The Fanconi anemia pathway supports hematopoietic stem cell survival in response to inflammatory and metabolic stress. We show that polyubiquitination and proteasome degradation of FANCL is inhibited by Akt1 activation, revealing a potentially important mechanism for the maintenance of stem cell function.
Abstract
Fanconi anemia hematopoietic stem cells display poor self-renewal capacity when subjected to a variety of cellular stress. This phenotype raises the question of whether the Fanconi anemia proteins are stabilized or recruited as part of a stress response and protect against stem cell loss. Here we provide evidence that FANCL, the E3 ubiquitin ligase of the Fanconi anemia pathway, is constitutively targeted for degradation by the proteasome. We confirm biochemically that FANCL is polyubiquitinated with Lys-48–linked chains. Evaluation of a series of N-terminal–deletion mutants showed that FANCL's E2-like fold may direct ubiquitination. In addition, our studies showed that FANCL is stabilized in a complex with axin1 when glycogen synthase kinase-3β is overexpressed. This result leads us to investigate the potential regulation of FANCL by upstream signaling pathways known to regulate glycogen synthase kinase-3β. We report that constitutively active, myristoylated-Akt increases FANCL protein level by reducing polyubiquitination of FANCL. Two-dimensional PAGE analysis shows that acidic forms of FANCL, some of which are phospho-FANCL, are not subject to polyubiquitination. These results indicate that a signal transduction pathway involved in self-renewal and survival of hematopoietic stem cells also functions to stabilize FANCL and suggests that FANCL participates directly in support of stem cell function.
INTRODUCTION
Fanconi anemia (FA) is a hereditary bone marrow failure syndrome associated with developmental defects and cancer predisposition. Fifteen human genes have been identified by biochemical and/or genetic models that demonstrate key cellular defects characteristic of FA (Kee and D'Andrea, 2012). These defects include hypersensitivity to DNA cross-linking agents such as mitomycin C in chromosome breakage assays and exaggerated arrest of cells in the G2/M phase of the cell cycle. The FA nuclear core complex consists of FANCA, B, C, E, F, G, L, and M. FANCL is a RING-type E3 ubiquitin ligase that monoubiquitinates FANCD2 and FANCI, a key functional role facilitated by other members of the nuclear core complex (Meetei et al., 2004; Seki et al., 2007; Alpi et al., 2008; Cole et al., 2010). The monoubiquitinated FANCD2–FANCI heterodimer complex in turn recruits key proteins involved in homologous recombination and DNA repair, such as FANCD1/BRCA2, FANCJ/BRIP1, FANCN/PALB2, and FANCO/RAD51C (Moldovan and D'Andrea, 2009). Loss of any one of the FA genes causes human disease. These findings suggest nonredundant biological and functional roles for the FANC genes in DNA damage responses.
The pathogenesis of bone marrow failure in FA is poorly defined. Qualitative and quantitative hematopoietic stem cell defects exist and occur in the absence of exogenous DNA-damaging agents (Gluckman et al., 1989; Auerbach et al., 1990; Haneline et al., 1999; Liu et al., 1999; Larghero et al., 2002; Kelly et al., 2007; Zhang et al., 2008; Jacome et al., 2009; Raya et al., 2010; Tulpule et al., 2010). Metabolic byproducts, such as reactive oxygen (Scarpa et al., 1985; Korkina et al., 1992; Zhang et al., 2007) and aldehyde species (Langevin et al., 2011; Rosado et al., 2011; Garaycoechea et al., 2012), are believed to contribute to hematopoietic stem cell loss. In addition, Fanconi anemia hematopoietic stem cells are hypersensitive to tumor necrosis factor-α (TNF-α), mip1-α, and interferon-γ (Broxmeyer et al., 1991; Dufour et al., 2003; Li et al., 2004, 2005, 2007; Si et al., 2006). Ex vivo exposure of murine Fancc-deficient hematopoietic stem cells to TNF-α increases stem cell loss and selects for TNF-α–resistant clonal variants (Li et al., 2005, 2007). These findings are particularly relevant to understanding the pathogenesis of marrow failure because cytokine levels in sera from FA patients’ blood and marrow are much higher than with normal controls (Schultz and Shahidi, 1993; Rosselli et al., 1994; Dufour et al., 2003). A key recurring defect described in FA hematopoietic stem cells is their poor self-renewal capacity when subjected to cellular stresses. Until recently, however, there has been little mechanistic and biochemical evidence that FA proteins directly regulate this function in hematopoietic stem cells.
We recently reported a biochemical and functional link between FANCL and β-catenin, a downstream effector of the Wnt pathway (Dao et al., 2012). FANCL ubiquitinates β-catenin with atypical ubiquitin chains (K11 linked), a modification that functionally enhances β-catenin activity. Suppression of FANCL significantly disrupted Wnt/β-catenin signaling and reduced transcription of downstream targets cyclinD1 and c-myc (Dao et al., 2012). These findings suggest that FANCL has a noncanonical function to regulate the Wnt/β-catenin signaling, a pathway with known functions in hematopoietic stem cell self-renewal. During the course of these studies, we sought to define how FANCL is itself posttranscriptionally regulated. Here we show that FANCL stability is regulated by the ubiquitin-proteasome pathway in a manner influenced directly by Akt1, suggesting that FANCL regulates stem cell function in part through its role as a downstream effector of phosphoinositide 3-kinase (PI3K)/Akt1 signaling.
RESULTS
FANCL expression is regulated in part by its own E3 ubiquitin ligase activity
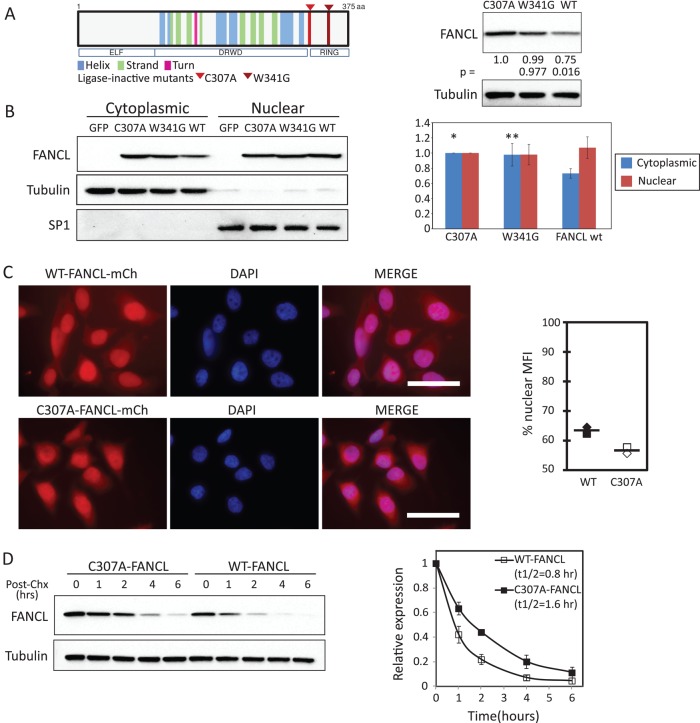
Many E3 ubiquitin ligases are known to autoubiquitinate and regulate their own protein levels (de Bie and Ciechanover, 2011). We examined the posttranscriptional regulation of FANCL in 293FT cells by quantifying human wild-type FANCL expression versus the ligase-inactive mutant forms, C307A- and W341G-FANCL. The missense mutations occur in the RING domain and abrogate key amino acids involved in the coordination of zinc (Figure 1A). The W341G-FANCL mutant retains some ability to assemble the nuclear core complex and partial E3 ubiquitin ligase activity, whereas the C307A-FANCL mutation is biochemically inactive (Meetei et al., 2003; Gurtan et al., 2006; Hodson et al., 2011). As shown, wild-type FANCL expression is lowest compared with the ligase-inactive mutants, suggesting that autoubiquitination may play a role in regulating FANCL if we assume that the activity of endogenous E3 ligases is not affected by wild-type or mutant FANCL overexpression. We quantified the relative cytoplasmic and nuclear expression of FANCL by immunoblot analysis. Data were normalized to tubulin and SP1 loading controls. Wild-type FANCL protein was proportionally more prominent in the nucleus than in the cytoplasm when compared with the ligase-inactive mutants (Figure 1B). These results were quantified from seven independent experiments and suggest that autoubiquitination may play an important role in the steady-state levels of FANCL, as well as in its relative subcellular localization. Similar findings were observed by microscopy analysis of HeLa cells stably expressing mCherry fusion proteins of wild-type FANCL or C307A-FANCL using automated software analysis (Figure 1C; n = 2). Wild-type FANCL is more localized to the nucleus (63.4%) than is the ligase-inactive C307A mutant (56.6%). Here we show representative images. In Supplemental Figure S1, we include a compilation of z-stack images as videos (Supplemental Movies 1–6). Using cycloheximide to inhibit new protein synthesis, we showed that the steady-state, reduced level of wild-type FANCL is due to increased protein turnover compared with C307A-FANCL (Figure 1D; n = 4). The estimated half-life is calculated as 0.8 h for wild-type FANCL versus 1.6 h for C307A-FANCL. We performed additional FANCL protein-turnover experiments in the nuclear and cytoplasmic fractions and found that the difference between wild-type and C307A-FANCL protein turnover is primarily in the cytoplasm (Supplemental Figure S2). The nuclear fraction of wild-type FANCL may be slightly more stable than that of C307A FANCL (p = 0.074). These experiments provide evidence that FANCL is regulated at the posttranscriptional level and its protein turnover and nuclear localization are dependent in part on its own E3 ubiquitin ligase activity.
FIGURE 1:
FANCL expression is regulated in part by its own E3 ubiquitin ligase activity. (A) Left, schematic of human FANCL protein and its domains. Highlighted by red arrowheads are the ligase-inactive FANCL mutants C307A and W341G. Right, steady-state expression of wild-type and ligase-inactive FANCL mutants as determined by immunoblot analysis. Relative expression (numbers below FANCL blot) was normalized to C307A and to tubulin loading controls from four experiments. These results were similarly observed in the untreated controls in the experiments shown in Figure 2A. (B) Nuclear and cytoplasmic expression of wild-type and ligase-inactive mutants in 293FT cells. A representative blot. Comparable purity of fractions was demonstrated by tubulin expression in the cytoplasmic fractions and SP1 in the nuclear fractions. The graph displays the quantitation of relative protein expression to C307A-FANCL for each of the nuclear and cytoplasmic fractions, relative to the respective loading control for each lane, from seven independent experiments (mean ± SD; *p = 0.033 and **p = 0.030 when we compared cytoplasmic wild type vs. C307A and vs. W341G mutants, respectively). Nuclear expression of wild type vs. mutants was not significantly different. (C) HeLa cells expressing mCherry-fusion proteins of FANCL were evaluated by microscopy. DAPI was used to stain nuclei. Scale bar, 50 μm. Nuclear and cytoplasmic mean fluorescence were quantified by software as detailed in Materials and Methods (average of n = 2). (D) Protein turnover experiments were performed in 293FT cells using cycloheximide. Cell lysates were harvested at the indicated time after the addition of cycloheximide. Right, graphical summary of four independent experiments and the calculated half-life (mean ± SEM). Densitometry of bands were normalized to time 0. Left, representative blot from one such experiment.
Constitutive mechanisms target FANCL for ubiquitination and degradation by the proteasome
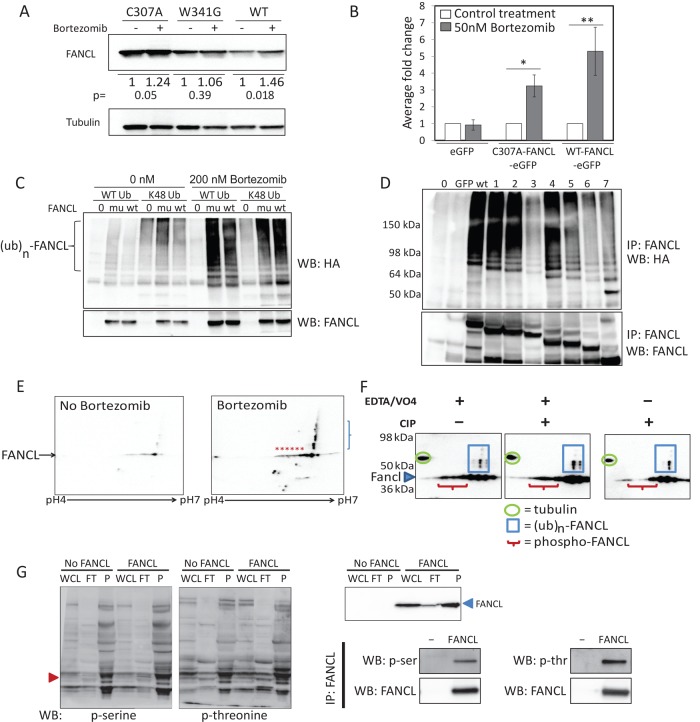
We next tested whether FANCL protein turnover is regulated by the proteasome. 293FT cells were treated with or without bortezomib, a 26S proteasome inhibitor. The effects on overexpressed FANCL were quantified by immunoblot analysis as shown in Figure 2A (n = 3 for mutants; n = 4 for wild type). Wild-type FANCL displays the greatest relative stabilization with proteasome inhibition compared with the ligase-inactive FANCL mutants, suggesting that mechanisms involving autoubiquitination of FANCL may be more responsive to proteasome inhibition. Similar findings were observed when we expressed FANCL–enhanced green fluorescent protein (eGFP) fusion proteins in HeLa cells and quantified protein levels by flow cytometry (Figure 2B; n = 4). We then performed ubiquitination assays to confirm that FANCL is marked for proteasome degradation by polyubiquitination. Here we expressed FANCL (wild type or C307A mutant) and hemagglutinin-tagged ubiquitin (HA-Ub). Wild-type HA-Ub has all seven lysines intact, whereas the Lys-48 HA-Ub mutant only has Lys-48 intact for ubiquitin chain extension. These studies revealed that FANCL is polyubiquitinated (Figure 2C; n = 2). As expected, proteasome inhibition greatly stabilized the polyubiquitinated forms of FANCL. Non–FANCL-expressing control cells provided confidence that the immunoprecipitated protein being evaluated was FANCL and not a nonspecific protein. The facts that the Lys-48 HA-Ub mutant can be used for ubiquitin chain extension and that these FANCL ubiquitinated species are highly responsive to proteasome inhibition provide evidence that FANCL is marked for degradation by Lys-48 polyubiquitination. These results are qualitative because the C307A mutation may affect FANCL's binding to the antibody in immunoprecipitation/nondenaturing conditions, and FANCL is immunoprecipitated from lysates with significantly different steady-state levels (e.g., Figure 1A). Furthermore, these results cannot be normalized with a high degree of confidence because polyubiquitinated FANCL presents as a smear on immunoblots, and some of the total FANCL may be much higher in molecular weight and are not evaluated. Compared to eGFP expression alone, there is rapid turnover of FANCL-eGFP expression, and the overall level is maintained at a very low level in real-time imaging experiments quantifying mean fluorescence in live cells over time (Supplemental Figure S3). Videos are provided as part of Supplemental Figure S3 (Movies 7 and 8). Evaluation of N-terminal–deletion FANCL mutants in similar ubiquitination assays revealed that the E2-like fold (ELF) domain may play a role in directing ubiquitination (Figure 1A, schematic, and Figure 2D; n = 4). Deletion of amino acids 78–114 resulted in a substantial loss of ubiquitinated FANCL species, whereas total del-114-FANCL-eGFP protein levels remained relatively stable. Further deletion of FANCL resulted in partial rescue of FANCL ubiquitination, suggesting that the region between amino acids 114 and 156 may play a role in inhibiting FANCL ubiquitination. Two-dimensional (2D) PAGE analysis of whole-cell extracts (Figure 2E; n = 2) demonstrated that treatment of cells with bortezomib stabilized total FANCL and ubiquitinated forms of FANCL (Figure 2E, blue bracket). We performed similar analyses of the ligase-inactive mutants, C307A and W341G, and detected that the mutants are also polyubiquitinated (Supplemental Figure S4). Acidic forms of FANCL (those species with a lower pI; red asterisks) were not subjected to ubiquitination, suggesting that phosphorylation protects FANCL from polyubiquitination. We confirmed that some of these acidic forms of FANCL are phosphorylated in experiments in which lysates were desalted and treated with calf intestinal alkaline phosphatase (Figure 2F; n = 2). Compared to control conditions (lysates spiked with alkaline phosphatase inhibitors EDTA and orthovanadate), the abundance of the acidic forms of FANCL was reduced with alkaline phosphatase treatment. In Figure 2G, we analyze FANCL abundance in whole-cell lysate, flowthrough, and phospho-enriched fractions (n = 2) and determine that FANCL is more abundant in the phospho-enriched fractions than in the flowthrough fractions (Figure 2G, top, right). We confirmed that the approach was efficient in enriching for phosphorylated proteins (Figure 2G, left). We demonstrate that FANCL is phosphorylated at serine and threonine residues by immunoprecipitation studies (Figure 2G, bottom, right). Together these results provide evidence that FANCL is regulated by phosphorylation and ubiquitin-mediated proteasome degradation.
FIGURE 2:
Constitutive mechanisms target FANCL for ubiquitination and proteasome degradation. (A) Cells expressing the wild-type or ligase-inactive FANCL mutant were treated with or without bortezomib, a 26S proteasome inhibitor. Numbers below the blots show the average change in expression relative to no-bortezomib lanes from independent experiments (n = 3 for mutants; n = 4 for wild type; p values displayed beneath blot). (B) Expression of eGFP-fusion proteins of wild-type and ligase-inactive FANCL mutants in HeLa cells was quantified by flow cytometry with or without bortezomib. Fold change was calculated relative to the untreated condition for each FANCL type, and the average was taken from four independent experiments and graphed as mean ± SD. (C) Ubiquitination assays were performed in 293FT cells with or without bortezomib and carried out with wild-type FANCL (wt) or C307A-FANCL (mu). The no-FANCL condition (0) demonstrates specificity of immunoprecipitation (IP) with FANCL antibody and ubiquitinated products as detected by HA antibody detecting HA-tagged wild-type or Lys-48 ubiquitin. Bracket denotes the polyubiquitinated FANCL species. This is a representative blot of two experiments. (D) The N-terminal truncation mutants of human FANCL were subjected to ubiquitination assays in 293FT cells as described. Deletion mutants 1–7 as follows: 1 (deleted amino acids 1–56), 2 (deleted amino acids 1–77), 3 (deleted amino acids 1–113), 4 (deleted amino acids 1–155), 5 (deleted amino acids 1–178), 6 (deleted amino acids 1–212), 7 (deleted amino acids 1–306). Top, IP with FANCL and probed for HA-tagged ubiquitinated products. Bottom, same blot probed for total FANCL. This is a representative blot of four experiments. (E) The 2D PAGE analysis of 293FT cell extracts under conditions of FANCL overexpression with or without bortezomib treatment. First dimension was separated by pI using the ZOOM strip with pH range of 4–7. The predicted pI of FANCL is 6.02 (ExPASy Compute pI/Mw tool). Ubiquitin molecules conjugated to FANCL are not predicted to shift its pI substantially. A ubiquitin molecule has a predicted theoretical pI/MW of 6.56/8564.84 using a similar ExPASy analysis. Second dimension was separated by SDS–PAGE and resolved by molecular weight. Blot was probed for FANCL. This blot is representative of two experiments. Ubiquitinated FANCL species marked by a blue bracket; acidic forms of FANCL marked by red asterisks. (F) Lysates were desalted and treated with calf intestinal alkaline phosphatase in the presence or absence of orthovanadate and EDTA, which completely inactivates alkaline phosphatase. Blots were probed for FANCL and tubulin. The overall abundance of acidic forms of FANCL was reduced in two independent experiments with alkaline phosphatase treatment. (G) Whole-cell lysate (WCL), flowthrough (FT, phospho-poor fraction), and phospho-enriched (P) fractions were probed for phosphoserine and phosphothreonine proteins (left). Red arrowhead denotes where FANCL is expected. There was a significant enrichment of phosphoproteins in the eluted fractions. The same cell lysates were evaluated for total FANCL (top, right; n = 2). FANCL was immunoprecipitated from 293FT cells overexpressing FANCL or not overexpressing FANCL (–) and probed with a phosphoserine or phosphothreonine antibody separately (n = 3).
FANCL and GSK3β directly interact in a complex involving axin1
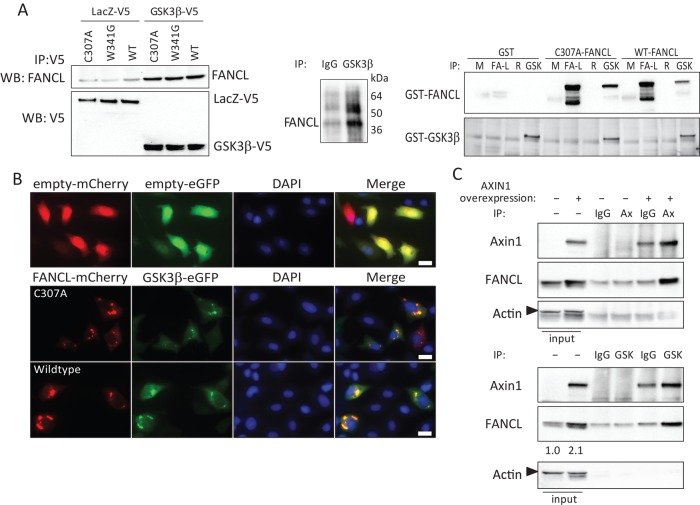
Because we previously observed a protein–protein interaction between FANCA, C, and G with β-catenin (Dao et al., 2012), we hypothesized that GSK3β, a serine-threonine kinase known to regulate a number of proteins by phosphorylation, might have a role in regulating FANCL ubiquitination by phosphorylation. We first tested whether FANCL binds to GSK3β in coimmunoprecipitation experiments. In cell extracts, we observed coimmunoprecipitation of FANCL and GSK3β when using a V5 or a GSK3β antibody but not with a FANCL antibody. We believe that this is due to the fact that the entire protein–protein binding domain was used to generate the mouse antibody. Therefore we believe the FANCL antibody cannot pull down GSK3β in the reverse immunoprecipitation because of epitope masking. The specificity of the immunoprecipitations was determined by carrying out parallel immunoprecipitations with V5-tagged LacZ control and isotype control antibodies as shown (Figure 3A, left; n = 4). We also demonstrated that the interaction is direct by performing coimmunoprecipitation studies with purified glutathione S-transferase (GST)–fusion proteins (Figure 3A, right). Coimmunoprecipitation of endogenous FANCL with GSK3β was also demonstrated (Figure 3A, middle; n = 3). Using microscopy, we observed colocalization of GSK3β-eGFP and FANCL-mCherry fusion proteins primarily in the cytoplasm (Figure 3B; n = 2). Pearson's coefficient approached 1.0 for select random cells evaluated. Their colocalization appeared in aggresome-like structures and disrupted FANCL's nuclear localization. We also tested whether the interaction between FANCL and GSK3β is facilitated by axin1, a scaffolding protein in the well-described destruction complex that regulates β-catenin phosphorylation and its subsequent polyubiquitination. Of interest, overexpression of axin1 stabilizes FANCL expression (Figure 3C, input lanes). We observed that axin1 coimmunoprecipitates with FANCL and GSK3β to a greater degree than isotype control antibody, which nonspecifically pulled down some axin1 (Figure 3C; n = 3). These results contrast with the roles of GSK3β and axin1 in regulating β-catenin and is opposite of what we had originally hypothesized. Instead of targeting FANCL for degradation, the FANCL-GSK3β-axin1 complex may stabilize FANCL at the posttranscriptional level.
FIGURE 3:
FANCL and GSK3β directly interact in a complex involving axin1. (A) 293FT cells overexpressing LacZ-V5 or GSK3β-V5 and FANCL were immunoprecipitated with anti-V5 and probed for FANCL (left; n = 4). We confirmed successful immunoprecipitation of V5-tagged proteins as shown in the blot. We did not observe coimmunoprecipitation when a FANCL antibody was used, possibly due to epitope masking (data not shown). Right, GST-fusion proteins were purified from Escherichia coli and subjected to immunoprecipitation studies. Isotype antibody control, mouse (M), and rabbit (R) were included as shown. Blot was probed for FANCL and for GSK3β. Middle, coimmunoprecipitation of endogenous FANCL and GSK3β was also demonstrated in HeLa cells (n = 3). (B) Microscopy analysis of HeLa cells expressing FANCL-mCherry and GSK3β-eGFP reveal colocalization primarily in the cytoplasm. Pearson's coefficient approached 1 when random cells were analyzed, indicating colocalization. Scale bar, 20 μm. (C) Immunoprecipitation was performed in cells expressing axin1, FANCL, and GSK3β with either isotype control antibody (IgG) or an axin (Ax) or GSK3β (Gsk) antibody and probed for axin and FANCL as shown (n = 3). The enhanced expression of FANCL by axin1 overexpression is quantified as shown, and the corresponding actin loading controls are included.
GSK3β stabilizes FANCL protein
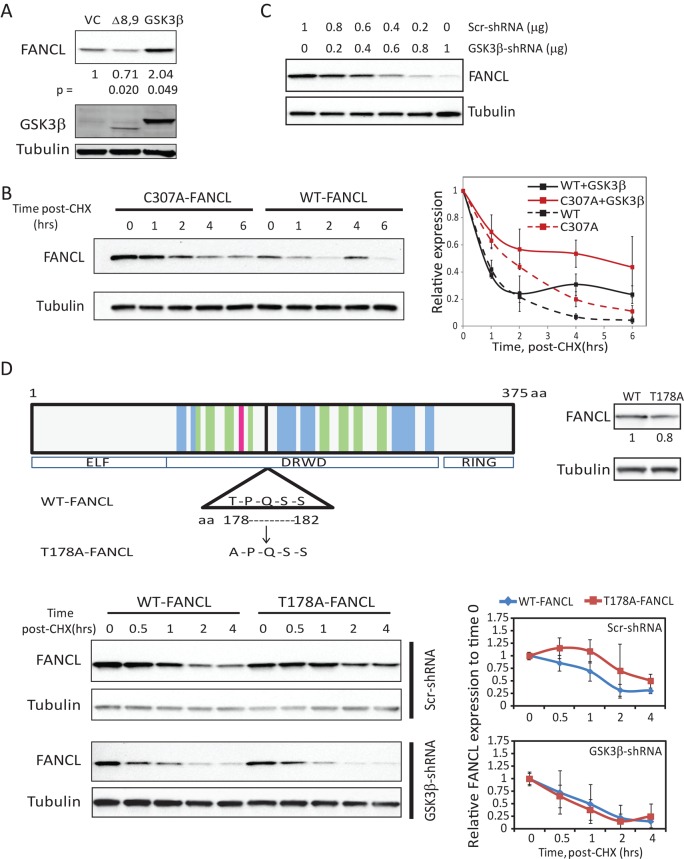
We then performed experiments to test the observation that FANCL is stabilized by GSK3β. When we overexpressed GSK3β compared with a vector-control or a kinase-dead version of GSK3β (exon 8/9-deleted GSK3β; Abrahamsson et al., 2009), we observed an increase in total FANCL levels (Figure 4A; n = 3). These results suggest that the stabilization of FANCL is dependent in part on the kinase activity of GSK3β. In cycloheximide experiments, we observed stabilization of both wild-type FANCL and ligase-inactive C307A-FANCL with GSK3β overexpression. A stable pool of FANCL remained at the 6-h time point with GSK3β overexpression compared with vector-control–transfected cells (Figure 4B; n = 4). The ligase-inactive C307A-FANCL mutant was stabilized to a higher degree than wild-type FANCL. These results suggest that GSK3β stabilizes FANCL in part by an E3-ligase–independent mechanism but that having intact ligase activity also plays a role in stabilizing FANCL (Figure 4B). Suppression of endogenous GSK3β by short hairpin RNA (shRNA) expression reduced the steady-state level of FANCL compared with control shRNA expression (Figure 4C; n = 3). We generated a T178A point mutation at a putative GSK3β phosphorylation site in FANCL identified by the NetPhosK1.0 and KinasePhos programs (Figure 4D, top, left). This site shares the S/T-X-X-X-pS/pT motif found in other known GSK3β substrates (Dajani et al., 2001). Our studies with the T178A mutant showed that this motif is important for the stabilization of FANCL. Steady-state expression of T178A-FANCL is consistently lower than that of the wild-type FANCL (Figure 4D, top, right). With complete knockdown of GSK3β by shRNA expression, however, which would inhibit the kinase activity and the scaffolding function of GSK3β, the T178A-FANCL mutant demonstrated similar protein turnover to wild-type FANCL (Figure 4D, bottom; n = 3). Together these results suggest that GSK3β stabilizes FANCL protein, potentially through a complex involving axin1 (Figure 3C), by phosphorylation. This is further supported by the 2D PAGE analysis of FANCL, which showed that acidic or phospho forms of FANCL are not subject to polyubiquitination (Figure 2E).
FIGURE 4:
GSK3β stabilizes FANCL expression. (A) FANCL was overexpressed with vector-control (VC), wild-type GSK3β, or a kinase-inactive, exon 8/9–deletion GSK3β version (Δ8,9) in 293FT cells, and steady-state levels of FANCL were analyzed by immunoblot analysis. Tubulin was used as a loading control. Numbers represent densitometric average of relative FANCL expression to VC in three experiments. (B) Protein turnover experiments using cycloheximide (CHX) to block new protein synthesis were performed in 293FT cells. Cells expressed wild-type or C307A-FANCL with GFP or GSK3β overexpression. A representative blot probing for FANCL and tubulin is shown for cells coexpressing GSK3β, and the quantification from four independent experiments is graphed (mean ± SD). Densitometries of bands were normalized to time 0. The half-life was not calculated, as the protein turnover did not meet first-order decay kinetics. Note that this experiment evaluates turnover of overexpressed FANCL, as endogenous FANCL is not detectable in these experiments. (C) FANCL protein levels were evaluated by immunoblot analysis of 293FT cell extracts with increasing concentrations of GSK3β shRNA (0–1 μg of DNA for a dish of 4-cm2 surface area). Scrambled shRNA was also added to maintain a similar amount of total DNA transfected in all conditions. A representative blot from three independent experiments. (D) A schematic of the putative S/T-X-X-X-pS/pT GSK3β phosphorylation motif in human FANCL. The T178A mutant was generated to test the role of the Thr-178 in FANCL protein stability. A representative blot is shown for protein turnover studies performed with GSK3β suppression (GSK3β-shRNA) or without (Scr-shRNA) in 293FT cells. Densitometries of bands from three independent experiments were normalized to time 0 and graphed (mean ± SD).
PI3K/Akt activation inhibits GSK3β by Ser-9 phosphorylation and increases FANCL expression
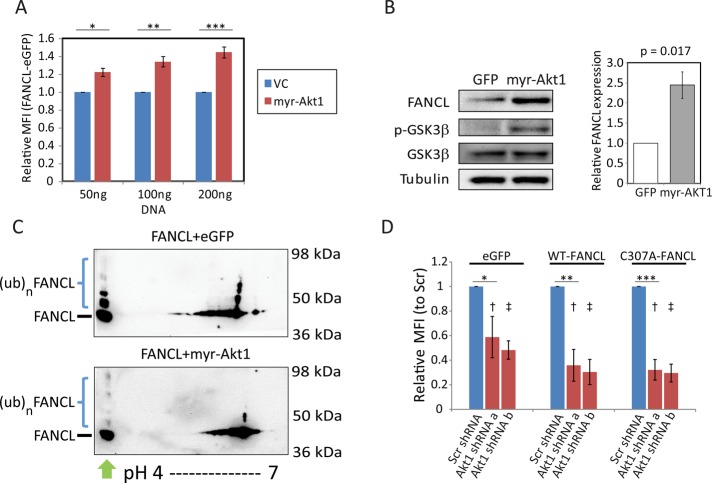
To explore the biological significance of our novel findings showing that GSK3β stabilizes FANCL via a scaffold that may facilitate its phosphorylation, we tested whether the PI3K/Akt pathway regulates this interaction. PI3K/Akt is a survival pathway that signals in part through GSK3β (Chappell et al., 2011). Activation of the PI3K/Akt pathway leads to phosphorylation of GSK3β at serine 9, resulting in a kinase-inactive form through a pseudosubstrate mechanism (Kockeritz et al., 2006). Our studies here reveal a more complex interaction between GSK3β and FANCL. Using flow cytometry, we observed that overexpression of a constitutively active Akt1 (myristoylated Akt1) stabilized FANCL-eGFP compared with vector-control conditions (Figure 5A; n = 4). We confirmed this regulation by conventional immunoblot analysis and quantified the average fold stabilization in three independent experiments (Figure 5B). There was ∼2.5-fold increase in total FANCL expression with activated Akt1 expression compared with control. As expected, overexpression of myristoylated Akt1 increased phosphoserine-9–GSK3β compared with vector-control conditions, whereas total GSK3β levels remained unaffected. The use of 2D PAGE showed that myr-Akt1 expression was associated with markedly reduced ubiquitinated FANCL forms compared with control-transfected cells (Figure 5C; n = 2). Therefore Akt1 stabilizes FANCL in part by directly or indirectly reducing FANCL ubiquitination. Using another approach to verify that Akt1 regulates FANCL posttranscriptionally, we suppressed Akt1 by shRNA expression and observed reduced steady-state levels of wild-type FANCL-eGFP and C307A-FANCL-eGFP (Figure 5D; at least three experiments for each shRNA). We noted, however, that eGFP control expression was also reduced but to an overall lesser degree than FANCL (p values are indicated in the figure legend for all comparisons). Interpretation of these results is limited by the fact that general suppression of Akt1 may cause these effects indirectly. Despite this limitation, the studies we describe here support the notion that Akt1 stabilizes FANCL at the posttranscriptional level and this is partly due to reduced ubiquitination of FANCL.
FIGURE 5:
PI3K/Akt activation inhibits GSK3β by Ser-9 phosphorylation and increases FANCL expression. (A) GFP was quantified by flow cytometry to reflect steady-state levels of FANCL-eGFP with coexpression of vector-control (VC) vs. myristoylated Akt1 (myr-Akt1) in HeLa cells. The graph is a summary of four independent experiments (mean ± SD; p < 0.003 for all three comparisons of VC vs. myr-Akt1 effects). (B) Cells expressing FANCL with GFP vs. myristoylated Akt1 were evaluated by immunoblot analysis. HeLa cell extracts were evaluated for FANCL, phosphoserine-9–GSK3β, total GSK3β, and tubulin. A representative blot. The graph depicts the quantification of relative FANCL expression from three independent experiments (mean ± SD; p value displayed). (C) Extracts from 293FT cells expressing FANCL with vector control or myr-Akt1 were analyzed by 2D PAGE and probed with a FANCL antibody (n = 2). Green arrow denotes lysates separated by PAGE (separation by size only). Blue brackets denote ubiquitinated FANCL forms. (D) Expression of eGFP (control), FANCL-eGFP, or C307A-FANCL-eGFP in 293FT was evaluated under conditions of scrambled-shRNA expression (Scr) vs. Akt1-shRNA expression (two different shRNAs, Akt1shRNA a and Akt1shRNA b). Mean fluorescence intensity was quantified by flow cytometry and normalized relative to control cells (Scr shRNA control for each eGFP construct). This experiment was performed three independent times, and the graph shows the mean ± SD. Compared to each individual Scr shRNA control, p values are *0.024, **0.0002, ***0.0004. Compared to eGFP control for each type of shRNA, p values are †<0.039 for Akt1shRNAa and ‡<0.023 for Akt1shRNAb for the FANCL-eGFP and C307A-FANCL-eGFP–expressing cells.
DISCUSSION
Our group is interested in investigating the alternative functions of the FA pathway and their potential interaction with other signaling pathways with known critical roles in maintaining hematopoietic stem cell function. FA hematopoietic stem cells display poor self-renewal capacity when subjected to a variety of cellular stresses (Scarpa et al., 1985; Korkina et al., 1992; Dufour et al., 2003; Li et al., 2004, 2005, 2007; Si et al., 2006; Zhang et al., 2007; Langevin et al., 2011; Rosado et al., 2011; Garaycoechea et al., 2012). Here we focused on dissecting the posttranscriptional regulation of FANCL, the E3 ubiquitin ligase of the FA pathway. Among the FA core complex members, we view FANCL as a key point of functional and signaling divergence, as most E3 ubiquitin ligases are known to have multiple protein targets.
Here we provide evidence that FANCL is constitutively targeted for degradation by the proteasome. FANCL, like other E3 ligases (de Bie and Ciechanover, 2011), likely regulates its own protein levels through autoubiquitination. We compared wild-type versus ligase-inactive FANCL protein turnover and showed that autoubiquitination may play a significant role in FANCL degradation (Figure 1D). This interpretation is based on the assumption that activity of endogenous E3 ligases is not different when wild-type versus ligase-inactive FANCL mutants are overexpressed in cells. In addition, our studies reveal that ubiquitination by another E3 ligase is mechanistically involved in marking FANCL for degradation by ubiquitination. We arrived at this conclusion based on the finding that ligase-inactive mutants also show some degree of protein stabilization with proteasome inhibition (Figure 2A). We have not identified the E3 ligase that might have this role. We confirm that polyubiquitination of FANCL occurs with Lys-48 ubiquitin chain extensions (Figure 2C), which typically mark proteins for proteasome degradation. It remains to be determined whether autoubiquitination by FANCL serves as a priming step to recruit other ubiquitin-binding protein/E3 ligase complexes. We also observed that autoubiquitination might have an additional role in promoting nuclear localization of FANCL (Figure 1B). Our results could also be explained by other mechanisms. For example, the C307A mutation might affect its nuclear transport or its ability to bind to chromatin or other E3 ligases, which might contribute to its more diffuse expression in the nucleus and cytoplasm compared with wild-type FANCL.
The recently solved crystal structure of FANCL has uncovered some unique features. FANCL possesses three domains, the N-terminal ELF, the tandem or double RWD domain involved in substrate binding (DRWD), and the C-terminal really interesting new gene (RING) involved in E2 binding (Cole et al., 2010). The ELF and the double RWD domains together form three folds; each fold is similar to the ubiquitin-conjugating (E2) superfamily. This is a novel structural arrangement not seen in other E3 ligases (Cole et al., 2010). We generated a series of N-terminal mutations of FANCL with the goal of understanding the role of the ubiquitin-conjugating folds in facilitating FANCL ubiquitination. Deletion of the N-terminus up to amino acids (aa) 114 in FANCL leads to an abrupt loss of ubiquitination (Figure 2D), suggesting that the region between aa 78 and 114 might direct ubiquitination or recruit proteins that enhance the efficiency of ubiquitination. Of interest, we found that additional truncation up to aa 179 resulted in partial recovery of ubiquitination, suggesting that this region might structurally inhibit ubiquitination. Crystal structural analyses of this mutant might reveal the basis for these observations.
There are many examples of proteins targeted for polyubiquitination and degradation by an initial phosphorylation signal. β-Catenin is one such example in which multiple phosphorylations at the N-terminal region leads to recruitment of a multiprotein E3 ligase complex SCF-β-TrCP (Aberle et al., 1997). We examined whether a similar mechanism could regulate FANCL, by performing 2D PAGE analysis. In contrast to what we expected, we observed that acidic forms of FANCL are not ubiquitinated, as shown in Figure 2E. These results raise the possibility that posttranscriptional regulation of FANCL is tightly regulated by kinases and that phosphorylation is protective against Lys-48–linked polyubiquitination. We confirmed that FANCL is indeed phosphorylated at serine and threonine residues (Figure 2G). Based on our previous publication (Dao et al., 2012), we tested the role of GSK3β, since we had observed that GSK3β modulates the protein level of FANCL.
We observed a direct interaction between FANCL and GSK3β by coimmunoprecipitation and by microscopy studies (Figure 3, A and B). Our studies also suggest a role for axin1 in this complex, as we coimmunoprecipitated FANCL and GSK3β using an axin antibody (Figure 3D). GSK3β expression stabilized FANCL, whether we evaluated FANCL steady-state levels or protein turnover (Figure 4, A and B). Conversely, suppression of GSK3β destabilized FANCL. We sought evidence to support the notion that this regulation is via phosphorylation through studies with the T178A-FANCL mutant, which has a disrupted putative GSK3β S/T-X-X-X-pS/pT motif. The steady-state expression of this mutant was reduced compared with wild-type FANCL, but its decay was similarly affected by GSK3β suppression (Figure 4C). The caveat is that our studies do not discern whether the mutation at this site leads to direct loss of phosphorylation itself, affects the phosphorylation at a different site, or indirectly affects ubiquitination due to conformational changes of FANCL. Multiple serine and threonine sites are likely phosphorylated in FANCL (based on Figure 2G and phosphorylation prediction programs), and there may be key functional roles of each phosphorylated residue and phosphorylated combinations. We identified six potential serine and threonine phosphorylation sites in FANCL. Collectively our results show that GSK3β plays a role in stabilizing a pool of FANCL potentially in a multiprotein complex involving axin1. The fact that axin1 expression alone stabilizes FANCL suggests a stoichiometric requirement in this complex. Although the precise components of this complex have yet to be defined, it will be interesting to determine whether the axin1 scaffold is able to recruit other kinases, in addition to GSK3β, to phosphorylate FANCL and protect it from degradation.
The PI3K/Akt signaling pathway regulates GSK3β and interfaces with multiple kinases (Chappell et al., 2011). We sought to determine the effects of these pathways on FANCL protein levels and ubiquitination. We found that in the cellular context of survival signals, FANCL protein levels are significantly stabilized (Figure 5, A–C). Accumulation of FANCL was associated with markedly reduced FANCL ubiquitination. These results are consistent with our finding that phosphorylation of FANCL is protective against proteasome-mediated degradation. These results suggest a role for FANCL as a downstream effector of PI3K/Akt. However, because these pathways inhibit GSK3β by Ser-9 phosphorylation, we believe that there are additional, GSK3β-independent mechanisms (e.g., other kinases) involved in regulating FANCL and its stabilization. Of importance, loss of Akt1 destabilizes FANCL (Figure 5D). Additional studies would be necessary to prove whether Akt1 has a role in directly phosphorylating FANCL. These studies are beyond the scope of this work.
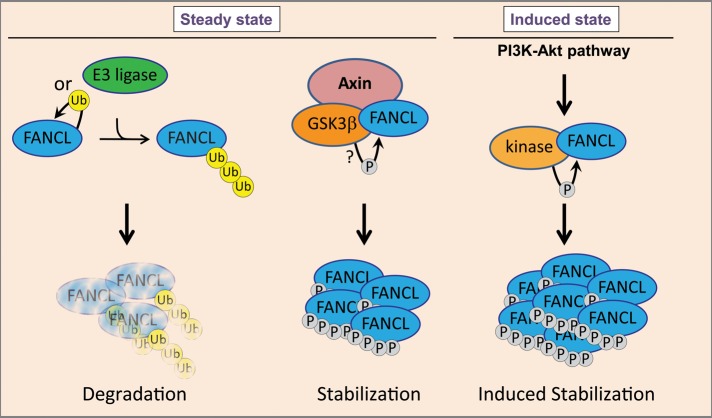
We characterized mechanisms of FANCL regulation and identified phosphorylation as one mechanism regulating ubiquitination of FANCL (Figure 6). Our finding that PI3K/Akt stabilizes FANCL protein level has several biological implications. PI3K/Akt has a role in cell cycle progression, survival, DNA-damage response, RNA export via REF/Aly, and tumorigenesis (Martelli et al., 2012). Phenotypic and functional similarities exist between normal stem cells and cancer stem cells (Yilmaz and Morrison, 2008). These findings suggest that in the context of pathological up-regulation of PI3K/Akt, FANCL may have an effect on tumor cell survival and chemotherapy resistance. Tumor cells displaying upregulated FANCL may correct DNA lesions more efficiently (Figure 6). In normal tissues and in tissue injury, the functions of FANCL may be protective against excessive cell death and genomic instability. This is apparent in FA patients, in whom loss of the FA pathway leads to progressive hematopoietic stem cell loss and a very high propensity for neoplastic transformation. The fact that PI3K/Akt has emerged as a key pathway regulating hematopoietic stem cell function further highlights the potential relevance of our finding that FANCL is a downstream effector of PI3K/Akt (Figure 6; Polak and Buitenhuis, 2012). Our studies are the first to characterize posttranslational modification of FANCL and to provide mechanistic insights into how they could regulate FANCL activity. In addition, we show that FANCL protein stabilization is responsive to upstream signaling pathways that regulate self-renewal and survival of hematopoietic stem cells.
FIGURE 6:
Mechanisms of FANCL regulation by phosphorylation and ubiquitination. Our studies showed that FANCL is highly regulated by ubiquitin-mediated proteolysis. We depict in this model that steady-state FANCL levels are maintained, at least in part, by a complex involving GSK3β and axin1. A subset of phosphorylated forms of FANCL is not subject to ubiquitination based on 2D PAGE analysis, suggesting that upstream kinases might play a role in stabilizing a pool of FANCL and enhancing the functions of FANCL. Our work showed that the PI3k/Akt1 pathway modulates FANCL protein levels and is involved in suppressing polyubiquitination of FANCL, directly or indirectly. These studies position FANCL as a downstream effector of the PI3K/Akt1 pathway and link the Fanconi anemia pathway to a key pathway known to regulate survival and self-renewal of hematopoietic stem cells.
MATERIALS AND METHODS
Cells and culture conditions
293FT and HeLa cells were maintained in 10% fetal bovine serum DMEM medium supplemented with nonessential amino acids and glutamine. All cells were grown in 5% CO2 humidified incubator at 37°C. Bortezomib was tested at a concentration ranging from 25 to 500 nM, depending on the cell type, for 16–24 h.
Constructs
Human cDNAs for FANCL, GSK3β, and axin1 were purchased from Open Biosystems (Huntsville, AL). The deletion exon 8/9-GSK3β mutant was kindly provided by Catriona Jamieson (University of California, San Diego). We used a pUSE-AMP-myristoylated-Akt1 vector for our Akt1 studies. cDNAs were cloned into the following mammalian expression vectors pCDNA6-CMV-V5/His (Invitrogen, Carlsbad, CA), pCMV-sport, or a modified pCDH1-EF1 vector (System Biosciences, Mountain View, CA). C-terminal eGFP and mCherry fusion proteins were cloned into the modified pCDH1-EF1 vector. pRK5-HA-Ub wild-type and mutant constructs were obtained from Addgene (Cambridge, MA) deposited by Ted Dawson (Johns Hopkins University) and Sandra Weller (University of Connecticut Health Center; Lim et al., 2005; Livingston et al., 2009). GST proteins were generated by cloning cDNAs into pGEX-4T constructs (GE Healthcare Life Sciences, Piscataway, NJ). Proteins were purified using glutathione cross-linked to Sepharose beads. The pLKO.1 FANCL and Akt1 shRNA sets generated by the RNAi Consortium were purchased from Open Biosystems. Missense mutations were generated by site-directed mutagenesis per the manufacturer's protocol (Agilent, Santa Clara, CA) and were confirmed by sequencing. The N-terminal deletion mutants of FANCL were generated with In-Fusion cloning (Clontech, Mountain View, CA).
Antibodies, Western blot analysis, and immunoprecipitation studies
FANCL, axin, GSK3β antibodies, and protein G plus agarose were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). V5 antibody was obtained from AbD Serotec (Kidlington, United Kingdom). Other antibodies include anti–α-tubulin (Sigma-Aldrich, St. Louis, MO) and anti-hemagglutinin (mouse, Covance, Princeton, NJ; rabbit, Santa Cruz Biotechnology). Whole-cell lysates were lysed in buffer containing 0.5% Triton X-100, 120 mM NaCl, 50 mM Tris (pH 8.0), 2 mM EDTA, 1 mM Na2VO4, and 1:300 protease Inhibitor cocktail (P8340; Sigma-Aldrich). Proteins were resolved by SDS–PAGE. Densitometric analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Nuclear extracts
Nuclear and cytoplasmic fractions from 293FT or HeLa cells were prepared essentially as described (Dao et al., 2012).
Phospho enrichment
A Pierce Phosphoprotein Enrichment Kit was used per the manufacturer's instructions (Thermo Scientific Pierce, Rockford, IL). From 20 to 40 μg of total protein from the various fractions was analyzed in immunoblots. Enrichment was determined by probing for phosphor-serine (Q5 antibody; Qiagen, Valencia, CA) and phosphor-threonine (Q7 antibody; Qiagen). FANCL was probed to determine the fraction of FANCL subject to phosphorylation. We also immunoprecipitated total FANCL and probed for phosphor-serine and phosphor-threonine. 293FT controls, without FANCL overexpression, were used to ensure that the protein being evaluated is indeed FANCL.
2D PAGE analysis
The ZOOM system was used for 2D PAGE analysis of cell extracts prepared by 2D solubilization buffer 1 (proprietary buffer purchased from the manufacturer) per the manufacturer's protocol (Invitrogen). Calf intestinal alkaline phosphatase was added to cell extracts at a concentration of 1 unit/1 μg of total protein in 10 μl at 37ºC for 1 h. Desalting and buffer exchange were carried out with Zeba 7K MWCO columns (Pierce).
Cell-based ubiquitination assays
293FT cells were transfected with wild-type FANCL, C307A-FANCL, N-term–deletion FANCL mutant, or a GFP control, along with either wild-type or mutant HA-ubiquitin, using Lipofectamine 2000. Whole-cell extracts were incubated with 1–2 μg of anti-FANCL antibody and incubated at 4°C with rocking overnight. From 20 to 40 μl of protein G plus agarose was added, and samples were incubated for an additional hour at 4°C with rocking. The beads were then washed four times with lysis buffer (as defined above). Proteins were resolved with SDS–PAGE, and ubiquitinated FANCL was detected with an anti-HA antibody.
Measurement of protein stability
Whole-cell lysates were harvested at the indicated time points after the addition of cycloheximide at 50 μg/ml. Densitometric analysis of bands from immunoblots were quantified using ImageJ and expressed relative to time 0 for each condition. The half-life was determined based on the time at which the densitometry signal reached 50% of the starting level.
Microscopy
For microscopy analysis, HeLa cells were fixed with 4% paraformaldehyde (pH 7.4 for 15 min at 37°C). Fixed cells were then permeabilized with 0.25% Triton X-100 in phosphate-buffered saline for 10 min at room temperature and stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei. Cells were viewed and analyzed on a Zeiss Axio Observer.Z1 microscope using a 20× objective (Carl Zeiss, Jena, Germany). The JACoP plug-in for ImageJ was used to calculate Pearson's coefficient for the analysis of colocalization of eGFP and the mCherry signals. For the z-stack imaging, 14 images were obtained with a 63× oil objective at 0.423-μm increments obtained above and below focused nuclei. Quantitative image cytometry was performed using an Olympus scan^R integrated imager and image analysis suite (Olympus-SIS, Münster, Germany) equipped with a Hamamatsu ORCA-R2 CCD digital camera (Hamamatsu Photonics, Tokyo, Japan; Rantala et al., 2013). Cells grown on coverslips were imaged with a 20× LUCPLFLN numerical aperture 0.40 objective using specific filter sets for DAPI, GFP, and red fluorescent protein (Semrock, Rochester, NY). The scan^R image analysis software suite was used for quantitation of nuclear and cytoplasmic localization and signal intensity. The IncuCyte machine was use for real-time evaluation of fluorescence in live cells (Essen BioScience, Ann Arbor, MI). Transiently transfected HeLa cells were imaged for 48–72 h.
Flow cytometry
A minimum of 5000 events was generally obtained for each condition using the Guava EasyCyte Plus (Millipore, Billerica, MA). Gating was generated on a negative population or relative to a control or untreated condition. Within each experiment, the same analysis was applied to all samples. For the experiments shown in Figure 5D we expressed the shRNA as indicated for 48 h and then selected for puromycin resistance at 1 μg/ml for 3 d before analyzing eGFP expression.
Statistical analysis
Averages are from independent experiments with SD or SEM calculated to display variability. Student's t test was performed using a two-tailed test.
Supplementary Material
Acknowledgments
We acknowledge Larry David, Fred Robinson, and Monika Davare for their expert advice on this project. K.T.D. and this project were largely supported by grants from the Knight Cancer Institute, the Aplastic Anemia and Myelodysplasia International Foundation, and the National Institutes of Health, National Heart, Lung and Blood Institute (1K08HL111280). J.W.T. is supported by grants from the Leukemia and Lymphoma Society, the William Lawrence and Blanche Hughes Fund, the V Foundation for Cancer Research, and the National Cancer Institute (4R00CA151457). B.J.D. is supported by the Howard Hughes Medical Institute. G.C.B. is supported by the National Cancer Institute (1R01CA138237), the Veteran's Affairs Merit Review, and the National Heart, Lung and Blood Institute (5P01 HL048546).
Abbreviations used:
- eGFP
enhanced green fluorescent protein
- FA
Fanconi anemia
- HA
hemagglutinin
- Ub
ubiquitin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0144) on June 19, 2013.
K.T.D, M.D.R, B.R.B, J.E.Y, J.R., and J.W.T. performed experiments and interpretation of data. The majority of the experiments and the data presented were performed by K.T.D. and M.D.R. K.T.D. supervised the design of the experiments, execution of the experiments, and data analyses and presentation and wrote the majority of the manuscript. C.T., B.J.D., and G.C.B. provided critical commentary on the hypotheses, experimental design, data analyses, and manuscript.
*These authors contributed equally.
REFERENCES
- Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson AE, et al. Glycogen synthase kinase 3beta missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci USA. 2009;106:3925–3929. doi: 10.1073/pnas.0900189106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Auerbach AD, Liu Q, Ghosh R, Pollack MS, Douglas GW, Broxmeyer HE. Prenatal identification of potential donors for umbilical cord blood transplantation for Fanconi anemia. Transfusion. 1990;30:682–687. doi: 10.1046/j.1537-2995.1990.30891020324.x. [DOI] [PubMed] [Google Scholar]
- Broxmeyer HE, Sherry B, Cooper S, Ruscetti FW, Williams DE, Arosio P, Kwon BS, Cerami A. Macrophage inflammatory protein (MIP)-1 beta abrogates the capacity of MIP-1 alpha to suppress myeloid progenitor cell growth. J Immunol. 1991;147:2586–2594. [PubMed] [Google Scholar]
- Chappell WH, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–164. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AR, Lewis LP, Walden H. The structure of the catalytic subunit FANCL of the Fanconi anemia core complex. Nat Struct Mol Biol. 2010;17:294–298. doi: 10.1038/nsmb.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105:721–732. doi: 10.1016/s0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Dao KH, Rotelli MD, Petersen CL, Kaech S, Nelson WD, Yates JE, Hanlon Newell AE, Olson SB, Druker BJ, Bagby GC. FANCL ubiquitinates beta-catenin and enhances its nuclear function. Blood. 2012;120:323–334. doi: 10.1182/blood-2011-11-388355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bie P, Ciechanover A. Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 2011;18:1393–1402. doi: 10.1038/cdd.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour C, Corcione A, Svahn J, Haupt R, Poggi V, Beka'ssy AN, Scime R, Pistorio A, Pistoia V. TNF-alpha and IFN-gamma are overexpressed in the bone marrow of Fanconi anemia patients and TNF-alpha suppresses erythropoiesis in vitro. Blood. 2003;102:2053–2059. doi: 10.1182/blood-2003-01-0114. [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- Gluckman E, et al. Hematopoietic reconstitution in a patient with Fanconi's anemia by means of umbilical-cord blood from an HLA-identical sibling. N Engl J Med. 1989;321:1174–1178. doi: 10.1056/NEJM198910263211707. [DOI] [PubMed] [Google Scholar]
- Gurtan AM, Stuckert P, D'Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006;281:10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- Haneline LS, Gobbett TA, Ramani R, Carreau M, Buchwald M, Yoder MC, Clapp DW. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94:1–8. [PubMed] [Google Scholar]
- Hodson C, Cole AR, Lewis LP, Miles JA, Purkiss A, Walden H. Structural analysis of human FANCL, the E3 ligase in the Fanconi anemia pathway. J Biol Chem. 2011;286:32628–32637. doi: 10.1074/jbc.M111.244632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacome A, et al. Lentiviral-mediated genetic correction of hematopoietic and mesenchymal progenitor cells from Fanconi anemia patients. Mol Ther. 2009;17:1083–1092. doi: 10.1038/mt.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest. 2012;122:3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PF, et al. Stem cell collection and gene transfer in Fanconi anemia. Mol Ther. 2007;15:211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- Kockeritz L, Doble B, Patel S, Woodgett JR. Glycogen synthase kinase-3—an overview of an over-achieving protein kinase. Curr Drug Targets. 2006;7:1377–1388. doi: 10.2174/1389450110607011377. [DOI] [PubMed] [Google Scholar]
- Korkina LG, Samochatova EV, Maschan AA, Suslova TB, Cheremisina ZP, Afanas'ev IB. Release of active oxygen radicals by leukocytes of Fanconi anemia patients. J Leukoc Biol. 1992;52:357–362. doi: 10.1002/jlb.52.3.357. [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- Larghero J, Marolleau JP, Soulier J, Filion A, Rocha V, Benbunan M, Gluckman E. Hematopoietic progenitor cell harvest and functionality in Fanconi anemia patients. Blood. 2002;100:3051. doi: 10.1182/blood-2002-07-2069. [DOI] [PubMed] [Google Scholar]
- Li J, et al. TNF-alpha induces leukemic clonal evolution ex vivo in Fanconi anemia group C murine stem cells. J Clin Invest. 2007;117:3283–3295. doi: 10.1172/JCI31772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang Y, Yuan J, Hong P, Freie B, Orazi A, Haneline LS, Clapp DW. Continuous in vivo infusion of interferon-gamma (IFN-gamma) preferentially reduces myeloid progenitor numbers and enhances engraftment of syngeneic wild-type cells in Fancc-/- mice. Blood. 2004;104:1204–1209. doi: 10.1182/blood-2004-03-1094. [DOI] [PubMed] [Google Scholar]
- Li X, et al. Ex vivo culture of Fancc-/- stem/progenitor cells predisposes cells to undergo apoptosis, and surviving stem/progenitor cells display cytogenetic abnormalities and an increased risk of malignancy. Blood. 2005;105:3465–3471. doi: 10.1182/blood-2004-06-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JM, Kim S, Read EJ, Futaki M, Dokal I, Carter CS, Leitman SF, Pensiero M, Young NS, Walsh CE. Engraftment of hematopoietic progenitor cells transduced with the Fanconi anemia group C gene (FANCC) Hum Gene Ther. 1999;10:2337–2346. doi: 10.1089/10430349950016988. [DOI] [PubMed] [Google Scholar]
- Livingston CM, Ifrim MF, Cowan AE, Weller SK. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 2009;5:e1000619. doi: 10.1371/journal.ppat.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli AM, Tabellini G, Bressanin D, Ognibene A, Goto K, Cocco L, Evangelisti C. The emerging multiple roles of nuclear Akt. Biochim Biophys Acta. 2012;1823:2168–2178. doi: 10.1016/j.bbamcr.2012.08.017. [DOI] [PubMed] [Google Scholar]
- Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak R, Buitenhuis M. The PI3K/PKB signaling module as key regulator of hematopoiesis: implications for therapeutic strategies in leukemia. Blood. 2012;119:911–923. doi: 10.1182/blood-2011-07-366203. [DOI] [PubMed] [Google Scholar]
- Rantala J, Kwon S, Korkola J, Gray J. Expanding the diversity of imaging-based RNAi screen applications using cell spot microarrays. Microarrays. 2013;2:97–114. doi: 10.3390/microarrays2020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Navarro S, Richaud-Patin Y, Guenechea G, Sanchez-Danes A, Consiglio A, Bueren J, Izpisua Belmonte JC. A protocol describing the genetic correction of somatic human cells and subsequent generation of iPS cells. Nat Protoc. 2010;5:647–660. doi: 10.1038/nprot.2010.9. [DOI] [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- Rosselli F, Sanceau J, Gluckman E, Wietzerbin J, Moustacchi E. Abnormal lymphokine production: a novel feature of the genetic disease Fanconi anemia. II. In vitro and in vivo spontaneous overproduction of tumor necrosis factor alpha. Blood. 1994;83:1216–1225. [PubMed] [Google Scholar]
- Scarpa M, Rigo A, Momo F, Isacchi G, Novelli G, Dallapiccola B. Increased rate of superoxide ion generation in Fanconi anemia erythrocytes. Biochem Biophys Res Commun. 1985;130:127–132. doi: 10.1016/0006-291x(85)90391-2. [DOI] [PubMed] [Google Scholar]
- Schultz JC, Shahidi NT. Tumor necrosis factor-alpha overproduction in Fanconi's anemia. Am J Hematol. 1993;42:196–201. doi: 10.1002/ajh.2830420211. [DOI] [PubMed] [Google Scholar]
- Seki S, et al. A requirement of FancL and FancD2 monoubiquitination in DNA repair. Genes Cells. 2007;12:299–310. doi: 10.1111/j.1365-2443.2007.01054.x. [DOI] [PubMed] [Google Scholar]
- Si Y, et al. Continuous in vivo infusion of interferon-gamma (IFN-gamma) enhances engraftment of syngeneic wild-type cells in Fanca-/- and Fancg-/- mice. Blood. 2006;108:4283–4287. doi: 10.1182/blood-2006-03-007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulpule A, Lensch MW, Miller JD, Austin K, D'Andrea A, Schlaeger TM, Shimamura A, Daley GQ. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115:3453–3462. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Morrison SJ. The PI-3kinase pathway in hematopoietic stem cells and leukemia-initiating cells: a mechanistic difference between normal and cancer stem cells. Blood Cells Mol Dis. 2008;41:73–76. doi: 10.1016/j.bcmd.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. Defective homing is associated with altered Cdc42 activity in cells from patients with Fanconi anemia group A. Blood. 2008;112:1683–1686. doi: 10.1182/blood-2008-03-147090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120:1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.