Abstract
In most organisms homologous recombination is vital for the proper segregation of chromosomes during meiosis, the formation of haploid sex cells from diploid precursors. This review compares meiotic recombination and chromosome segregation in the fission yeast Schizosaccharomyces pombe and the distantly related budding yeast Saccharomyces cerevisiae, two especially tractable microorganisms. Certain features, such as the occurrence of DNA breaks associated with recombination, appear similar, suggesting that these features may be common in eukaryotes. Other features, such as the role of these breaks and the ability of chromosomes to segregate faithfully in the absence of recombination, appear different, suggesting multiple solutions to the problems faced in meiosis.
The cardinal feature of meiosis is the generation of haploid gametes from diploid precursor cells by the proper segregation of chromosomes. In most species this crucial event is intimately associated with homologous recombination and the generation of genetic diversity. The faithful segregation of homologous chromosomes at the first (reductional) division of meiosis generally requires recombination. In this case meiotic recombination is vital: in its absence homologs missegregate and the resulting aneuploid gametes give rise to defective or inviable progeny. Recombination between homologs during meiosis also generates genetic diversity among the gametes and resultant progeny. Such diversity may be vital to the long-term survival of the species.
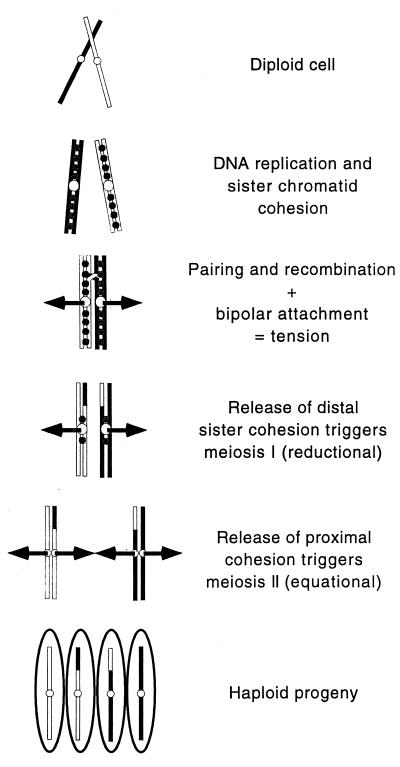
In the conventional view of meiosis, chromosome segregation at the first meiotic division (MI) depends on recombination in the following way (Fig. 1) (reviewed in ref. 1). After replication, a chromatid of one homolog recombines with a chromatid of the other homolog, thereby precisely joining homologous chromosomes. As the centromeres of the homologs are pulled to opposite sides of the cell by the spindle apparatus, tension between the recombined homologs signals that homologs are oriented for proper segregation. Without recombination homologs are not joined, tension is not generated, and homologs eventually move to the poles at random. The second meiotic division (MII) is not preceded by replication, and the sister chromatids separate equationally. Thus, in meiosis one diploid cell produces four haploid cells, which then differentiate into specialized gametes—eggs and sperm in animals, ovules and pollen in plants, or spores in fungi.
Figure 1.
Segregation of recombined chromosomes during the two meiotic divisions. Black and white lines represent homologous chromosomes. Gray circles represent cohesin; open circles represent kinetechores. Arrows represent the meiotic spindle. See text for explanation.
An especially tractable organism for the study of meiotic recombination and chromosome segregation is the fission yeast Schizosaccharomyces pombe (2). Like other ascomycetes it encloses the four haploid products of each meiosis (spores) in a sac, called an ascus. Analysis of these four spores reveals the recombinational and segregational fate of each of the four chromatids (Fig. 1). After germination the spores can be propagated indefinitely as haploid cells, or these haploids can be mated to form stable diploids, which also multiply indefinitely. Having only three chromosomes, S. pombe forms a substantial number of viable spores in the absence of recombination, thereby facilitating studies of meiotic recombination-deficient (Rec−) mutants. Biochemical analyses are aided by a mutant, described below, that undergoes rapid, synchronous meiosis when the temperature is raised. Finally, the nucleotide sequence of the S. pombe genome is essentially complete (www.sanger.ac.uk/projects/S_pombe), and the near isogenicity of the commonly used S. pombe strains aids comparisons between different studies.
S. pombe is only distantly related to the budding yeast Saccharomyces cerevisiae, in which meiosis also has been extensively studied (1–3). Some features of meiosis are the same in the two yeasts, suggesting that they may be common among eukaryotes. Other features, however, appear to be distinctly different and illustrate the diversity of meiosis. In this review we shall emphasize some of these similarities and differences. To place meiotic recombination and chromosome segregation in context, we briefly discuss the changes in physiology, gene expression, and DNA replication during meiosis.
Genetic and Physiologic Control of the Entry into Meiosis
Haploid S. pombe cells of opposite mating type, designated mat1-P (or h+) and mat1-M (or h−), mate only upon starvation, and normally the diploid zygotes directly undergo meiosis. But upon return to a growth medium, the zygotes resume mitotic divisions as diploids, which undergo meiosis when subsequently starved.
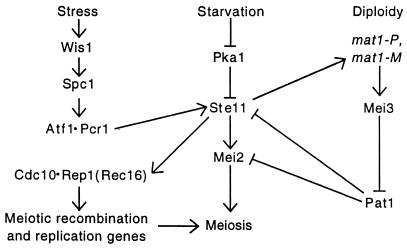
Starvation and mat1 heterozygosity, a sign of diploidy, activate two key regulators of meiosis, Ste11 and Mei2, via three interacting pathways (Fig. 2; reviewed in ref. 4). First, starvation activates the “stress”-induced Wis1-Spc1 protein kinase cascade to phosphorylate the transcription factor Atf1⋅Pcr1. Second, starvation lowers the cAMP level, which inactivates the protein kinase Pka1. These two changes induce ste11 transcription. Third, Ste11 in conjunction with a pheromone signaling pathway induces expression of the heterozygous mat1-P and mat1-M genes; together, their products induce Mei3, an inhibitor of the critical protein kinase Pat1. Ste11 also induces other meiotic genes, including Mei2, which activates multiple meiotic events.
Figure 2.
Control of the entry into meiosis. “Stress” includes starvation, DNA damage, high osmolarity, or heat shock, each of which can activate Atf1⋅Pcr1. Arrowheads indicate activation of the indicated protein or its gene or process; straight lines indicate inhibition or repression. See text for explanation.
In the absence of starvation and mat1 heterozygosity, active Pat1 kinase prevents meiosis by inhibiting Ste11 and Mei2. Thermal inactivation of the Pat1-114 temperature-sensitive mutant protein leads to synchronous meiosis even in haploids: premeiotic DNA replication begins at ∼2 h, MI occurs at ∼5 h, and spores appear at ∼7 h. Meiotic events are similar in thermally induced haploid or diploid pat1-114 mutants and in starvation-induced pat1+ diploids, except that only ∼2% of the spores from pat1-114 haploids are viable, due to insufficient copies of the chromosomes (5, 6).
Control of Meiotic rec Gene Expression
Among the many genes induced in meiosis are those whose products promote recombination (rec genes and others described below). This induction is responsible, at least in part, for the high level of meiotic recombination. Induction of many analyzed rec genes requires Rep1(Rec16), perhaps in a complex with Cdc10, a transcriptional activator that regulates the mitotic cell cycle (Fig. 2; refs. 7 and 8). The rep1(rec16) gene was first identified by a strong meiotic Rec− mutation rec16-125 (9) and later as a high-copy suppressor (rep1+) of a cdc10 mutation (8, 10). Several such high-copy suppressors have been identified, and two (Res1 and Res2) form complexes with Cdc10 (11). Rep1(Rec16) also may complex with Cdc10 to form a meiosis-specific transcriptional activator that induces the other analyzed rec genes and genes required for meiotic replication. The rec6, 7, 8, 10, 11, 12, and 15 genes have nearby MluI sites (5′-ACGCGT-3′; MCB or MluI cell cycle box) or closely related sequences, to which Cdc10 complexes bind (12–17). Induction of rep1(rec16) by Ste11 early in meiosis renders the putative Cdc10⋅Rep1 complex meiosis-specific (10). In addition, Ste11 appears to directly activate some meiotic recombination genes, such as dmc1 (18).
Rep1(Rec16) Links Meiotic Replication and Recombination
That these two processes are closely connected is manifest by the phenotype of rep1(rec16) mutations. The rec16-125 mutation delays meiotic replication by about 2 h and only about half of the cells complete replication; this mutation reduces recombination by a factor of about 50 (7, 9). The rep1∷ura4+ null allele essentially abolishes both meiotic replication and recombination (8, 10). These observations led to the proposal that these two events are mechanistically connected, as in prokaryotes (7, 19, 20). The connection via Rep1(Rep16) is most simply explained, however, by Rep1(Rec16) inducing two sets of meiotic genes: one for replication and one for recombination (ref. 8; Fig. 2). Recent evidence, however, indicates that DNA replication is a necessary prelude to meiotic DNA breakage in S. cerevisiae (21).
Gene Products Required for Meiotic Recombination
The products of more than two dozen identified genes are required for meiotic recombination in S. pombe (Table 1 and references therein). Mutations in these genes confer a wide range of deficiencies in recombination, from a modest reduction (∼3-fold) to near abolition (>1,000-fold reduction), suggesting that some steps are more critical than others or that there are redundant means for some steps. Some of these mutations are specific for meiotic recombination; others affect additional meiotic or mitotic events, suggesting a close interrelation between recombination and other events such as meiotic replication and chromosome segregation or mitotic DNA repair.
Table 1.
Meiotic recombination proteins in S. pombe
| Proteina | Extent of reduction by mutationb | Putative S. cerevisiae homologc | Inferred primary activityd | References |
|---|---|---|---|---|
| Control of rec gene expression | ||||
| Rep1(Rec16) | ↓↓↓ | — | Meiosis-specific partner for Cdc10 | (7, 8, 10) |
| M26 hotspot activation | transcriptional activator | |||
| Atf1⋅Pcr1 (Mts1⋅Mts2, Gad7⋅Pcr1) | ↓ | Sko1 | Heterodimeric transcriptional activator; binds M26 and related sequences | (45–48) |
| Spc1 (Sty1) | ↓ | Hog1 | Protein kinase; phosphorylates Atf1 | (45, 47, 48) |
| Wis1 | ↓ | Pbs2 | Protein kinase; phosphorylates Spc1 | (45) |
| Meiotic DNA breakagee | ||||
| Rec6 | ↓↓↓ | — | (14, 25) | |
| Rec7 | ↓↓↓ | Rec114 | (13, 25, 83) | |
| Rec12 | ↓↓↓ | Spo11 | Active site protein | (9, 14, 30) |
| Rec14 | ↓↓↓ | Ski8 (Rec103) | (9, 86) | |
| Rec15 | ↓↓↓ | — | (9, 16) | |
| Putative action after DNA breakage | ||||
| Rad50 | Rad50 | Processing of DNA breaksf | ||
| Rad32 | ↓ | Mre11 | Processing of DNA breaks | (29) |
| Dmc1 | ↓ | Dmc1 | Strand exchange | (18) |
| Rqh1 (Rad12, Hus2, Rec9) | ↓g | Sgs1 | DNA helicase | (87, 88) |
| Nuclear movement, telomere clustering, or chromosome pairing | ||||
| Kms1 | ↓ | Nuf1? | Spindle pole body component | (65) |
| Dhc1 | ↓ | Dyn1 | Dynein heavy chain | (68) |
| Taz1 | ↓ | Smc2? Tbf1? | Telomere-binding protein | (63, 89) |
| Meu13 | ↓ | Hop2 | K. Nabeshima, personal | |
| SCC | communication | |||
| Rec8 | ↓↓h | Rec8 | SCC; LE formation | (13, 22, 24, 25, 72) |
| Rec10i | ↓↓h | — | (15, 22, 24, 25) | |
| Rec11 | ↓↓h | Irr1(Scc3)? | SCC | (17, 22, 24, 25) |
| Mismatch repair | ||||
| Pms1 | * | Pms1 | Mismatch-binding | (40) |
| Msh2 | * | Msh2 | Mismatch-binding | (41) |
| Swi10 | * | Rad10 | Strand incision | (42) |
| ExoI | * | ExoI | 5′→3′ exonuclease | (90, 91) |
Only proteins whose genes are assigned to a nucleotide sequence are listed. Other genes, mutations in which reduce meiotic recombination, include rec13 and rec17 to 21 (9) and swi5 (9, 92). Genes controlling the induction of meiosis are not listed (see text and Fig. 2).
Alternate designations are in parentheses.
↓↓↓, reduction by a factor of ∼1,000; ↓↓, by ∼100; ↓, by ∼3–10; ∗, major effect is the increase of PMS, as recombinant frequencies may be increased or decreased by a factor of ≤5.
Homology is based on a low probability (<10−4) of a random match in a blast search and on similarity of mutant phenotypes. —, no clear homolog; ?, homology is less extensive or phenotypes differ, making functional homology less certain.
Primary activity is inferred from biochemical, cytological, and mutant phenotype analyses in one or both organisms. A blank indicates absence of sufficient information for an inference.
From ref. 30 and J. Young and R. Schreckhise (personal communication).
Based on homology between the two Rad50 proteins, the phenotype of a Rad50S mutant (E. Hartsuiker and J. Kohli, personal communication), and the accumulation of meiotic DNA breaks in a Rad50S mutant (R. Schreckhise and J. Young, personal communication).
J. Young and G. R. S., unpublished data.
Ranges from <∼3-fold to >100-fold, depending on the genetic interval examined.
There is no direct evidence that Rec10 is involved in SCC. It is placed here based on its regional specificity of recombination similar to that of Rec8 and Rec11 (22).
Meiotic Rec− mutants were identified in multiple ways. A direct screen for such mutants revealed 16 complementation groups, rec6–rec21, 10 of which have been assigned to sequenced genes (Table 1). Certain mutants identified on another basis subsequently were found to be Rec−; these include the radiation-sensitive mutant rad32 and the mating type switching-defective mutant swi5. Some meiotically induced genes, such as dmc1 and meu13, were found to be Rec− when mutated. A search for biochemical activities relevant to recombination revealed the M26 hotspot-activating protein Atf1⋅Pcr1 and the mismatch repair exonuclease ExoI.
The rec8, rec10, and rec11 mutants display an unusual regional specificity (22–24). Recombination in some intervals of the genome is reduced as much as 100- to 300-fold, whereas in other intervals the reduction is ∼3-fold or less. The strongly affected intervals are in the central regions of the chromosomes (encompassing the centromeres), and those less affected are nearer the ends. Rec8 and Rec11 encode meiosis-specific sister chromatid cohesins, which may be predominantly localized in the central regions of meiotic chromosomes; the role of cohesins in meiotic recombination is an intriguing problem discussed later.
Studies of meiotic Rec− mutants have revealed both similarities and differences between S. pombe and S. cerevisiae. In some cases there are close homologs that appear to have the same function; for example, Rec12 and Spo11 are homologs intimately involved in meiotic DNA breakage, discussed later. But in other cases, such as Rec6, Rec10, and Rec15, S. cerevisiae has no obvious homologs; similarly S. pombe has no obvious homologs of several S. cerevisiae meiotic recombination proteins, including Rec102, Rec104, Mei4, Sae2, Xrs2, Hop1, Red1, Msh4, and Msh5.
In still other cases there are clear amino acid sequence homologs that appear to function differently. For example, S. pombe rqh1 (rec9) and S. cerevisiae sgs1 mutants lack closely related DNA helicases; although the rqh1 mutants are reduced ∼5-fold for meiotic recombination (ref. 25, J. Young and G.R.S., unpublished data), the sgs1 mutants are not significantly reduced (26). Conversely, S. pombe dmc1 mutants complete sporulation and are only modestly Rec− (∼3-fold reduction) (18), whereas S. cerevisiae dmc1 mutants are blocked before the first meiotic division and have a stronger reduction of recombination (27). S. cerevisiae Mre11 is essential for both formation and repair of meiotic DNA breaks (28), whereas its S. pombe homolog Rad32 appears to be involved only in the repair of breaks: the sporulation deficiency of rad32 mutants (29) is suppressed by a rec6 mutation, which blocks break formation (ref. 30; J. Bedoyan and G.R.S., unpublished data). S. cerevisiae rad51, 52, 54, and 55 mutants are blocked in the repair of meiotic DNA breaks, recombination, and sporulation (1), but S. pombe mutants lacking distinct homologs of these proteins are only mildly deficient in meiotic recombination, if at all, although the spore viability of the rhp51 mutant is only ∼5% of wild type (31, 32). Further studies may reveal whether these apparent similarities and differences are reflected in the basic mechanism of meiotic recombination in the two organisms.
The M26 Meiotic Recombination Hotspot
A genetic interval contains a recombination hotspot if the frequency of exchange per unit physical distance is greater than that of the genome average (reviewed in refs. 33 and 34). Hotspots presumably stimulate a rate-limiting step in recombination and therefore identify a crucial step for study. In fungi hotspots were first recognized by the high frequency of gene conversion (nonreciprocal recombination) of markers at or near the hotspot; these markers occasionally segregate as three wild-type spores and one mutant spore (3+:1−) or the reverse (1+:3−) in an ascus, rather than the normal 2+:2− Mendelian segregation. In S. cerevisiae the frequency of convertant asci ranges widely from 0.6% to 18%, depending on the marker and locus studied (35); several loci with a high frequency of gene conversion are near sites of frequent DNA double-strand breaks (33), as discussed later. In S. pombe the frequency of gene conversion of most markers is much lower, about 0.25%, and seems more uniform, although the data are limited (P. Munz, personal communication).
The S. pombe ade6-M26 mutation, discovered by Gutz (36), creates a hotspot called M26. This mutation converts in about 5% of meioses, about 10 times more frequently than other ade6 mutations. The M26 mutation is a single base pair change creating a nonsense codon near the 5′ end of the gene (GGA→T GA) (37). The ade6-M375 mutation, frequently used as a nonhotspot control, is an identical change in the preceding GGA codon. M26 recombines with other ade6 mutations to produce Ade+ spores about 10 times more frequently than does M375 (36). This simple assay for M26 hotspot activity has been used in many subsequent studies and showed, for example, that M26 is meiosis-specific (38). In addition to conversion, M26 stimulates crossing-over (reciprocal recombination) between flanking markers (39). M26 is at least as active when homozygous as when heterozygous (38), showing that M26 hotspot activity is not simply a reflection of unusual mismatch correction.
Mismatch correction does, however, influence the outcome of M26-stimulated recombination. In otherwise wild-type cells the convertant asci are either 3+:1− or 1+:3− (36). In cells deficient for mismatch repair, however, the M26 mutation frequently manifests postmeiotic segregation (PMS), a reflection of heteroduplex DNA containing a base mismatch at the M26 site. In this event a single spore germinates, divides, and produces an ade6+ cell and an ade6-M26 cell; counting the eight cells present after the four spores divide, the ascus is said to be 5+:3− or 3+:5− if one spore shows PMS or to be aberrant 4+:4− if two spores show PMS. In pms1 or msh2 mismatch repair-deficient mutants 56% or 85%, respectively, of the non-Mendelian events manifest PMS (40, 41). In a pms1 swi10 double mutant background, in which mismatch repair is more severely impaired but perhaps not eliminated, a mutation very near the (homozygous) M26 hotspot manifests 91% PMS; 5+:3−, 3+:5−, and aberrant 4+:4− asci are equally frequent (42). Thus, heteroduplex DNA at the M26 site is usually formed on one chromatid and at least 30% of the time on two chromatids, reflected in the aberrant 4+:4− asci. In mismatch repair-proficient cells M26 produces about eight times more 3+:1− asci than 1+:3− asci (36). Thus, M26 is a recipient of genetic information, but this disparity may reflect a bias either in the direction of mismatch repair or in the basic mechanism of the M26 hotspot, such as the formation of DNA breaks and strand exchange; it may reflect both.
When M26 converts, nearby markers are frequently co-converted; the conversion tracts can extend to the left of M26 or to the right or both (36). The frequency of co-conversion of a marker decreases roughly exponentially with its distance from M26, approximately a factor of 2 for every 500 bp (43). Thus, the observed recombinational exchange points are typically within about 1 kb of M26.
The M26 hotspot is one of a family of hotspots with closely related nucleotide sequences. The ade6-M26 mutation creates the sequence 5′-ATGACGT-3′; any single bp mutation within this heptamer strongly reduces or abolishes hotspot activity, whereas mutations outside this heptamer have little or no effect (44). A recent study of multiple bp changes, however, showed that the related sequences 5′-(C/T/G)TGACGT(A/C)-3′ have hotspot activities similar to that of M26 and that 5′-GTGACGTG-3′ has partial activity (45).
The earlier finding of a unique sequence required for M26 hotspot activity (44) led to a search for a protein that binds and activates the hotspot. A “gel shift” assay for a protein specifically binding M26 DNA identified a heterodimeric protein Mts1⋅Mts2 (46). The binding of this protein to M26 and single bp mutant DNA fragments correlates well with the hotspot activities of these sequences, implying that the protein is required for hotspot activity. Further analysis (47) showed that Mts1⋅Mts2 is the transcription factor Atf1⋅Pcr1 important in the induction of meiosis (Fig. 2) but not essential for it (48). atf1 and pcr1 null mutants lack hotspot activity but have nearly wild-type levels of recombination in the absence of the M26 hotspot (in crosses using M375) (47). Thus, Atf1⋅Pcr1 is indispensable for the M26 hotspot but is not required for basal level recombination, at least at ade6. One class of S. cerevisiae hotspots also requires transcription factors and their binding sites (49). The ability of Atf1⋅Pcr1 to bind to the M26-related sequences 5′-(C/T/G)TGACGT(A/C)-3′ correlates well with the hotspot activities of these sequences (45). The one M26-related hotspot tested further (5′-CTGACGTA-3′) requires Atf1⋅Pcr1 and the Wis1-Spc1 protein kinase cascade, is meiosis-specific, and alters the local chromatin structure (see below).
The M26 family of hotspot sequences is closely related to the mammalian cAMP response element (CRE) consensus sequence 5′-NTGACGT(A/C)-3′, which is activated by the transcription factor ATF1. The S. pombe homolog Atf1, with Pcr1, binds CRE sites and activates transcription of stress-induced genes (50). The meiotic specificity of M26 may be due in part to the activation of Atf1⋅Pcr1 during meiosis, but other meiosis-specific factors such as the Rec proteins must be involved, because stresses that activate Atf1⋅Pcr1 but not meiosis do not activate M26 (M. Fox, personal communication).
The protein kinase cascade Wis1-Spc1 required for activation of the Atf1⋅Pcr1 transcription factor in response to stress (Fig. 2) also is required for M26 hotspot activity (45, 48). This observation and others discussed above raise the question of whether transcription is activated by Atf1⋅Pcr1 near M26 and whether such transcription might be related to M26 hotspot activity. The available evidence is not conclusive but suggests not. The levels of ade6 transcripts are not significantly different whether the hotspot is active or not in the following situations: meiosis vs. mitosis, M26 vs. M375, atf1+ vs. atf1 null mutant (47, 48). However, a 500-bp deletion of the putative promoter region of ade6 inactivates the hotspot when in coupling (cis) with M26 but not when in repulsion (trans) (51). Similarly, replacement of the ade6 promoter with the stronger adh1 promoter increases both ade6 transcription and ade6 meiotic recombination (52). These changes in recombination might be due to changes of transcription at or near M26 or to alteration of the chromosomal context of M26, which may indirectly affect its activity.
The activity of the M26 hotspot clearly depends on its context within the genome. When DNA fragments 0.4–5.9 kb long and containing ade6-M26 near their center are moved to other locations in the genome, in most cases M26 is inactive (53–55). For example, only four of 16 ade6-M26 fragments inserted into the ura4 locus, about 1 Mb away from ade6, manifest M26 hotspot activity. These results demonstrate the genomic context-dependence of M26, but the basis of this dependence remains unclear. There is no obvious difference between the active and inactive transplacements, such as length or orientation, that can account for the marked differences in their activity. Presumably, some complex aspect of chromosomal organization, such as chromatin structure, has a crucial influence on M26 hotspot activity.
The reciprocal influence is clear: M26 alters the local chromatin structure. Isolated M26 chromatin has a micrococcal nuclease (MN)-sensitive site very near M26 not seen in M375 chromatin. The sensitivity of this site increases during meiosis and requires Atf1⋅Pcr1, the Wis1 and Spc1 protein kinases, and an active M26 heptamer or related sequence (refs. 45 and 56; K. Ohta, personal communication). These observations are consistent with the view that activated Atf1⋅Pcr1 binds to M26 and “opens” the chromatin structure during meiosis, thereby allowing recombination-promoting proteins access to the DNA, perhaps facilitated by direct interaction with Atf1⋅Pcr1. However, there is much greater MN sensitivity at two sites in the putative ade6 promoter region, about 250 bp and 400 bp from M26, than at M26 itself; these sensitivities increase during meiosis but are not M26-dependent (45, 56). Thus, the total amount of open ade6 chromatin is not much different in M26 and M375 chromatin. Perhaps the open chromatin at the M26 site is especially receptive to recombination-promoting proteins.
The relation between M26 and chromatin structure suggests that the inactivity of M26 in most transplacements, described above, may be due to a gross alteration of chromatin structure in those transplacements; such alterations have been observed for transplaced hotspots in S. cerevisiae (33). To test this suggestion, the M26 heptamer was created by 1- to 3-bp changes at four widely spaced sites in ade6 and one in ura4 in both orientations; in each case an active hotspot was created (57). This result suggests that the wild-type chromatin at ade6 and ura4 is receptive to M26 action, provided that only minimal alterations are made. A corollary is that at least some of the hundreds of M26 heptamers and related sequences in wild-type S. pombe are active meiotic recombination hotspots. This prediction has not been rigorously tested.
DNA Breaks Associated with Meiotic Recombination
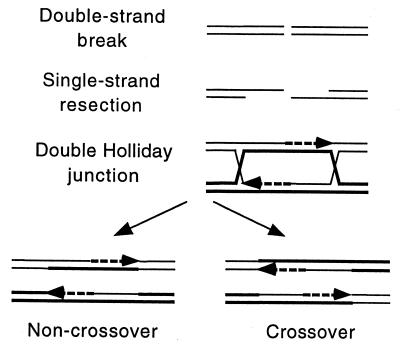
In S. cerevisiae DNA double-strand breaks appear to be precursors of most meiotic recombination (1, 33). These breaks occur at hotspots of recombination and are made, in conjunction with other proteins, by the Spo11 protein, which becomes covalently attached to the 5′ ends at the break. Resection of the 5′-ended strands, perhaps by the Rad50⋅Mre11⋅Xrs2 complex, exposes 3′-ended single strands (Fig. 3). Aided by Rad51, Dmc1 and perhaps other strand-exchange proteins, these single strands are thought to invade duplex DNA of the homolog to form displacement loops (D loops). These joint molecules apparently are converted into double Holliday junctions, which are processed into recombinant molecules with either the parental (noncrossover) or nonparental (crossover) configuration of DNA flanking the Holliday junctions. Heteroduplex DNA between the Holliday junctions usually is corrected by mismatch repair enzymes to produce a gene conversion (3+:1− or 1+:3−) or a restoration (2+:2−). Resection of the 5′ ends is limited to about 1 kb; the distance between the two Holliday junctions and, hence, the distance between the breaks and the recombinational exchange points is about 1–2 kb. These features are largely in accord with the double-strand break repair model proposed by Szostak et al. (58), except that conversion occurs by mismatch repair, not by double-strand gap repair as originally proposed.
Figure 3.
Meiotic recombination initiated by a DNA double-strand break [after Szostak et al. (58)]. Thin and thick lines indicate single DNA strands of the parental chromatids. Dashed lines indicate newly synthesized DNA; arrowheads indicate 3′ ends. Alternative resolutions at the last step are not shown. See text for explanation.
The similarities between the genetic properties of the M26 hotspot in S. pombe and meiotic hotspots in S. cerevisiae encouraged the search for DNA breaks at M26. These searches were initially fruitless, but the methods used may have been inadequate to detect breaks (see below). The homology between Rec12 and Spo11, including a conserved tyrosine residue essential for meiotic recombination in both yeasts, and the likely direct involvement of this residue in DNA break formation (30, 59, 60), reignited the search for DNA breaks in S. pombe.
To expand the search beyond the M26 hotspot, whole chromosome DNA was examined by pulsed-field gel electrophoresis (ref. 30; Fig. 5A, which is published as supplemental material on the PNAS web site, www.pnas.org). Before meiotic induction the three chromosomes are largely intact. Shortly after replication intact DNA mostly disappears and is replaced with faster migrating (broken) DNA. At about the time of MI the broken DNA disappears and intact DNA reappears. This is the behavior expected of DNA broken and subsequently repaired by recombination.
This meiotic DNA breakage is closely associated with recombination, because it depends on multiple rec gene products. Meiosis-specific breakage is not detectable in rec6, 7, 12, 14, or 15 mutants and is strongly reduced in rec8, 10, and 11 mutants (ref. 30; J. Young and R. Schreckhise, personal communication). There is, thus, a strong correlation between the deficiency of meiotic recombination and the deficiency of meiotic DNA breakage in all seven rec mutants. This outcome renders it unlikely that the rec gene products promote two separate processes—recombination and DNA breakage—that happen to be mutationally inactivated in each case. The simplest interpretation is that recombination depends on DNA breakage. The alternative, that breakage depends on recombination, is not ruled out but is not easily accommodated by current models of recombination.
In S. pombe, meiotic DNA breakage occurs at a limited number of sites. This is seen globally as the occurrence of discernible bands, rather than continuous smears, of ethidium bromide-stained (bulk) DNA on electrophoretic gels that separate DNA molecules in the range of 0.1 to 1.5 Mb (30). Southern blot hybridizations of such gels reveal prominent meiotic break sites ∼100–200 kb apart (ref. 30; Fig. 5B). A more detailed analysis of a 501-kb segment of chromosome I (NotI fragment J) indicates six sites, or tight clusters of sites, of meiosis-specific, Rec12-dependent breakage spaced ∼40 kb to ∼135 kb apart (J. Young and R. Schreckhise, personal communication). This spacing appears to be much greater than that in S. cerevisiae, which has about 3–5 kb between break sites in most regions of its chromosome III (61).
As noted above, meiotic recombination depends on DNA breakage in both S. pombe and S. cerevisiae. The differences in spacing of the breaks and in required proteins suggest, however, that some aspect of the breaks, such as their regulation or role in recombination, is different in the two organisms. In S. pombe the breaks may promote recombination both near to and far from the sites of breakage. Our laboratory has recently detected meiosis-specific, hotspot sequence-dependent breakage at or near two M26 heptamer sites in ade6 (R. Schreckhise and W. Steiner, personal communication). The factors required for this breakage and the locations of the breaks have not been completely determined. Nevertheless, there appears to be meiotic DNA breakage associated with the M26 hotspot. These results suggest that recombination can be stimulated near the site of M26 breakage, perhaps as diagramed in Fig. 3.
Recombination may occur far from other meiotic breaks. The intensity of recombination (centimorgans per kb) differs by a factor of ∼2 or less for many tested genetic intervals, with or without prominent break sites, in the 501-kb segment of chromosome I containing six widely spaced break sites (J. Young and G.R.S., unpublished data). The currently available data thus suggest that recombination is more uniformly distributed than are the prominent DNA breaks. This view is consistent with the fairly uniform frequency of gene conversion at most loci in S. pombe and the lack of tight clusters of genes on the S. pombe genetic map. Recombination thus appears frequently to occur far from these prominent meiotic DNA breaks, but low-level, as-yet-undetected breaks may account for some recombination. The prominent breaks may relieve torsion in a chromosomal domain and thereby allow recombination at a distance, or they may allow the entry of a traveling entity, such as a protein complex or a DNA structure, that promotes recombination at a distance.
Nuclear Reorganization and Movement
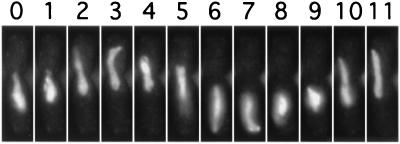
As a prelude to meiotic recombination and chromosome segregation, there is in many organisms considerable reorganization of the nuclear architecture as cells switch from the mitotic to the meiotic program. During mitotic growth the centromeres cluster near the microtubule organizing center. At the onset of meiosis the chromosomes reorient and the telomeres cluster near the microtubule organizing center, called the spindle pole body (SPB) in yeast. In S. pombe, but apparently not S. cerevisiae, dramatic nuclear movements accompany this reorganization (reviewed in ref. 62). The nucleus oscillates from end to end within the cell led by the SPB in a microtubule-dependent manner during prophase of meiosis I. This is termed horsetail movement because of the characteristic morphology of the nucleus (Fig. 4).
Figure 4.
Horsetail movement observed in live meiosis. Photomicrographs are of a single cell whose DNA was stained with Hoechst 33342. The numbers at the top are time in minutes. [Reproduced with permission from ref. 1 (Copyright 1994, American Association for the Advancement of Science).]
Telomere clustering and horsetail movement are required for efficient homolog pairing and meiotic recombination. The telomere binding protein Taz1 and the spindle pole body component Kms1 both are required for proper meiotic telomere clustering and normal horsetail movement (63–67). Deletion of dhc1, which encodes the heavy chain of the microtubule motor protein dynein, eliminates horsetail movement and reduces homolog pairing (68). Consistent with a pairing defect, all three mutants reduce meiotic recombination from 2- to 10-fold (63, 65, 67, 68).
These results indicate that efficient pairing and recombination of homologous chromosomes in meiosis require proper telomere clustering and horsetail movement. These processes may help align homologs much as swinging ropes (horsetail movement) held up by their ends (telomere clustering) would tend to align those of equal length and limit the interaction of those of unequal length. Consistent with this view, minichromosome III derivatives do not efficiently recombine with full-length chromosome III, and their recombination rate is increased in kms1 and taz1 mutants (66).
Linear Elements not Synaptonemal Complex
In most eukaryotes paired homologs are found in the context of a tripartite proteinaceous structure called the synaptonemal complex (SC) (reviewed in ref. 1). SC formation begins with the elaboration of two axial elements, which form between sister chromatids and extend the length of the aligned homologs. Synapsis is complete when the axial elements are brought together by a central core to form the SC. S. pombe cells lack SC but possess structures called linear elements (LE) that appear to be analogous to the axial elements of true SC (69). Unlike SC, LE do not appear to form continuously along the length of the chromosomes.
The roles of SC and LE in recombination may differ. Rec8 is required for normal SC and LE formation in S. cerevisiae and S. pombe, respectively (70, 71). In S. cerevisiae rec8 mutants, wild-type levels of meiotic DNA breaks are formed, although they remain unrepaired (70). In S. pombe rec8 mutants, however, only low levels of meiotic DNA breaks are formed, at least in the regions of the genome analyzed (30). Nonetheless, some regions of the genome recombine at appreciable levels (22–24). Together with the high viability of rec8 mutant spores (72), these results suggest that Rec8 is not required for repair of meiotic DNA breaks in S. pombe. Thus, the LE of S. pombe may be important for formation of breaks, and the SC of S. cerevisiae important for their repair. Alternatively, Rec8 may have dual but independent roles in each organism.
Sister Chromatid Cohesion (SCC), Recombination and Meiotic Segregation
In meiosis two successive nuclear divisions follow a single round of DNA replication (Fig. 1). Homologous chromosomes segregate to opposite poles at MI, then sister chromatids segregate at MII. At MI, unlike mitosis or MII, one functional kinetochore, the proteinaceous bridge linking the chromosomes to the spindle microtubules, is associated with each homolog rather than with each chromatid. At MII, like mitosis, each sister chromatid has one functional kinetochore.
In most organisms SCC and reciprocal recombination provide the physical connection between homologs required to ensure proper positioning of the chromosomes on the MI spindle. SCC then must be released on the chromosome arms, distal to all reciprocal recombination events, to allow homologs to segregate. But SCC must be maintained proximal to the centromere until MII to properly orient the chromosomes on the MII spindle (Fig. 1). Centromere-proximal cohesion is then released, allowing sister chromatids to segregate.
Our current understanding of SCC, mostly from mitosis in S. cerevisiae, indicates that a specialized protein complex called cohesin forms a physical connection between sister chromatids. Cohesin is actively loaded during late G1 and activated during S phase (reviewed in refs. 73 and 74). Cohesin components Scc1, Scc3, Smc1, and Smc3 are conserved among eukaryotes. Although there are meiosis-specific homologs of cohesin subunits (see below), the complete composition of cohesin in meiosis has not been determined.
While SCC is required for proper alignment of chromosomes on the spindle, cohesion must be released at the proper time to allow their segregation to opposite poles (reviewed in ref. 75). In mitotic S. cerevisiae cells, separin triggers the release of cohesion by proteolytic cleavage of the cohesin Scc1. The anaphase-promoting complex indirectly controls this reaction by directing the degradation of securin, an inhibitor of separin. In meiosis, securin is degraded at anaphase of both MI and MII, and separin-dependent cleavage of Rec8 is required for the segregation of recombined homologs at MI (76). The factors required to maintain cohesion proximal to the centromeres until MII are unknown.
Interestingly, SCC is linked genetically to DNA repair and meiotic recombination. Mutations that perturb SCC generally inhibit mitotic repair, meiotic recombination, or both (reviewed in ref. 77). For instance rad21, which encodes the S. pombe Scc1 homolog, was first identified by a radiation-sensitive mutant (78). In mitotic cells the template for repair of a DNA lesion is usually the sister chromatid, perhaps because SCC ensures the proximity of the lesion and the template. Because meiotic recombination occurs between homologs, not sisters, a similar role for SCC in meiosis is harder to understand. SCC proteins may have a role in meiotic recombination distinct from their role in promoting cohesion.
S. pombe Rec8, which is required for wild-type levels of meiotic DNA breaks and recombination (see above), is a meiosis-specific homolog of the cohesin subunit Scc1 (23). Consistent with the region-specific reduction of recombination in rec8 mutants that, to a first approximation, reduces recombination near the center of chromosomes most severely (see above), Rec8 is concentrated near the centromeres in the meiotic nucleus (72). A role in SCC is shown by the premature sister chromatid separation in rec8 mutant meioses (71). Additionally, a rec8 mutant undergoes a completely equational MI in which sister chromatids segregate to opposite poles (72). This indicates that each chromatid is associated with a functional kinetochore. Perhaps Rec8 promotes a specialized cohesive structure, such as LE, that prevents the activation of both sister kinetechores on a homolog at MI.
In S. pombe, inactivation of Pat1 in G2 cells induces entry directly into meiosis with already replicated homologs and bypasses meiotic DNA replication. These cells exhibit phenotypes similar to those of rec8 mutants: reduced meiotic recombination and equational segregation at MI (79). This implies that Rec8 must be either loaded onto chromosomes or activated during replication to be functional.
Mutations in rec10 and rec11, like rec8, affect meiotic recombination in a region-specific manner (see above). Genetic and cytological analysis demonstrate elevated levels of meiotic chromosome missegregation in rec10 and rec11 mutants (24). Together with the significant sequence similarity shared by Rec11 and the cohesin subunit Scc3, these results suggest that Rec11, and perhaps Rec10, are required for meiotic SCC. Strangely, the predominant class of viable abnormal meiotic products from rec10 and rec11 mutants, unlike those from rec8, is indicative of errors in MII not MI (24). The phenotypic differences between rec10 or 11 mutants and rec8 mutants indicate that their functions do not completely overlap. Perhaps Rec10 and Rec11 are required for cohesion only proximal to the centromere and thus are required only at MII.
Meiotic Segregation in the Absence of Recombination
In several species, perhaps including S. pombe, there are exceptions to the rule that recombination is necessary for the MI reductional division. For instance, in Drosophila female meiosis chromosome IV never recombines and the X chromosome fails to recombine ≈5% of the time. Nonetheless, chromosomes segregate properly in over 99% of meioses (reviewed in refs. 80 and 81). Mutations that are specifically defective in the segregation of recombinationless chromosomes define two distinct achiasmate segregation systems. One of these determines which chromosomes segregate from each other based on homology, and the other on size (81). Despite the ability to properly segregate naturally occurring achiasmate chromosomes, mutations that abolish meiotic recombination display significantly increased levels of MI nondisjunction, presumably because the system cannot accommodate all four pairs of homologous chromosomes (80). A similar system has been demonstrated in S. cerevisiae. Nonhomologous artificial chromosomes segregate properly in 90% of meioses even though they do not recombine (82). Presumably, the achiasmate segregation system can be overloaded, because natural chromosomes segregate randomly when recombination is eliminated in a spo11 mutant (70). Lastly, Drosophila males undergo no detectable meiotic recombination, yet their chromosomes properly segregate, indicating a highly efficient achiasmate segregation system (80).
S. pombe also may have an achiasmate segregation system. In rec7 mutants there is no detectable meiotic recombination, and spore viability is reduced to ∼25% (22, 25, 83). Unexpectedly, a large fraction of the viable spores (39% vs. <1.4% in wild type) are homozygous diploids (83). Most of these arise from two-spored asci, which comprise ∼30% of the total asci in rec7 and only ∼1% of total asci in wild type. Dissection of the two-spored asci from the rec7 mutant meiosis revealed that in 46% both spores were viable homozygous diploids, the result of a single reductional (MI) division in which all chromosomes segregated properly. Assuming that the three chromosomes segregate equally well, these data suggest that in the absence of recombination each chromosome segregates properly >75% of the time, more frequently than random. Recombination thus seems to facilitate reductional segregation but not to be essential for it. Our limited analysis of rec6, 14, and 15 mutants and extensive analysis of rec12 mutants have given results similar to those of rec7. This suggests that the segregation pattern described above is a general characteristic of Rec− meiosis in S. pombe.
Although analysis of the meiotic segregation of two minichromosomes in Rec+ cells does not indicate an achiasmate segregation system (84), the fact that the minichromosomes studied were homologous to full-length chromosome III makes this experiment difficult to interpret. A homology-dependent achiasmate segregation system might have difficulty in properly segregating four chromosomes that share extensive homology. Perhaps, like Drosophila females, S. pombe has a homology-dependent achiasmate segregation system that is inefficient in the complete absence of recombination.
Perspectives
All sexually reproducing organisms face the problem of reductional chromosome segregation in meiosis. Many species use recombination between homologs to accomplish this vital task, but some succeed without recombination. Others, including S. pombe, appear to use both recombination-dependent and recombination-independent mechanisms. The mechanism of recombination also appears to differ between some species, including the two model organisms discussed here, S. pombe and S. cerevisiae. The ease of genetic and biochemical manipulation of these unicellular organisms promises to speed the elucidation of the molecular mechanisms of meiotic recombination and chromosome segregation. This knowledge will facilitate investigations in other organisms, including humans, and allow an assessment of the diversity of meiotic mechanisms among eukaryotes.
Supplementary Material
Acknowledgments
We thank Jirair Bedoyan, Mary Fox, Edgar Hartsuiker, Jürg Kohli, Peter Munz, Kentaro Nabeshima, Paul Nurse, Kunihiro Ohta, Randall Schreckhise, Walter Steiner, Yoshinori Watanabe, and Jennifer Young for unpublished information; Yasushi Hiraoka for permission to reprint Fig. 4; Sue Amundsen, Susan Biggins, Walter Steiner, Andrew Taylor, and Jeff Virgin for helpful comments on the manuscript; and Karen Brighton and Jennifer Young for help in preparing it. Research in our laboratory is supported by National Institutes of Health Grants GM31693 and GM32194 to G.R.S. and National Research Service Award F32 GM20125 and National Institutes of Health Traineeship T32 CA09437 to L.D.
Abbreviations
- MI
first meiotic division
- MII
second meiotic division
- Rec−
meiotic recombination-deficient
- PMS
postmeiotic segregation
- SC
synaptonemal complex
- LE
linear elements
- SCC
sister chromatid cohesion
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Roeder G S. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 2.Nasim A, Young P, Johnson B F. Molecular Biology of the Fission Yeast. San Diego: Academic; 1989. [Google Scholar]
- 3.Forsburg S L. Trends Genet. 1999;15:340–344. doi: 10.1016/s0168-9525(99)01798-9. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto M, Imai Y, Watanabe Y. In: Yeast III. Pringle J R, Jones J W, Broach J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 1037–1106. [Google Scholar]
- 5.Iino Y, Yamamoto M. Mol Gen Genet. 1985;198:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 6.Bähler J, Schuchert P, Grimm C, Kohli J. Curr Genet. 1991;19:445–451. doi: 10.1007/BF00312735. [DOI] [PubMed] [Google Scholar]
- 7.Li Y F, Smith G R. Genetics. 1997;146:57–67. doi: 10.1093/genetics/146.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding R, Smith G R. Mol Gen Genet. 1998;258:663–670. doi: 10.1007/s004380050780. [DOI] [PubMed] [Google Scholar]
- 9.DeVeaux L C, Hoagland N A, Smith G R. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugiyama A, Tanaka K, Okazaki K, Nojima H, Okayama H. EMBO J. 1994;13:1881–1887. doi: 10.1002/j.1460-2075.1994.tb06457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakashima N, Tanaka K, Strum S, Okayama H. EMBO J. 1995;14:4794–4802. doi: 10.1002/j.1460-2075.1995.tb00161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowndes N F, McInerny C J, Johnson A L, Fantes P A, Johnston L H. Nature (London) 1992;355:449–453. doi: 10.1038/355449a0. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y, Larson K L, Dorer R, Smith G R. Genetics. 1992;132:75–85. doi: 10.1093/genetics/132.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, Smith G R. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Smith G R. Curr Genet. 1995;27:440–446. doi: 10.1007/BF00311213. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Smith G R. Mol Microb. 1995;17:439–448. doi: 10.1111/j.1365-2958.1995.mmi_17030439.x. [DOI] [PubMed] [Google Scholar]
- 17.Li Y F, Numata M, Wahls W P, Smith G R. Mol Microb. 1997;23:869–878. doi: 10.1046/j.1365-2958.1997.2691632.x. [DOI] [PubMed] [Google Scholar]
- 18.Fukushima K, Yoshimi T, Nabeshima K, Yoneki T, Tougan T, Tanaka S, Nojima H. Nucleic Acids Res. 2000;28:2709–2716. doi: 10.1093/nar/28.14.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith G R. Cell. 1991;64:19–27. doi: 10.1016/0092-8674(91)90205-d. [DOI] [PubMed] [Google Scholar]
- 20.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borde V, Goldman A S H, Lichten M. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 22.DeVeaux L C, Smith G R. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- 23.Parisi S, McKay M J, Molnar M, Thompson M A, van der Speck P J, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers J H J, Kohli J. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krawchuk M D, DeVeaux L C, Wahls W P. Genetics. 1999;153:57–68. doi: 10.1093/genetics/153.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponticelli A S, Smith G R. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watt P M, Hickson I D, Borts R H, Louis E J. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 28.Johzuka K, Ogawa H. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tavassoli M, Shayeghi M, Nasim A, Watts F Z. Nucleic Acids Res. 1995;23:383–388. doi: 10.1093/nar/23.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cervantes M D, Farah J A, Smith G R. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 31.Muris D F R, Vreeken K, Schmidt H, Ostermann K, Clever B, Lohman P H M, Pastink A. Curr Genet. 1997;31:248–254. doi: 10.1007/s002940050202. [DOI] [PubMed] [Google Scholar]
- 32.Khasanov F K, Savchenko G V, Bashkirova E V, Korolev V G, Heyer W-D, Bashkirov V I. Genetics. 1999;152:1557–1572. doi: 10.1093/genetics/152.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lichten M, Goldman A S H. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 34.Smith G R. Experientia. 1994;50:234–241. doi: 10.1007/BF01924006. [DOI] [PubMed] [Google Scholar]
- 35.Fogel S, Mortimer R, Lusnak K, Tavares F. Cold Spring Harbor Symp Quant Biol. 1979;43:1325–1341. doi: 10.1101/sqb.1979.043.01.152. [DOI] [PubMed] [Google Scholar]
- 36.Gutz H. Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szankasi P, Heyer W D, Schuchert P, Kohli J. J Mol Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 38.Ponticelli A S, Sena E P, Smith G R. Genetics. 1988;119:491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuchert P, Kohli J. Genetics. 1988;119:507–515. doi: 10.1093/genetics/119.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schär P, Baur M, Schneider C, Kohli J. Genetics. 1997;146:1275–1286. doi: 10.1093/genetics/146.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph C, Kunz C, Parisi S, Lehmann E, Hartsuiker E, Fartmann B, Kramer W, Kohli J, Fleck O. Mol Cell Biol. 1999;19:241–250. doi: 10.1128/mcb.19.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleck O, Lehmann E, Schär P, Kohli J. Nat Genet. 1999;21:314–317. doi: 10.1038/6838. [DOI] [PubMed] [Google Scholar]
- 43.Grimm C, Bähler J, Kohli J. Genetics. 1994;135:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuchert P, Langsford M, Käslin E, Kohli J. EMBO J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fox M F, Yamada T, Ohta K, Smith G R. Genetics. 2000;156:59–68. doi: 10.1093/genetics/156.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahls W P, Smith G R. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- 47.Kon N, Krawchuk M D, Warren B G, Smith G R, Wahls W P. Proc Natl Acad Sci USA. 1997;94:13756–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kon N, Schroeder S C, Krawchuk M D, Wahls W P. Mol Cell Biol. 1998;18:7575–7583. doi: 10.1128/mcb.18.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirkpatrick D T, Fan Q, Petes T D. Genetics. 1999;152:101–115. doi: 10.1093/genetics/152.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones R H, Jones N C. Proc Natl Acad Sci USA. 1989;86:2176–2180. doi: 10.1073/pnas.86.7.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zahn-Zabal M, Lehmann E, Kohli J. Genetics. 1995;140:469–478. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grimm C, Schär P, Munz P, Kohli J. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ponticelli A S, Smith G R. Proc Natl Acad Sci USA. 1992;89:227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Virgin J B, Metzger J, Smith G R. Genetics. 1995;141:33–48. doi: 10.1093/genetics/141.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virgin J B, Bailey J P. Genetics. 1998;149:1191–1204. doi: 10.1093/genetics/149.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizuno K-i, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 57.Fox M E, Virgin J B, Metzger J, Smith G R. Proc Natl Acad Sci USA. 1997;94:7446–7451. doi: 10.1073/pnas.94.14.7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 59.Bergerat A, de Massy B, Gadelle D, Varoutas P-C, Nicolas A, Forterre P. Nature (London) 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 60.Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 61.Baudat F, Nicolas A. Proc Natl Acad Sci USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hiraoka Y. Genes Cells. 1998;3:405–413. doi: 10.1046/j.1365-2443.1998.00205.x. [DOI] [PubMed] [Google Scholar]
- 63.Nimmo E R, Pidoux A L, Perry P E, Allshire R C. Nature (London) 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 64.Hiraoka Y, Ding D-Q, Yamamoto A, Tsutsuni C, Chikashige Y. Chromosoma. 2000;109:103–109. doi: 10.1007/s004120050417. [DOI] [PubMed] [Google Scholar]
- 65.Shimanuki M, Miki F, Ding D-Q, Chikashige Y, Hiraoka Y, Horio T, Niwa O. Mol Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- 66.Niwa O, Shimanuki M, Miki F. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cooper J P, Watanabe Y, Nurse P. Nature (London) 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto A, West R R, McIntosh J R, Hiraoka Y. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bähler J, Wyler T, Loidl J, Kohli J. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein F, Mahr P, Galova M, Buonomo S B C, Michaelis C, Nairz K, Nasmyth K. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- 71.Molnar M, Bähler J, Sipiczki M, Kohli J. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe Y, Nurse P. Nature (London) 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 73.Koshland D E, Guacci V. Curr Opin Cell Biol. 2000;12:297–301. doi: 10.1016/s0955-0674(00)00092-2. [DOI] [PubMed] [Google Scholar]
- 74.Uhlmann F. Curr Biol. 2000;10:R698–R700. doi: 10.1016/s0960-9822(00)00709-0. [DOI] [PubMed] [Google Scholar]
- 75.Cohen-Fix O. Curr Biol. 2000;10:R816–R819. doi: 10.1016/s0960-9822(00)00799-5. [DOI] [PubMed] [Google Scholar]
- 76.Buonomo S B, Clyne R K, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- 77.van Heemst D, Heyting C. Chromosoma. 2000;109:10–26. doi: 10.1007/s004120050408. [DOI] [PubMed] [Google Scholar]
- 78.Birkenbihl R P, Subramani S. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. Nature (London) 2000;409:359–363. doi: 10.1038/35053103. [DOI] [PubMed] [Google Scholar]
- 80.Hawley R S. In: Genetic Recombination. Kucherlapati R, Smith G R, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 497–527. [Google Scholar]
- 81.Hawley R S, Theurkauf W E. Trends Genet. 1993;9:310–317. doi: 10.1016/0168-9525(93)90249-h. [DOI] [PubMed] [Google Scholar]
- 82.Dawson D S, Murray A W, Szostak J W. Science. 1986;234:713–717. doi: 10.1126/science.3535068. [DOI] [PubMed] [Google Scholar]
- 83.Molnar M, Parisi S, Kakihara Y, Mojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J. Genetics. 2001;157:519–532. doi: 10.1093/genetics/157.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niwa I T, Matsumoto Y, Chikashige Y, Yanagida M. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 86.Evans D H, Li Y F, Fox M E, Smith G R. Genetics. 1997;146:1253–1264. doi: 10.1093/genetics/146.4.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stewart E, Chapman C R, Al-Khodairy F, Carr A M, Enoch T. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davey S, Han C S, Ramer S A, Klassen J C, Jacobson A, Eisenberger A, Hopkins K M, Lieberman H B, Freyer G A. Mol Cell Biol. 1998;18:2721–2728. doi: 10.1128/mcb.18.5.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cooper J P, Nimmo E R, Allshire R C, Cech T R. Nature (London) 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 90.Szankasi P, Smith G R. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 91.Rudolph C, Fleck O, Kohli J. Curr Genet. 1998;34:343–350. doi: 10.1007/s002940050405. [DOI] [PubMed] [Google Scholar]
- 92.Schmidt H. Curr Genet. 1993;24:271–273. doi: 10.1007/BF00351803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.