS2 delayed proliferative arrest caused by inhibition of C-MYC in melanoma cells. Overexpression of C-MYC in normal melanocytes enhanced expression of the above enzymes and increased individual dNTP pools. Analysis of in vivo C-MYC interactions with TS, IMPDH2 and PRPS2 genes confirmed that they are direct C-MYC targets. Moreover, all three proteins express at higher levels in cells from several metastatic melanoma lines compared to normal melanocytes. Our data establish a novel functional link between C-MYC and dNTP metabolism and identify its role in proliferation of tumor cells.
Keywords: C-MYC, melanoma, nucleotide metabolism
Introduction
An elevated expression of C-MYC has been detected in more than 50% of human malignancies.1,2 In experimental settings, C-MYC overexpression was shown to induce tumors in several transgenic mouse models,3 whereas inactivation of C-MYC in the majority of these models led to tumor regression, suggesting that proliferation and/or survival of C-MYC-induced tumors continue to rely upon sustained expression of C-MYC.4
A conventional model of C-MYC function suggests that it is a transcription factor, modulating the expression of downstream target genes.5,6 C-MYC contains an N-terminal transcription activation domain and a C-terminal helix-loop-helix/leucine zipper domain.7 The leucine zipper domain interacts with the protein Max, which is a prerequisite for C-MYC binding to DNA via the helix-loop-helix domain at a subset of E-box elements in the promoter of target genes (transcriptional activation)8 or at the initiator sequences (transcriptional repression).9
Multiple studies utilizing different approaches and technologies (from guesses to high-throughput screenings) have been performed with the goal of discovering the MYC-responsive genes.5 Collectively, these studies have identified well over 2,000 genes whose products participate in a variety of cellular processes, including, but not limited to cell cycle control, apoptosis, cell adhesion, biosynthesis of ribosomal and transfer RNAs, protein biosynthesis and metabolism.5 Interestingly, transcriptional modulation of the majority of the identified genes, including bona fide C-MYC targets, was not high: two-to-fourfold on average in logarithmically growing cells.10 These observations suggest that, unlike other transcription factors (e.g., NFκB or p53) that induce significant changes in the expression of defined sets of genes,11,12 C-MYC performs its function by modest regulation of a large gene pool. Indeed, C-MYC is capable of binding promoters of up to 10%–15% of all transcriptionally active genes, according to some estimates.13
On the other hand, only a small number of C-MYC-responsive genes has been shown to be capable of partial reconstitution of some of the C-MYC-dependent phenotypes in the absence of MYC-family proteins.14-17 Given the wide spectrum of cellular processes controlled by C-MYC, the list of such genes is clearly incomplete.
Here, in order to identify C-MYC-target genes rate-limiting for proliferation of tumor cells, we stably inhibited C-MYC in cells from several human melanoma lines to the levels detected in normal melanocytes using siRNA-based methodology. The inhibition of C-MYC resulted in a repression of several enzymes rate-limiting for nucleotide metabolism, the depletion of intracellular dNTPs, and the suppression of proliferation. Further analysis confirmed that genes encoding the above enzymes are bona fide C-MYC targets whose products are functionally involved in C-MYC-mediated maintenance of proliferation of tumor cells. Therefore, our data provide a connection between C-MYC-dependent control of dNTP metabolism and proliferation of tumor cells.
Results
shRNA-mediated depletion of C-MYC affects proliferation of melanoma cells
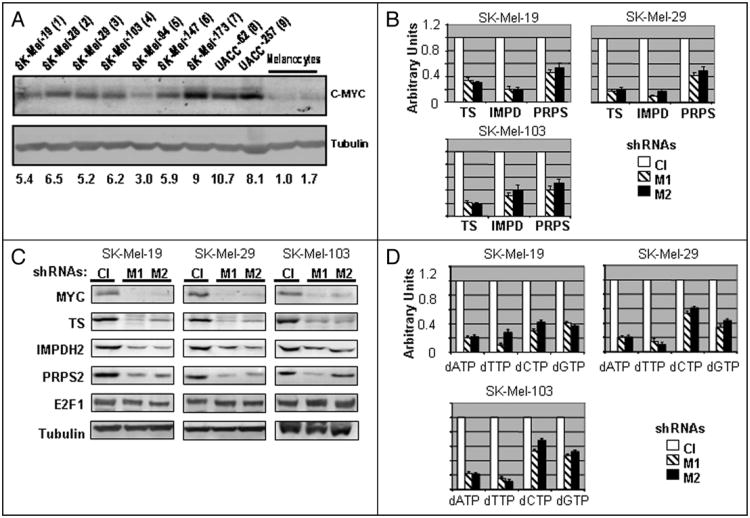
We analyzed C-MYC levels in nine human melanoma cell lines established from metastases and in two independently isolated populations of primary human skin melanocytes. Total protein extracts obtained from cells in the logarithmic growth phase were probed in Western blotting with antibodies specific to C-MYC or tubulin. As shown in Figure 1A, C-MYC protein levels were significantly (approximately 3- to 10-fold) higher in all examined melanoma cell lines than in normal melanocytes.
Figure 1.
C-MYC suppression results in depletion of dNTP pools. (A) Total cellular extracts from two independently isolated populations of normal melanocytes and indicated metastatic melanoma cells were probed in Western blotting with antibodies designated on the right. (B) Quantitative Reverse-Transcription PCR was performed on RNA isolated from indicated melanoma cells infected with control (Cl) or independently with two C-MYC-shRNAs (M1 and M2). Empty boxes—control shRNA, dashed boxes—C-MYC-shRNA-1 (M1) black boxes—C-MYC-shRNA-2 (M2). (C) Melanoma cells described in (B) were harvested 72 hours post-infection in the SDS-containing buffer and probed in Western blotting with the antibodies designated on the left. (D) dNTPs were extracted and quantified from indicated melanoma cells described in (B) 72 hours after infection. All data are normalized by the amounts of corresponding dNTP in extracts from the cells infected with control shRNA. Empty boxes—control shRNA, dashed boxes—C-MYC-shRNA-1 (M1) black boxes—C-MYC-shRNA-2 (M2).
Recently, we have shown that shRNA-mediated depletion of C-MYC in normal cells and the majority of human tumor cell lines led to a growth arrest.18 Here, we confirmed these observations for three melanoma cell lines (SK-Mel-19, SK-Mel-29 and SK-Mel-109) using two lentivirus-delivered C-MYC-specific shRNAs: C-MYC-shRNA-1 (M1) and C-MYC-shRNA-2 (M2) (see Materials and Methods), (Suppl. Fig. 1). In agreement with the results shown previously,18 a significant inhibition of C-MYC (approximately 3- to 10-fold) was obtained with the use of either of the shRNAs, resulting in a complete growth arrest between days 4 and 6 post-infection (Suppl. Fig. 1).
C-MYC depletion affects expression of several enzymes essential for nucleotide biosynthesis
To detect changes in global gene expression in melanoma cells undergoing C-MYC depletion, we utilized microarray technology. To this end, SK-Mel19 cells were infected with control or MYC-shRNA-1 in duplicate, followed by mRNA isolation 72 hours post-infection. RNA conversion into cDNA, preparation of double-stranded DNA, preparation of biotinylated cRNA, hybridization to an Affymetrix GeneChip Array and statistical analysis were performed at an in-house facility (see Materials and Methods for details). In the course of data analysis, we identified several genes whose expression was affected by C-MYC depletion more than twofold, and which were previously characterized as MYC targets including CDNK1B (p27KIP1), cyclin B1, CDC2, PAX3, INHBA, etc.,19 (Suppl. Table 1). According to the microarray data, expression of other bona-fide C-MYC targets, such as telomerase reverse transcriptase,20 cytosolic and mitochondrial serine hydroxymethyltransferases (cSHMT and mSHMT),16 ornithine decarboxylase21 and E2F122 was affected by 1.5- to 2-fold, while the expression of some other known C-MYC target genes, including cyclin-dependent kinase 4 (CDK4),15 carbamoylphosphate synthetase 2, aspartate transcarbamylase, dihydroorotase (CAD)23 and Cyclin D224 was not altered.
Additionally, we identified a group of genes whose products are involved in nucleotide metabolism and whose expression was decreased two- or more fold in C-MYC-depleted SK-Mel-19 cells compared to control cells (Table 1). Some of these genes have been previously identified as MYC-dependent ones via various high-throughput methodologies;19 however, no analysis of their individual pattern of expression or functional involvement in C-MYC-dependent phenotypes has been provided. Although genes involved in nucleotide metabolism were not statistically over-represented among all identified potential MYC-responsive genes, we have noticed that proteins encoded by several of the identified genes (indicated by asterisk in Table 1) have been suggested as rate-limiting steps for nucleotide biosynthesis. Thus, regulation of such genes could be important for the total cellular nucleotide metabolism. These include: (i) thymidylate synthase (TS), an enzyme that converts dUMP to dTMP and, therefore, serves as a major source of de novo dTTP biosynthesis;25 (ii) inosine monophosphate dehydrogenase 2 (IMPDH2), an enzyme that converts inosine monophosphate (IMP) to xanthosine monophosphate which is a rate-limiting step in the biosynthesis of guanosine and deoxyguanosine nucleotides,26 and (iii) phosphoribosyl pyrophosphate synthetase 2 (PRPS2), an enzyme generating phosphoribosyl pyrophosphate, which is required for several rate-limiting steps of de novo and salvage nucleotide synthesis.27
Table 1. C-MYC DEpletion affects expression of genes whose products are involved in nucleotide metabolism.
| # | Gene symbol | Regulation |
|---|---|---|
| de novo and salvage nucleotide synthesis | ||
| 1 | PRPS2* | D |
| de novo purine biosynthesis | ||
| 2 | PAICS | D |
| 3 | PPAT | D |
| 4 | GART | D |
| 5 | IMPDH2* | D |
| de novo thymidine biosyntesis | ||
| 6 | TYMS* | D |
| salvage nucleotide synthesis | ||
| 7 | TK1 | D |
|
| ||
(microarray-based analysis of SK-Mel-19 cells expressing control or Myc-shRNA-1).
Rate-limiting enzymes; D, downregulated ≥2 fold.
We were interested in whether C-MYC could maintain the proliferation of studied melanoma cells through control of nucleotide metabolism by direct regulation of TS, IMPDH2 and PRPS2. As an initial step in testing this hypothesis, we determined the mRNA and protein levels of the identified genes in three melanoma lines depleted of C-MYC independently by each of the C-MYC shRNAs described above. As shown in Figure 1B and C, mRNA and protein levels of TS, IMPDH2 and PRPS2 were affected by the inhibition of C-MYC in all tested melanoma lines, suggesting that C-MYC plays a critical role in the maintenance of expression of TS, IMPDH2 and PRPS2 in studied cells.
One of the genes examined, TS, has been characterized as a direct target of E2F1.28 Additionally, sequence analysis of the IMPDH2 promoter revealed several potential E2F-binding sites (not shown). E2F1 has been previously described as a direct target of C-MYC,22 and expression of its mRNA was mildly downregulated in C-MYC-depleted SK-Mel-19 cells according to the microarray data. Consequently, we tested whether decreased expression of TS, IMPDH2 and PRPS2 in C-MYC-depleted melanoma cells could be due to the inhibition of E2F1 by assessing its levels via Western blotting in control and C-MYC-depleted SK-Mel-19 melanoma cells. As shown in Figure 1C, no significant differences in the levels of E2F1 were detected in C-MYC-depleted cells compared to control cells, suggesting that E2F1 is not a major mediator of MYC-dependent transcriptional regulation of TS, IMPDH2 and PRPS2 in studied cells.
C-MYC inhibition correlates with depletion of intracellular dNTP pools
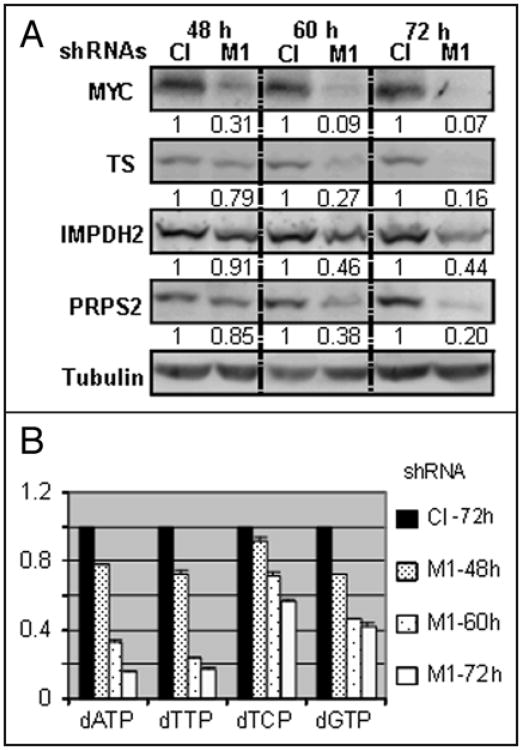
The above data suggest that inhibition of C-MYC in melanoma cells should affect levels and/or ratios of intracellular dNTP pools. To determine whether that was the case, we quantified individual dNTP pools using either a DNA template assay or HPLC analysis. (see Materials and Methods for details). Both methods produced similar results with regard to fold changes in the amounts of individual dNTPs between control and C-MYC-depleted cells. As shown in Figure 1D, amounts of dNTP in C-MYC depleted cells from three melanoma cell lines (SK-Mel-19, 29 and 103) were lower than in the respective control cells, with the amounts of dATP, dTTP, dCTP and dGTP decreased approximately 80%, 80%, 40% and 60%, respectively. Measurements of the amounts of TS, IMPDH2, PRPS2 and levels of dNTP in C-MYC-depleted and control cells during 48–72 hour interval post-infection (Fig. 2) revealed a correlation between (i) C-MYC, (ii) TS, IMPDH2 PRPS2 and (iii) intracellular dNTP pools. Based on these data, we conclude that shRNA-mediated inhibition of C-MYC correlates with a downregulation of several key enzymes involved in dNTP metabolism and a depletion of dNTP levels.
Figure 2.
dNTP levels correlate with C-MYC expression. (A) SK-Mel-19 melanoma cells infected with control or M1 shRNAs were harvested at time points shown on top, lysed in the SDS-containing buffer and probed in Western blotting with the antibodies designated on the left. Quantification was performed using ImageQuant 5.1 software. (B) dNTPs were extracted from the cells described in (A) and quantified 48, 60 or 72 hours after infection according to the procedures described in the Material and Methods. Data were normalized by the amounts of corresponding dNTP in extracts from the cells expressing control shRNA. Black boxes correspond to cells infected with control shRNA and collected 72 hours after infection. Densely dotted, sparsely dotted and empty boxes correspond to cells infected with M1 shRNA and collected at 48, 60 or 72 hours post-infection, respectively.
Inhibition of TS, IMPDH2 and PRPS2 affects dNTP pools and proliferation of melanoma cells
To identify whether suppression of TS, IMPDH2 and PRPS2 could affect proliferation of melanoma cells in a manner similar to the depletion of C-MYC, we individually decreased the amounts of the above enzymes via shRNAs (two per gene) in SK-Mel-19 melanoma cells approximately to the levels detected in C-MYC-depleted cells (Fig. 3A) and determined the impact of such inhibition on the proliferation of melanoma cells and their cell cycle progression. As shown in Figure 3B and Supplemental Figure 2, shRNA-mediated inhibition of TS, IMPDH2 and PRPS2 resulted in changes of corresponding dNTP pools. Amounts of dTTP were most severely affected (up to 80%) in TS depleted cells. Depletion of IMPDH2 affected mostly dGTP and dATP levels, while depletion of PRPS2 resulted in predominant downregulation of dATP, dTTP and dCTP (Fig. 3B).
Figure 3.
Inhibition of TS, PRPS2 or IMPDH2 in melanoma cells mimics the effects caused by C-MYC depletion. All experiments were performed in SK-Mel-19 cells. (A) Cells expressing shRNAs shown on the top were harvested at 3 days post-infection, lysed in the SDS-containing buffer and probed in Western blotting with the antibodies designated on the left. (B) dNTPs were extracted from cells expressing control shRNA (empty boxes) or designated shRNA (filled boxes) and quantified. All data are normalized by the amounts of corresponding dNTPs in extracts from the cells expressing control shRNA. (C) Cells infected with designated shRNAs were plated the day after infection in triplicates in 24-well plates (2 × 103 cells per well) and counted every day for five days. The numbers below the graph correspond to the days post-infection. (D) Cells were infected during log phase growth with lentiviral vectors expressing indicated shRNAs. Five days post-infection, cells were harvested, stained with propidium iodide and analyzed by flow cytometry.
Suppression of TS, IMPDH2 or PRPS2 also affected proliferation of SK-Mel-19 cells and resulted in the cell cycle retardation in the G0/G1 stage (Fig. 3C and D). Similar effects of studied shRNAs on cell proliferation and cell cycle profile were also observed in SK-Mel-29 melanoma cells (Suppl. Fig. 3). Significantly, individual suppression of TS, IMPDH2 or PRPS2 resulted in phenotypes similar to those induced by the depletion of C-MYC in SK-Mel-19 and SK-Mel-29 cells, although C-MYC depletion affected proliferation more severely, as evidenced by cell counting and more profound G0/G1 arrest (Fig. 3, Suppl. Fig. 3). This could be due to the fact that C-MYC-depleted melanoma cells express at reduced levels all three studied enzymes. We were unable to test this hypothesis by simultaneous shRNA-mediated depletion of all three genes due to the interference among different shRNAs resulted in only marginal suppression of individual genes.
Therefore, our data suggest that alterations in nucleotide levels and cell cycle progression caused by C-MYC depletion in SK-Mel-19 and -29 cells could be mediated by the suppression of TS, IMPDH2 and PRPS2.
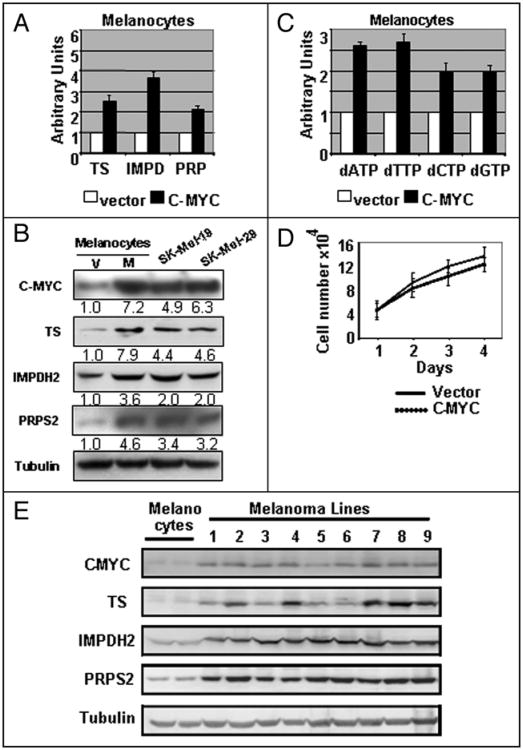
C-MYC overexpression upregulates TS, IMPDH2, PRPS2 and dNTP pools in normal melanocytes
A decrease in mRNA levels of TS, IMPDH2 and PRPS2 in response to C-MYC depletion implies a C-MYC-dependent pattern of expression of the above genes. To further test this possibility, we overexpressed C-MYC cDNA in parallel with control vector in normal melanocytes via lentivirusmediated infection and assessed levels of C-MYC, TS, IMPDH2 and PRPS2 in three days post-infection. As shown in Figure 4A and B, ectopic overexpression of C-MYC approximately to the levels detected in melanoma cells, resulted in a 3- to 8-fold induction of mRNA and protein levels of TS, IMPDH2 and PRPS2. Accordingly, dNTP pools in melanocytes overexpressing C-MYC were increased by 2.5- to 3-fold (Fig. 4C). On the other hand, C-MYC overexpression did not result in significant changes in the proliferation rates of normal melanocytes during this time: 1–4 days post infection (Fig. 4D). These data further confirm a C-MYC-dependent pattern of expression of TS, IMPDH2 and PRPS2 and demonstrate the ability of C-MYC to increase the levels of intracellular dNTP pools independently from activation of cellular proliferation. As was demonstrated above (Fig. 1) C-MYC protein amounts are significantly higher in tumor-derived melanoma cells compared to normal melanocytes.
Figure 4.
C-MYC overexpression in melanocytes increases expression of TS, IMPDH2 and PRPS2 and upregulates nucleotide levels. (A) Normal melanocytes were infected with empty lentiviral vector (Vector) or with lentiviral vector encoding human C-MYC cDNA (C-MYC). 72 hours after infection, the total RNA was isolated from melanocytes, followed by reverse transcription reaction and Q-PCR with probes shown on the bottom. (B) Total cellular extracts from normal melanocytes described in (A) were probed in Western blotting with antibodies designated on the left. Signal quantification was done using ImageJ software. (C) dNTPs were extracted from melanocytes described in (A) and quantified. All data are normalized by the amounts of corresponding dNTP in the extracts from cells expressing control vector. (D) Cells described in (A) were plated in 24-well plates (2 × 104 cells per well) in triplicates. Cells were counted every day for 4 days after infection. Numbers below the graph indicate days post infection. (E) Total cellular extracts from two independently isolated populations of normal melanocytes and metastatic melanoma cells we probed in Western blotting with antibodies designated on the left. Numbers on the top correspond to melanoma cell lines described in Figure 1A.
Western blot analysis of TS, IMPDH2 and PRPS2 in melanoma cells and normal melanocytes revealed that these proteins, like C-MYC, express at higher levels in melanoma cells (Fig. 4).
Taken together, these results suggest that TS, IMPDH2 and PRPS2 play an important role in mediation of C-MYC-dependent proliferation during tumor progression of normal melanocytes towards malignant melanomas.
Endogenous C-MYC interacts with the promoters of TS, IMPDH2 and PRPS2 genes in vivo
To find a direct role of C-MYC in the regulation of expression of TS, IMPDH2 and PRPS2 we set up experiments to determine whether there was a direct binding of C-MYC to the promoters of the above genes. A computer-based analysis (DNA Strider V), performed on the sequences 1.5–2 kb upstream and downstream from the transcription start sites of the above genes, has identified several canonical (CACGTG) and noncanonical29 MYC-binding sequences. As shown in Supplemental Figure 4, two canonical MYC binding sites (MBS) in human PRPS2 gene and one non-canonical MBS in the promoter of human IMPDH2 gene were also present in the promoters of their mouse orthologs (filled triangles).
An E-box 5′-CACTTG-3′ has been previously described within 28 bp direct repeats in the promoter of human TS gene (Suppl. Fig. 4).30,31 Additionally, it has been shown that the number of 28-bp repeats and the integrity of 5′-CACTTG-3′ site were important for mRNA and protein expression of human TS,30,31 suggesting an important role of these sequences in control of TS gene transcription.
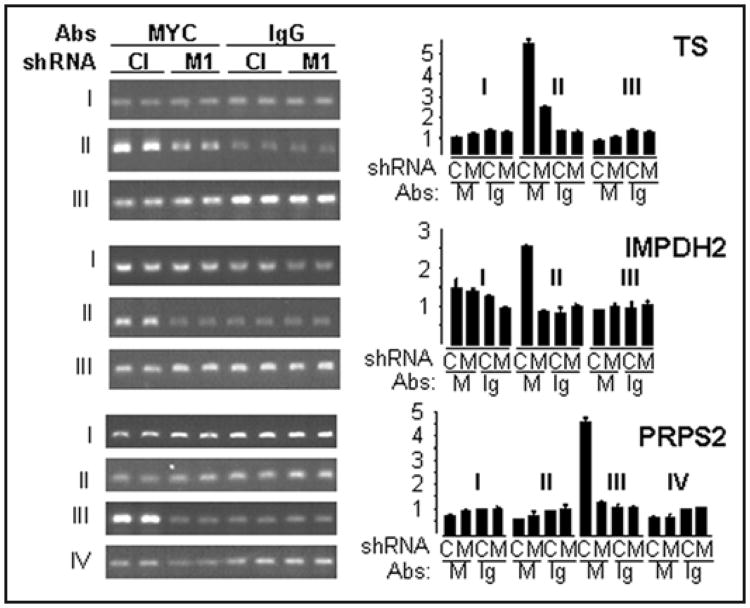
To test physical interactions between C-MYC and the above genes in vivo, we performed Chromatin Immunoprecipitation (ChIP) of cross-linked chromatin from control and C-MYC-depleted SK-Mel-19 cells with anti-C-MYC rabbit antibodies or control IgG rabbit antibodies (see Materials and Methods). After reversion of the cross-linking, immunoprecipitated DNA was purified and subjected to PCR with three or four pairs of primers encompassing regions of approximately 200 bps within the area of 1.5–2 kbs upstream or downstream from the mRNA start sites of the above genes (see schematic in Suppl. Fig. 4).
As shown in Figure 5, the regions within all three genes have been identified that were substantially more enriched with the chromatin precipitated with C-MYC antibodies compared to control IgG antibodies. Moreover, the amounts of DNA corresponding to these regions have been reduced in chromatin precipitated from cells depleted of C-MYC (Fig. 5). Thus, our data have identified areas of C-MYC binding in the regulatory areas of studied genes. Taken together, these data confirm that TS and IMPDH2 are direct C-MYC target genes.
Figure 5.
C-MYC interacts with the promoters of TS, IMPDH2 and PRPS2 genes in SK-Mel-19 cells. SK-Mel-19 cells infected with control shRNA (Cl) or Myc-shRNA-1 (M1) were cross-linked, lysed, and chromatin was immunoprecipitated with C-MYC-specific (“MYC”) or non-specific (“IgG”) antibodies, followed by the reversal of the cross-linking and DNA isolation. Isolated DNA was used in PCR with oligonucleotides flanking areas designated by Roman numbers to the left of the gels as described in (Suppl. Fig. 4). PCR products were resolved on 1% agarose gel containing ethidium bromide. Quantification of the DNA bands was performed using ImageJ software.
Overexpression of TS, IMPDH2 and PRPS2 delays proliferative arrest of C-MYC-depleted melanoma cells
In order to determine the functional involvement of TS, IMPDH2 and PRPS2 in C-MYC-mediated control of proliferation of melanoma cells, we set up experiments to complement C-MYC-depletion in melanoma cells by concurrent ectopic expression of the above genes. To this end, cDNAs for TS, IMPDH2 and PRPS2 were commercially obtained and cloned into lentiviral or retroviral expression vectors (see Materials and Methods). Following sequence verification of the inserts, these vectors in parallel with control empty vectors were delivered into melanoma cells SK-Mel-19.
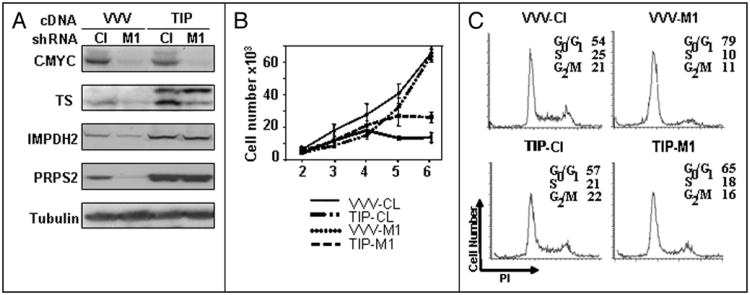
Overexpression of the above proteins was confirmed by Western blotting (Fig. 6A). Melanoma cells overexpressing all three cDNAs (hereafter described as TIP cells for TS, IMPDH2 and PRPS2) or control cells infected with empty vectors three times (hereafter described as VVV cells for triple Vector infection) were superinfected with control (Cl) or C-MYC-1 (M1)shRNAs. Expression of C-MYC, TS, IMPDH2 and PRPS2 was followed by Western analysis. As shown in Figure 6A, the depletion of C-MYC in TIP cells did not significantly change total (exogenous and endogenous) amounts of TS, IMPDH2 and PRPS2.
Figure 6.
Ectopic expression of TS, IMPDH2 and PRPS2 delays proliferative arrest caused by C-MYC depletion. (A) SK-Mel-19 cells were infected with control vectors three times (designated as VVV) or vectors expressing TS, IMPDH2 and PRPS2 cDNAs (designated as TIP). Cells were superinfected with control shRNA (Cl) or MYC-shRNA-1 (M1), harvested 72 hours post-infection, lysed in the SDS-containing buffer and probed in Western blotting with the antibodies to the protein designated on the left. Note that the exogenous TS contains FLAG tag and therefore migrates on the gel slower than the endogenous TS. Exogenous IMPDH2 and PRPS2 do not contain tags and migrate on the gel identically to the corresponding endogenous proteins. (B) Cells treated like in (A) were plated the day after infection in triplicates in 24 well plates (2 × 103 cells per well) and counted every day for 5 days. Numbers below the graph correspond to the days post-infection. (C) Cells treated like in (A) were collected 5 days post-infection, stained with propidium iodide and analyzed by flow cytometry.
The proliferation of control or MYC-depleted VVV and TIP cells was assessed daily by cell counting. As shown in Figures 6B, at earlier time points after infection, the proliferation of TIP cells was less affected by depletion of C-MYC than was the proliferation of their control counterparts. These observations were further confirmed by the cell cycle analysis of studied cells performed at day 5 post infection. As shown in Figure 6C, MYC-depleted TIP cells are undergoing less profound growth arrest compared to MYC-depleted VVV as evidenced by the differences in the proportion of cells in S and G0/G1 phases of the cell cycle. Similar observations were made using cells from another melanoma line— SK-Mel-29 (Suppl. Fig. 5).
On the other hand, higher proliferation rates were not sustained in TIP-cells depleted of C-MYC, and eventually they ceased to proliferate. Thus, ectopic overexpression of TS, IMPDH2 and PRPS2 could delay proliferation arrest caused by C-MYC inhibition in melanoma cells demonstrating functional involvement of these genes in C-MYC-dependent maintenance of proliferation of melanoma cells.
Discussion
Multiple approaches have been performed to discover genes whose expression is modulated by C-MYC in normal or transformed cells.5 However, very few studies provided data on a possibility of functional substitution of C-MYC by identified genes in any C-MYC-dependent phenotypes. In part, that could be attributed to the highly pleiotrophic nature of C-MYC functioning, resulting in simultaneous regulation of several pathways each encompassing multiple gene products.
C-MYC-dependent control of nucleotide metabolism has not been studied in detail, even though among over 2000 genes collectively identified as C-MYC-dependent, there were several whose products participate in de novo or salvage pathways of nucleotide biosynthesis.19 In the present work, we established the first direct link between C-MYC and control of the production of deoxynucleoside triphosphates, the end products of nucleotide biosynthesis, as precursors to DNA synthesis. We have identified three genes encoding rate-limiting enzymes for nucleotide metabolism as MYC targets: TS, IMPDH2 and PRPS2. Expression of these genes depended on changes in C-MYC levels (Figs. 1 and 4) and C-MYC was shown to specifically interact with the regulatory sequences of these genes in vivo (Fig. 5). We have provided two groups of data suggesting that TS, IMPDH2 and PRPS2 are functionally important for the MYC-dependent maintenance of the proliferation of melanoma cells. First, shRNA-mediated suppression of identified C-MYC targets resulted in a downregulation of specific dNTP(s) and an inhibition of proliferation in a manner similar to that of C-MYC depletion (Fig. 3 and Suppl. Fig. 4). As was mentioned above, we were unable to simultaneously deplete all three genes due to the interference among different shRNAs. However, it is conceivable that downregulation of all three enzymes (which actually occurs in MYC-depleted cells) would affect melanoma cell proliferation even more severely. Therefore, transcriptional repression of the studied genes could be a mediator of the effects induced by C-MYC depletion. Second, ectopic overexpression of the cDNAs for the above genes was capable of partial and temporal substitution of C-MYC, evidenced by the delay in the proliferative arrest caused by C-MYC depletion (Fig. 6 and Suppl. Fig. 5).
As C-MYC controls expression of hundreds of genes involved in dozens of cellular pathways,5 it is understandable that the overexpression of three C-MYC target genes could not completely substitute for C-MYC in melanoma cells. Indeed, partial reconstitution of C-MYC-dependent proliferation with efficiency similar to that described here has also been reported for overexpression of CDK4,15 or mitochondrial serine hydroxymethyltransferase (mSHMT)16 in HO15.19 cells (both genes are bona-fide C-MYC targets). It is noteworthy that studied MYC-depleted melanoma cells cease to proliferate in 4–6 days, unlike myc-null HO15.19 cells which proliferate continuously at lower rates.32 This could be a reason for only temporal complementation of proliferation of MYC-depleted melanoma cells by TS, IMPDH2 and PRPS2 in contrast to the continuous partial rescue of proliferation of HO15.19 cells by CDK4 or mSHMT.
We demonstrate that overexpression of C-MYC in normal melanocytes results in a near-symmetrical 2- to 2.8-fold increase in dNTP levels independently of proliferation rates. Previously, it has been shown by one of our groups that mutational rates increased several fold when the amounts of dNTPs added to the in vitro replication system with HeLa cytoplasmic extracts, were symmetrically increased 3 to 4-fold.33 Until now, C-MYC has been implicated in the induction of genomic instability largely via its ability to induce intracellular reactive oxygen species.34 Since it is unclear whether the induction of ROS completely accounts for C-MYC induced mutagenesis,35 our data raise an intriguing possibility that C-MYC could promote mutagenesis through a nearly symmetrical increase in the amounts of intracellular dNTPs.
Taken together, our data create a novel functional link between the regulation of cell proliferation, nucleotide metabolism and C-MYC transactivation activity.
Materials and Methods
Cell lines
Melanoma cell lines were originally obtained from Memorial Sloan Kettering Cancer Center. Cells were cultured in Dulbecco's modified Eagle's essential minimal medium as recommended by the supplier. Supplements included fetal calf serum (10–20%), 2 mM glutamine, and 100 units/ml penicillin G + 100 μg/ml streptomycin. All cell culture agents were purchased from Invitrogen, Inc., (Carlsbad, CA). Normal melanocytes were isolated from neonatal foreskins as reported before36 and maintained in medium 254 supplemented with 0.2 mM CaCl2, 16 nM TPA and melanocyte growth factors (Cascade Biologics).
Lentiviral and retroviral constructs and infection
Lentiviral expression vector FG12-GFP was described previously.36Lentiviral vectors pLV-SV40-hygro and pLV-SV40-hygro-human-C-MYC were gifts from Dr. Andrei Gudkov (Roswell Park Cancer Institute). pLKO1-control vector (expressing control shRNA) and pLKO1 vectors expressing shRNAs specific to the following human genes TS, IMPDH2, PRPS were purchased from Sigma-Aldrich. shRNAs for C-MYC gene were described previously.18
Lentiviral packaging reactions were performed in the 293-FT cell line in the presence of packaging plasmids VSG and ΔDR using Superfect Transfection Reagent (Qiagen). Viral supernatants were collected 48 hr after transfection, filtered through disposable 0.45 μM cellulose acetate filters and frozen in individual aliquots at -80°C. For infection cells were plated in 60 or 100 mm tissue culture dishes and allowed to achieve 40–50% confluence before adding viral supernatant in the presence of 8 μg/ml polybrene for 24 hours (Sigma, St. Louis, MO).
Retroviral expression vectors pLHCX and pLPCX were purchased from BD Bioscience. cDNA clones for human TS, IMPDH2 and PRPS2 were purchased from American Tissue Culture Collection. Regions of TS, IMPDH2 and PRPS2 cDNAs corresponding to the open reading frames of the above genes were cloned into FG12-GFP pLPCX and pLHCX vectors, respectively. Retroviral infection was performed as described by us earlier.16
Microarray analysis
SK-Mel-19 cells were infected with control or M1 shRNAs in two parallels each. Total cellular RNA was isolated 72 hours post-infection using RNeasy Mini Kit (Qiagen). 7 μg from each samples was used for production of biotinylated cRNA as described in the Affymetrix GeneChip analysis instruction manual (Affymetrix, Santa Clara, Calif, USA). RNA conversion into cDNA, preparation of double-stranded DNA, preparation of biotinylated cRNA and hybridization to Affymetrix GeneChip Array was done at the University of Michigan Microarray Core Facility. The raw experimental microarray data were normalized with the Affymetrix Microarray Suite (MAS 5.0) based on the housekeeping gene method. Expression values obtained were adjusted to the intensity of the corresponding expression value of 100 housekeeping genes. A linear model specifically designed for microarray analysis was applied to the data37 and then differentially expressed probesets were detected using a nested F-test approach. An adjusted p-value of 0.005 was used to assess significance, adjusting for multiple comparisons using false discovery rate.
Reverse-transcription and quantitative real-time PCR (Q-PCR)
cDNA synthesis was performed with 1–5 mg of total cellular RNA using SuperScript III First-Strand Synthesis System for RT-PCR. Real-time PCR was performed using a Taqman Universal PCR Master Mix kit (Applied Biosystems) and 7700 Sequence Detector (Applied Biosystems). PCR primers and probes for the following human genes: C-MYC, β-actin TS, IMPDH2 and PRPS2 were purchased from Applied Biosystems. Target gene mRNA levels were quantified based on standard and normalized to β-actin mRNA levels.
Chromatin immunoprecipitation
Direct binding of c-MYC tothe promoter of studied genes was assessed using the EZ-Chip kitfrom Millipore according to the manufacturer's recommendations.Briefly, 2 × 107 SK-Mel-19 cells were fixed in 1% formaldehyde.Chromatin was sheared to an average DNA size of 200–500 bps bysonication using Microson ultrasonic cell disruptor (10 times of 10second pulses, 50% output). Sonicated chromatin was incubated overnight with 5 μg of anti-C-MYC (N262, Santa Cruz) or rabbit IgG(Upstate) antibodies. Immunoprecipitated DNA was de-crosslinked,purified using columns from the kit and used in PCR (GC-RichPCR System, Roche), with the following primers (all shown in 5′–3′orientation). hTS-1boxFwd:CTGGGTGAGAGAGCGAGACTand hTS-1boxRev:AGGTTGCAGTGAGCCAAGAT; hTS-2boxFwd:CCGTGGCTCCTGCGTTTCC,hTS-2boxRev: TGCATGCCGAATACCGACAG; hTS-3boxFwd: GCCACACCGTCCTGCCGT, hTS-3boxRev:CAGCTGCTATGTATTCGTC-TCT; h-IMPDH2-1BoxFwd:GCCAG-GAAAAACCAGACAGA, h-IMPDH2-1BoxRev: CACTCAAGGAAACCCAGAGC; h-IMPDH2-2BoxFwd: CTATACGCATGCGCTGTTTC, h-IMPDH2-2BoxRev: TGCCCCCACTAATCAGGTAG; h-IMPDH2-3BoxFwd: CTCCCTTCCCTCGCAGTGA, h-IMPDH2-3BoxRev: GACAAACGTCAACCAGTGTG; h-PRPS2-1BoxFwd: CCTGGTTGCTGAACTTCTTC, h-PRPS2-1BoxRev: GCTCGGAG-CTCTCTCGC; h-PRPS2-2BoxFwd: CCTCTGGATAACGCAGTG, h-PRPS2-2BoxRev: GCT-GAACCTCAGACCAGT; h-PRPS2-3BoxFwd: TACTTCAGTGTCTGAGTGAGT, h-PRPS2-3BoxFwd: TTTGTTCAGAAGGTGTCGTCA. PCR products were resolved on 1.5% agarose gel and stained with ethidium bromide.
Protein immunoblots
Total cell lysates were obtained by extraction in Laemmli buffer. Protein samples were separated on 8% or 10% SDS-containing polyacrylamide gels and transferred to PVDF membranes (Millipore, Bedford, MA). Antibodies used in this study were: E2F-1 (sc-193), c-MYC (SC-42) from Santa Cruz Biotechnology, TS (ab7398-1) from Abcam, IMPDH2 (HPA001400) from Atlas Antibodies, PRPS2 (H00005634) from Abnova, tubulin (T5168) from Sigma. Protein levels were estimated by densitometry and normalized with respect to tubulin used as loading controls.
dNTP quantification assays
For the HPLC-based method cells were harvested by trypsinization, extracted using 0.4 N perchloric acid and neutralized. Ribonucleotides were removed from the extracts using a boronate affinity column.34 Cellular dNTPs were separated and quantified using a strong anion exchange column (Whatman, Hillshore, OR) with a gradient HPLC system (Waters Milford, MA) equipped with a photodiode array detector and controlled by Millennium 2010 software. Nucleotides were eluted at 2 ml/min with a linear gradient of ammonium phosphate buffer (0.15 M, pH 2.8 to 0.60 M, pH 2.9 or 3.4). Nucleotides were identified based on their UV absorbance spectrum and quantified at either 254 or 281 nm by comparison to the absorbance of a known amount of authentic standard. For the enzymatic assay, dNTPs were extracted and assayed by the DNA polymerase-based method as described in Song et al.36 For convenience data are presented in the respective figures in terms of arbitrary units. However, the actual pool sizes determined, in pmol/cell, were comparable to values previously reported for cultured mammalian cells.33,38,39
FACS analysis
Cells were pelleted, and the pellet was resuspended in 500 μl of phosphate-buffered saline containing 0.5% fetal calf serum and fixed in 70% ethanol at -20°C overnight. The fixed cells were pelleted and washed once in 500 μl of phosphate-buffered saline containing 0.5% fetal calf serum, pelleted again and resuspended in 800 μl of phosphate-buffered saline containing 0.5% fetal calf serum supplemented with 8 μl of propidium iodide (1 mg/ml) and 4 μl of RNase A (DNase free, 10 mg/ml). Cells were incubated for 30 min at 37°C and analyzed on a BD Biosciences FACScan fluorescence activated cells sorter.
Supplementary Material
Acknowledgments
We are grateful to Laure Rittie (University of Michigan) for critical reading of the manuscript. We thank Elena Tuneva (Roswell Park Cancer Institute) and Suresh Patil (University of Michigan) for technical assistance. This work was supported by Dermatology Research Foundation Carrier Development Award to M.A.N, NIH grant R01-CA120244-A1 to M.A.N., and (in part) by the University of Michigan's Cancer Center Support Grant (5 P30 CA46592). M.A.N is a Melanoma Research Foundation Scholar.
Footnotes
References
- 1.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–5. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 2.Nesbit CE, Tersak JM, Prochownik EV. Myc oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–16. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 3.Hermeking H. The MYC oncogene as a cancer drug target. Curr Cancer Drug Targets. 2003;3:163–75. doi: 10.2174/1568009033481949. [DOI] [PubMed] [Google Scholar]
- 4.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–7. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–4. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–30. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 7.Nair SK, Burley SK. Structural aspects of interactions within the Myc/Max/Mad network. Curr Top Microbiol Immunol. 2006;302:123–44. doi: 10.1007/3-540-32952-8_5. [DOI] [PubMed] [Google Scholar]
- 8.Lee LA, Dang CV. Myc target transcriptosomes. Curr Top Microbiol Immunol. 2006;302:145–68. doi: 10.1007/3-540-32952-8_6. [DOI] [PubMed] [Google Scholar]
- 9.Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- 10.Cole MD, Nikiforov MA. Transcriptional activation by the Myc oncoprotein. Curr Top Microbiol Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- 11.Basseres DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–30. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- 12.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–61. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 13.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–68. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 14.Cappellen D, Schlange T, Bauer M, Maurer F, Hynes MF. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 2007;8:70–6. doi: 10.1038/sj.embor.7400849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, Dang CV, Sedivy JM, Eick D, Vogelstein B, Kinzler KW. Identification of CDK4 as a target of c-Myc. Proc Natl Acad Sci USA. 2000;97:2229–34. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothermund K, Rogulski K, Fernandes E, Whiting A, Sedivy J, Pu L, Prochownik EV. c-Myc-independent restoration of multiple phenotypes by two c-Myc target genes with overlapping functions. Cancer Research. 2005;65:2097–107. doi: 10.1158/0008-5472.CAN-04-2928. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Mannava S, Grachtchouk V, Zhuang D, Soengas MS, Gudkov AV, Prochownik EV, Nikiforov MA. c-Myc depletion inhibits proliferation of human tumor cells at various stages of the cell cycle. Oncogene. 2008;27:1905–15. doi: 10.1038/sj.onc.1210823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. 2003 doi: 10.1186/gb-2003-4-10-r69. http://genomebiology.com/2003/4/10/R69. [DOI] [PMC free article] [PubMed]
- 20.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–4. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 21.Pena A, Reddy CD, Wu S, Hickok NJ, Reddy EP, Yumet G, Soprano DR, Soprano KJ. Regulation of human ornithine decarboxylase expression by the c-Myc:Max protein complex. J Biol Chem. 1993;268:27277–85. [PubMed] [Google Scholar]
- 22.Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, Thompson MA, Yang H, Fujiwara Y, Greenberg ME, Orkin S, Smith C, Nevins JR. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell. 2001;8:105–13. doi: 10.1016/s1097-2765(01)00275-1. [DOI] [PubMed] [Google Scholar]
- 23.Miltenberger RJ, Sukow KA, Farnham PJ. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–35. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–33. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montfort WR, Weichsel A. Thymidylate synthase: structure, inhibition and strained conformations during catalysis. Pharmacol Ther. 1997;76:29–43. doi: 10.1016/s0163-7258(97)00099-5. [DOI] [PubMed] [Google Scholar]
- 26.Hedstrom L. IMP dehydrogenase: mechanism of action and inhibition. Curr Med Chem. 1999;6:545–60. [PubMed] [Google Scholar]
- 27.Becker MA. Phosphoribosylpyrophosphate synthetase and the regulation of phosphoribosylpyrophosphate production in human cells. Prog Nucleic Acid Res Mol Biol. 2001;69:115–48. doi: 10.1016/s0079-6603(01)69046-9. [DOI] [PubMed] [Google Scholar]
- 28.Voeller DL, Rahman M, Zajac-Kaye M. Elevated levels of thymidylate synthase linked to neoplastic transformation of mammalian cells. Cell Cycle. 2004;3:1005–7. [PubMed] [Google Scholar]
- 29.Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–24. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calascibetta A, Cabibi D, Martorana A, Sanguedolce G, Rausa L, Feo S, Dardanoni G, Sanguedolce R. Thymidylate synthase gene promoter polymorphisms are associated with TSmRNA expressions but not with microsatellite instability in colorectal cancer. Anticancer Research. 2004;24:3875–80. [PubMed] [Google Scholar]
- 31.Marcuello E, Altes A, del Rio E, Cesar A, Menoyo A, Baiget M. Single nucleotide polymorphism in the 5′ tandem repeat sequences of thymidylate synthase gene predicts for response to fluorouracil-based chemotherapy in advanced colorectal cancer patients. Int J Cancer. 2004;112:733–37. doi: 10.1002/ijc.20487. [DOI] [PubMed] [Google Scholar]
- 32.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–48. [PubMed] [Google Scholar]
- 33.Martomo SA, Mathews CK. Effects of biological DNA precursor pool asymmetry upon accuracy of DNA replication in vitro. Mutat Res. 2002;499:197–211. doi: 10.1016/s0027-5107(01)00283-4. [DOI] [PubMed] [Google Scholar]
- 34.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–44. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 35.Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Research. 2006;66:6598–605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez Y, Verhaegen M, Miller TP, Rush JL, Steiner P, Opipari AW, Jr, Lowe SW, Soengas MS. Differential regulation of noxa in normal melanocytes and melanoma cells by proteasome inhibition: therapeutic implications. Cancer Res. 2005;65:6294–304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- 37.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 38.Shewach DS. Quantitation of deoxyribonucleoside 5′-triphosphates by a sequential boronate and anion-exchange high-pressure liquid chromatographic procedure. Anal Biochem. 1992;1:178–82. doi: 10.1016/s0003-2697(05)80030-2. [DOI] [PubMed] [Google Scholar]
- 39.Song S, Wheeler LJ, Mathews CK. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J Biol Chem. 2003;278:43893–6. doi: 10.1074/jbc.C300401200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.