Spatiotemporal dynamics of condensins I and II in the primitive red alga Cyanidioschyzon merolae is surprisingly similar to that observed in mammalian cells. Condensin II is not essential for mitosis under laboratory growth conditions but is required for sister centromere resolution in the presence of a microtubule drug.
Abstract
Condensins are multisubunit complexes that play central roles in chromosome organization and segregation in eukaryotes. Many eukaryotic species have two different condensin complexes (condensins I and II), although some species, such as fungi, have condensin I only. Here we use the red alga Cyanidioschyzon merolae as a model organism because it represents the smallest and simplest organism that is predicted to possess both condensins I and II. We demonstrate that, despite the great evolutionary distance, spatiotemporal dynamics of condensins in C. merolae is strikingly similar to that observed in mammalian cells: condensin II is nuclear throughout the cell cycle, whereas condensin I appears on chromosomes only after the nuclear envelope partially dissolves at prometaphase. Unlike in mammalian cells, however, condensin II is confined to centromeres in metaphase, whereas condensin I distributes more broadly along arms. We firmly establish a targeted gene disruption technique in this organism and find, to our surprise, that condensin II is not essential for mitosis under laboratory growth conditions, although it plays a crucial role in facilitating sister centromere resolution in the presence of a microtubule drug. The results provide fundamental insights into the evolution of condensin-based chromosome architecture and dynamics.
INTRODUCTION
Chromosomes are the principal carrier of genetic information in all living organisms. Despite such fundamental functions assigned to chromosomes, their size and number in individual organisms are astonishingly variable. For instance, the average chromosome length in a eukaryotic species ranges from ∼0.26 Mb (Encephalitozoon cuniculi, a parasitic fungus) to ∼1800 Mb (Triturus cristatus, a newt). It is therefore highly puzzling, as well as thought-provoking, how higher-order architecture and dynamics of chromosomes might be regulated by an evolutionarily conserved set of protein components whose sizes are not radically different from species to species.
Among the structural components constituting chromosomes, condensins are of particular interest. Recent studies established that this class of protein complexes plays central roles in chromosome condensation and segregation during mitotic and meiotic cell divisions (Cuylen and Haering, 2011; Hirano, 2012). The canonical condensin complex (condensin I) is composed of five subunits, all of which are conserved in all eukaryotes examined (Hirano et al., 1997; Sutani et al., 1999). In contrast, when the second condensin complex (condensin II) was first discovered from vertebrate cells (Ono et al., 2003; Yeong et al., 2003), none of the corresponding subunits specific to condensin II was found in fungi. This observation lead to the hypothesis that condensin II might be a new invention in evolution that provided an additional level of organization and rigidity with large chromosomes (Ono et al., 2003). This hypothesis turned out to be wrong, however, based on sequencing of an ever-growing number of eukaryotic genomes. It is now clear that the subunits of both condensins I and II are widely conserved among eukaryotes (Hirano, 2012). The implication is that the last eukaryotic common ancestor possessed both condensins I and II and some species, such as fungi, lost condensin II during evolution (Supplemental Figure S1).
The notion that most eukaryotes have two different types of condensin complexes raises a number of fundamental questions in chromosome biology. For example, why do two condensins exist? Do they each have unique functions? How are they regulated, and how do they work at a mechanistic level? Early studies in human tissue culture cells demonstrated that condensins I and II are subjected to differential spatiotemporal regulation during the cell cycle and make distinct contributions to mitotic chromosome architecture and dynamics (Ono et al., 2003). Condensin II localizes to the nucleus or chromosomes throughout the cell cycle and participates in the early stage of chromosome condensation within the prophase nucleus. In contrast, condensin I is sequestered into the cytoplasm during interphase and gains access to chromosomes only after the nuclear envelope breaks down in prometaphase (Hirota et al., 2004; Ono et al., 2004; Gerlich et al., 2006). More recent studies show that condensin II initiates its function as early as S phase (Ono et al., 2013) and that the relative ratio of condensin I to II is one of the critical factors that determine the shape of metaphase chromosomes (Shintomi and Hirano, 2011; Green et al., 2012). Despite such progress, understanding of the mechanisms, functions, and regulation of the two condensin complexes is very limited.
To address these fundamental and diverse set of questions, an evolutionary point of view is of great importance. It has been noted that there is no apparent relationship between the possession of condensin II and the size of the genome among eukaryotic species. This is best exemplified by the finding that all candidate subunits comprising condensins I and II are encoded by the very compact genome (∼16.5 Mb) of the primitive red alga Cyanidioschyzon merolae (Matsuzaki et al., 2004; Hirano, 2005). C. merolae is a small and simple unicellular organism composed of a single nucleus, mitochondrion, and plastid. Its nucleus contains 20 chromosomes, whose average length (∼0.83 Mb) is comparable to that of Saccharomyces cerevisiae chromosomes (∼0.76 Mb; Matsuzaki et al., 2004; Nozaki et al., 2007). Thus C. merolae arguably represents the smallest and simplest organism predicted to possess both condensins I and II.
In the present study, we report the first and comprehensive set of analyses of C. merolae condensins that combines biochemistry, cytology, and genetics. We show that the primitive red alga indeed has two biochemically defined condensin complexes, both of which are expressed during the mitotic cell cycle. Remarkably, the localization and dynamics of condensins I and II in C. merolae are very similar to those in vertebrate cells, except that C. merolae condensin II is enriched at centromeres and absent along arms in metaphase. A modified targeted gene disruption technique, firmly established in the present study, shows that condensin II is not essential for cell division under laboratory growth conditions but is required for resolving sister centromeres in the presence of oryzalin. We also discuss the evolutionary implications of these findings for the chromosome organization/segregation machinery.
RESULTS
Redefining the cell cycle of C. merolae using stage-specific markers
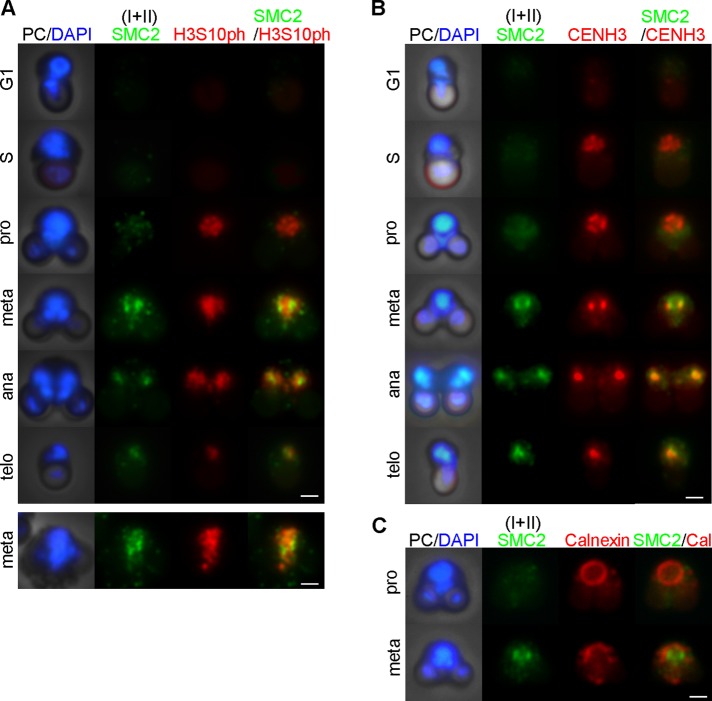
The cell cycle of C. merolae had been defined primarily on the basis of its relationship to the division cycles of the mitochondrion and the chloroplast (reviewed by Imoto et al., 2011; typical G1- and M-phase cells of C. merolae are depicted in Figure 1A; also see Supplemental Figure S2). In the present study, we first sought to establish stage-specific immunofluorescence markers for defining the cell cycle of this organism. On the basis of previous microarray data (Fujiwara et al., 2009), we selected proliferating cell nuclear antigen (PCNA), a processivity factor for DNA polymerase δ, as a candidate for S phase–specific markers (Celis and Celis, 1985; Supplemental Figure S3, A and B) and also tested the phosphoepitope of histone H3 at serine 10 (H3S10ph) for a potential M-phase maker (Hendzel et al., 1997; Supplemental Figure S3C). Immunofluorescence analyses of C. merolae cells in synchronized cultures showed that the signals of PCNA and H3S10ph were both detectable within the nucleus but in distinct phases of the cell cycle (Figure 1B). The signals of PCNA were intensely observed within the nucleus in cells with a single spherical chloroplast (S phase) but diminished before the chloroplast division plane initiated contraction (G2 phase). In contrast, the signals of H3S10ph became detectable within the nucleus in cells with a pair of divided chloroplasts (M phase) and persisted until cytokinesis was completed. We therefore concluded that PCNA and H3S10ph can be used as specific and reliable markers for S and M phases, respectively, of the C. merolae cell cycle (Figure 1C; Supplemental Figure S3D). Of importance, we also noticed that it is possible to distinguish between prophase and metaphase cells on the basis of 4′,6-diamidino-2-phenylindole (DAPI)–stained and H3S10ph-labeled images: the nucleus had an ellipsoidal shape in prophase and became more compact by metaphase (Figure 1B, top, pro and meta). Furthermore, when viewed from a chloroplast-proximal angle, the H3S10ph signals in the metaphase nucleus displayed a characteristic rod-like shape (Figure 1B, bottom, compare pro and meta). These results implicate that the bulk of C. merolae chromosomes undergo substantial structural changes during mitosis, although individual chromosomes are indiscernible from each other.
FIGURE 1:
Characterization of cell-cycle markers in C. merolae. (A) Schematic diagrams of C. merolae cells at G1 and M phases. Chl, chloroplast; Chn, chloroplast nucleoid; Mt, mitochondrion; Mtn, mitochondrial nucleoid; Nu, nucleus. (B) Immunofluorescence labeling of C. merolae cells with antibodies against PCNA (green) and H3S10ph (red). Phase-contrast (PC) and DAPI-stained (blue) images are also shown. The images are arranged according to the predicted stages of the cell cycle. Bottom, prophase and metaphase cells viewed from a different angle. Bar, 1 μm. (C) Time course of frequencies of cells positive for PCNA and H3S10ph in a synchronized culture (shown in B; n > 300). Error bars represent the SD. L indicates a light period. (D) Immunofluorescence labeling with antibodies against PCNA (green) and CENH3 (red). Bar, 1 μm. (E) Time course of frequencies of cells at different stages of the cell cycle as judged by PCNA and CENH3 labeling in a synchronized culture (shown in D; n > 180). The sum of non-G1 cells (from S phase through telophase) is also shown by the gray line. (F) Immunoblotting analysis for protein/modification levels in a synchronized culture. A part of the gel stained with Coomassie brilliant blue (CBB) is shown as a loading control. (G) Prophase and metaphase cells labeled with antibodies specific to calnexin (red) and CENH3 (green). Bar, 1 μm. (H) Prophase and metaphase cells labeled with antibodies specific to nuclear pore complex proteins (NPC; green) and calnexin (red). Bar, 1 μm. (I) Schematic diagrams depicting “semiopen” mitosis in C. merolae.
We then performed colabeling of PCNA and the centromere-specific histone H3 variant CENH3 (also known as CENP-A; Maruyama et al., 2007). The signals of CENH3 were faintly detectable in early S phase and became intense by mid S phase (Figure 1D). The speckle-like signals of CENH3 persisted from G2 through prophase and were then converted into two discrete clusters in metaphase. The CENH3 clusters, each of which would contain separating sister centromeres of 20 chromosomes, were partitioned into daughter nuclei in telophase. Figure 1E shows the progression of the cell cycle on the basis of the CENH3/PCNA labeling patterns. Immunoblotting analyses further confirmed that the protein levels of PCNA, CENH3, and H3S10ph increased and decreased during the cell cycle in a manner consistent with the immunofluorescence analyses (Figure 1F).
To examine whether the nuclear envelope dissolves during mitosis in C. merolae, we colabeled mitotic cells with an antibody against calnexin, a marker for nuclear endoplasmic reticulum (representing the nuclear envelope in C. merolae; Yagisawa et al., 2012), and anti-CENH3. In prophase cells, anti-calnexin smoothly decorated the whole surface of the nucleus. In metaphase cells, in which the CENH3 signals were converted into two discrete clusters, the calnexin signals lost its continuity, implicating a partial breakdown of the nuclear envelope (Figure 1G). We then colabeled mitotic cells with a monoclonal antibody recognizing a panel of nuclear pore complex proteins (NPCs; Aris and Blobel, 1989) and anti-calnexin and found that dotty signals of NPCs detected around the prophase nucleus were largely dispersed by metaphase (Figure 1H). We therefore suggest that C merolae undergoes so-called “semiopen” mitosis (Figure 1I).
Biochemical characterization of condensin complexes in C. merolae
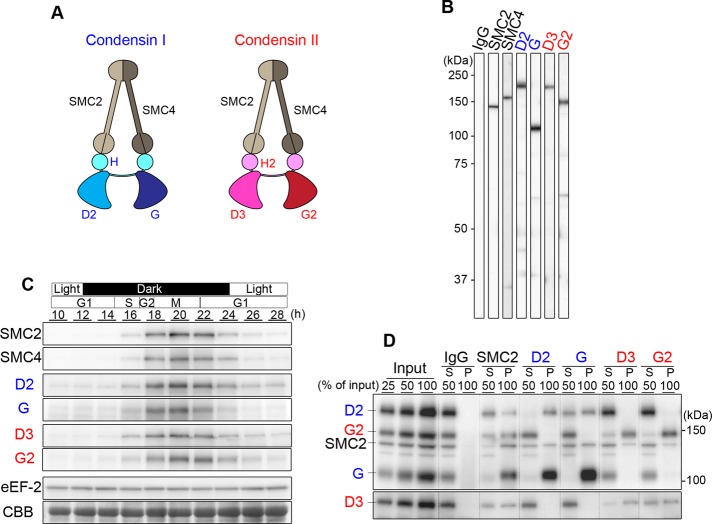
As summarized in Figure 2A, condensins I and II share a pair of SMC core subunits (SMC2 and SMC4) and contain different sets of non-SMC subunits (CAP-D2, CAP-G, and CAP-H for condensin I; CAP-D3, CAP-G2, and CAP-H2 for condensin II; Ono et al., 2003). We prepared a panel of specific antibodies against C. merolae SMC2, SMC4, CAP-D2, CAP-G, CAP-D3, and CAP-G2, each of which recognized a band with the predicted molecular weight of the target subunit, as judged by immunoblotting against a total lysate (Figure 2B). Immunoblotting against total lysates prepared from different time points of a synchronized culture showed that the protein levels of condensin subunits peaked in M phase (Figure 2C). No noticeable difference in the kinetics of protein levels was detected among different subunits of condensins I and II.
FIGURE 2:
Biochemical characterization of condensin complexes in C. merolae. (A) Subunit compositions of condensins I and II. (B) Immunoblotting analysis of a total lysate with affinity-purified antibodies against SMC2, SMC4, CAP-D2, CAP -G, CAP-D3, and CAP-G2. (C) Time course of levels of condensin subunits in a synchronized culture. Immunoblotting with anti–eukaryotic elongation factor-2 and a CBB-stained part of the gel is shown as loading controls. (D) Immunoprecipitation of condensin complexes. A lysate prepared from C. merolae cells synchronized at M phase (input) was subjected to immunoprecipitations using the antibodies indicated at the top. We analyzed 50% of each supernatant (S) and 100% of each immunoprecipitate (P) by immunoblotting using the antibodies indicated at the left.
We then performed a set of reciprocal immunoprecipitation analyses to test whether C. merolae indeed has two biochemically separable condensin complexes, as shown in vertebrate cells (Ono et al., 2003; Lee et al., 2011). We found that anti-SMC2 precipitated the CAP-D2 and CAP-G subunits of condensin I, as well as the CAP-D3 and CAP-G2 subunits of condensin II, along with SMC2 itself (Figure 2D). Anti-CAP-D2 or anti-CAP-G precipitated SMC2, CAP-D2, and CAP-G but not CAP-D3 or CAP-G2. Conversely, anti-CAP-D3 or anti-CAP-G2 precipitated SMC2, CAP-D3, and CAP-G2 but not CAP-D2 or CAP-G. Although we do not have antibodies against the remaining two subunits (i.e., CAP-H and CAP-H2), the present results strongly indicate that C. merolae has two conventional types of condensins (condensins I and II), each of which is composed of five subunits. Moreover, as judged by the amounts of SMC2 precipitated with the different antibodies, we estimate that condensin I is more abundant than condensin II in C. merolae.
Localization of SMC2, a common subunit of condensins I and II, during the cell cycle in C. merolae
To examine the subcellular localization of condensins in C. merolae, we performed immunofluorescence labeling with an antibody against SMC2, a common subunit of condensins I and II, along with other cell-cycle markers. Although the signals of SMC2 were hardly detectable in G1 and S phase cells, faint signals of SMC2 were observed within the nucleus in prophase when the H3S10ph epitope appeared (Figure 3A). In metaphase, brighter signals of SMC2 decorated the whole nucleus, but a pair of discrete clusters, reminiscent of CENH3-positive centromeres, often dominated (Supplemental Figure S4A). When metaphase cells were viewed from a chloroplast-proximal angle, SMC2 largely overlapped with the H3S10ph-positive, rod-shaped region, yet the pair of centromere-like signals were also clearly discernible (Figure 3A, bottom). Colabeling with anti-CENH3 confirmed that the bright pair of SMC2 signals was indeed colocalized with CENH3-enriched centromeres (Figure 3B). The SMC2 signals persisted on chromosomes from anaphase through telophase (Figure 3, A and B).
FIGURE 3:
Subcellular localization of SMC2 during the cell cycle of C. merolae. (A) Immunolocalization of SMC2 (green) and H3S10ph (red). Phase-contrast (PC) and DAPI-stained (blue) images are also shown. The images are arranged according to the predicted stages of the cell cycle. Bars, 1 μm. (B) Immunolocalization of SMC2 (green) and CENH3 (red). Bar, 1 μm. (C) Immunolocalization of SMC2 (green) and calnexin (red). Bar, 1 μm.
Colabeling of SMC2 and calnexin demonstrated that the appearance of the bulk of SMC2 signals was coincident with the dispersion of calnexin in metaphase (Figure 3C), indicating that a large portion of SMC2 entered into the nucleus upon partial dissolution of the nuclear envelope.
Differential distributions of condensins I and II on chromosomes during the cell cycle
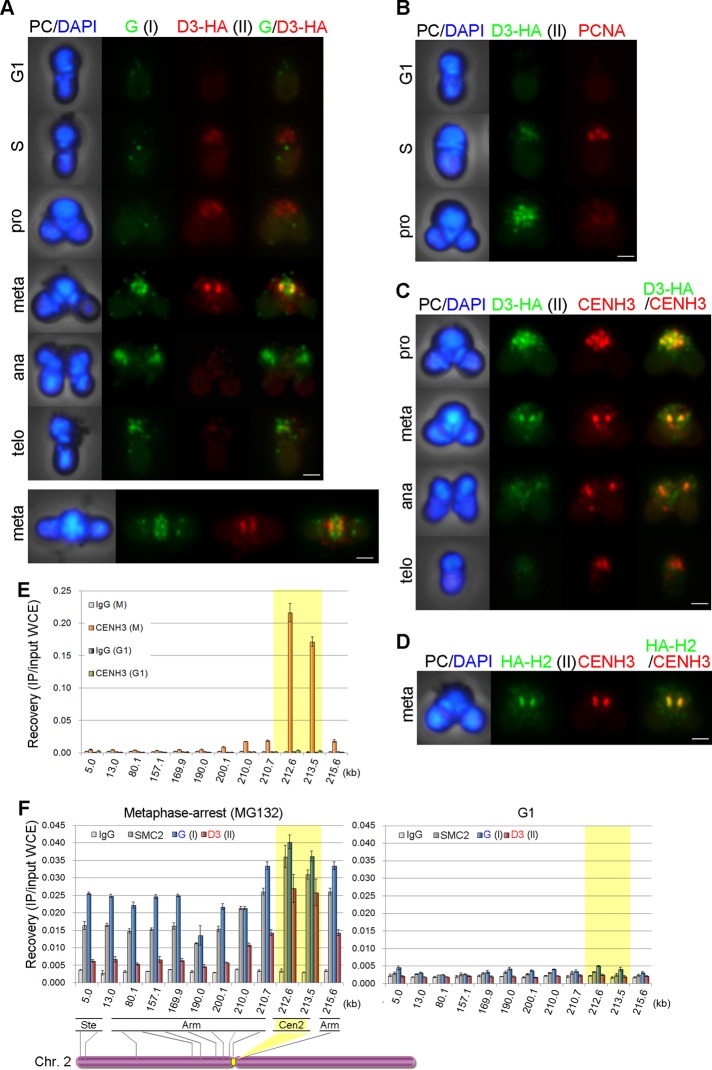
We then sought to differentially localize condensin I– and condensin II–specific subunits in C. merolae cells. Because our antibodies against condensin II–specific subunits (CAP-D3 and CAP-H2) failed to work for immunofluorescence analysis, we decided to construct strains in which the genomic copy of the CAP-D3 gene was replaced with a hemagglutinin (HA)–tagged version. To this end, we modified previously reported methods of targeted gene replacement (Imamura et al., 2009, 2010) by using the wild-type C. merolae URA5.3 gene (rather than the chimera of C. merolae URA5 and Galdieria sulphuraria URA3 [referred to as URACm-Gs]) as a selection marker. As fully described in Supplemental Figure S5, our modified method guaranteed single-copy insertions, resulting in one-step replacement of the targeted genes by homologous recombination at a high frequency. In this way, transformation of the uracil-auxotrophic strain M4 (Minoda et al., 2004), followed by selection for uracil-autotrophic cells, allowed us to recover strains expressing a C-terminally HA-tagged version of CAP-D3 (D3-HA; see Supplemental Materials and Methods). It should be emphasized that the level of D3-HA expressed in this strain was exactly the same as that of endogenous CAP-D3 expressed in M4 (Supplemental Figure S5D, right). Reciprocal immunoprecipitation analyses confirmed that D3-HA was incorporated into the condensin II complex (Supplemental Figure S5E).
To determine the localization of condensins I and II, we colabeled the D3-HA cells with an affinity-purified rabbit antibody against CAP-G and a rat monoclonal antibody against HA (Figure 4A). Neither CAP-G nor D3-HA signals were detectable in G1 cells. From S phase to prophase, diffuse or speckled signals of D3-HA were observed within the nucleus. Then the signals of D3-HA became highly concentrated at centromeres in metaphase and quickly disappeared in anaphase. In contrast, the signals of CAP-G were hardly detectable within the nucleus from S phase through prophase and became apparent on chromosomes only after metaphase. Unlike D3-HA, the chromosomal signal of CAP-G persisted through telophase. A metaphase image viewed from a different angle also indicated that D3-HA was specifically localized at centromeres, whereas CAP-G was distributed broadly on whole chromosome regions (Figure 4A, bottom; also see Supplemental Figure S4, B and C). The timing of the appearance of D3-HA signals in S phase and their enrichment at metaphase centromeres were further confirmed by colabeling with anti-PCNA (Figure 4B) or anti-CENH3 (Figure 4C). Moreover, we also constructed strains expressing an N-terminally HA-tagged version of CAP-H2 (see HA-H2; Supplemental Figure S6) and confirmed its colocalization with CENH3 in metaphase (Figure 4D and Supplemental Figure S7).
FIGURE 4:
Differential distributions of condensins I and II during the cell cycle of C. merolae. (A) Immunolocalization of CAP-G (green) and D3-HA (red). Phase-contrast (PC) and DAPI-stained (blue) images are also shown. Bar, 1 μm. (B) Immunolocalization of D3-HA (green) and PCNA (red). Bar, 1 μm. (C) Immunolocalization of D3-HA (green) and CENH3 (red). Bar, 1 μm. (D) Immunolocalization of HA-H2 (green) and CENH3 (red). Bar, 1 μm. (E) ChIP-qPCR analysis for CENH3 on chromosome 2 in M- and G1-phase cells. Error bars represent the SD (n = 3). (F) ChIP-qPCR analysis for condensin subunits along chromosome 2 in metaphase-arrested and G1-phase cells. Bottom, schematic diagram of chromosome 2 (457 kb long) with the positions analyzed by ChIP-qPCR (Cen, centromere; Ste, subtelomere). Error bars represent the SD (n = 3).
A genome-wide chromatin immunoprecipitation (ChIP)-chip analysis had identified a single CENH3-enriched region in each of 20 chromosomes in C. merolae. The details of this analysis will be described elsewhere. In the present study, we performed ChIP-quantitative PCR (ChIP-qPCR) analyses using 12 pairs of primers designed along one arm of chromosome 2 and confirmed a specific enrichment of CENH3 within the putative centromeric regions in M phase cells but not in G1 cells (Figure 4E, 212.6 and 213.5 kb). In the present study, we defined this region as the centromere of chromosome 2. We then performed the same set of ChIP-qPCR analyses to reveal the chromosomal distributions of SMC2 (I + II), CAP-G (I), and CAP-D3 (II). In this experiment, a synchronous culture was further treated with MG132, a proteasome inhibitor, to enrich metaphase-arrested cells (Supplemental Figure S8). We found that in these cells the occupancy of CAP-D3 peaked at the centromeric region, decreased gradually toward both sides of the centromere, and reached a very low level at the subtelomeric region (Figure 4F, left, red bars). In contrast, the occupancies of SMC2 and CAP-G were detectable broadly along the entire length of the chromosome arm, although highest peaks were found at the centromeric region (Figure 4F, left, gray and blue bars). The same set of analyses using G1 cells offered a negative control (Figure 4F, right). Thus the ChIP-qPCR analyses nicely complemented the immunolocalization data, demonstrating that condensin II is enriched around centromeres at metaphase, whereas condensin I distributes more broadly along chromosome arms in C. merolae.
Targeted gene disruption of condensin subunits in C. merolae
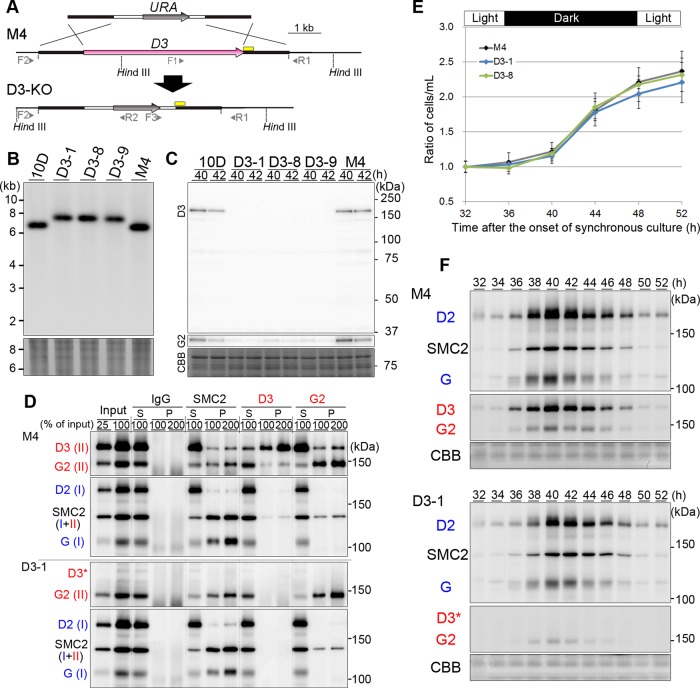
To test how condensins might contribute to chromosome architecture and segregation in C. merolae, we set up targeted gene disruption of genes encoding condensin subunits (see Supplemental Materials and Methods). Figure 5A shows the basic strategy for one-step disruption of the CAP-D3 gene by homologous recombination, which allowed us to recover three independent strains, named D3-1, D3-8, and D3-9. We verified the occurrence of the predicted recombination events by PCRs and Southern blotting (Supplemental Figure S9A and Figure 5B) and further confirmed the loss of CAP-D3 in the mutant lysates by immunoblotting (Figure 5C). These results clearly demonstrated that the predicted recombination by a double-crossover reaction successfully disrupted the CAP-D3 gene in these CAP-D3–knockout (CAP-D3-KO) strains. We then performed immunoprecipitation analyses of the lysates prepared from M4 and D3-1 with antibodies against SMC2, CAP-D3, and CAP-G2. As expected, CAP-D3 was not detectable in all immunoprecipitates prepared from the D3-1 lysate (Figure 5D). Anti-SMC2 coprecipitated the condensin I subunits CAP-D2 and CAP-G from the D3-1 lysate, indicating that the condensin I complex is intact in this mutant strain. Whereas the level of CAP-G2 was substantially reduced in D3-1 compared with M4 (Figure 5C, anti-G2), the residual fraction of CAP-G2 was found to associate with SMC2, as judged by reciprocal immunoprecipitations (Figure 5D). It was therefore most likely that a defective form of condensin II missing only CAP-D3 exists in D3-1 cells, albeit at a reduced level. Moreover, ChIP-qPCR analyses demonstrated that the occupancy of CAP-D3 at the centromeric region was no longer detectable in D3-1 (Supplemental Figure S10).
FIGURE 5:
Construction and characterization of the CAP-D3-KO strains. (A) Schematic diagram of targeted gene disruption by homologous recombination. The gray and magenta arrows indicate the URA5.3 (URA) and CAP-D3 genes, respectively. The thick white lines indicate the upstream and downstream regions of the URA5.3 gene. The black lines indicate the upstream and downstream regions of the CAP-D3 gene. The positions of primers for PCRs are shown by the arrowheads. The yellow bars indicate the probe used for Southern blotting analysis. (B) Southern blotting analysis of three CAP-D3-KO strains isolated independently (D3-1, D3-8, and D3-9), along with 10D (wild-type strain) and M4 (parental strain used for gene targeting). A part of the ethidium bromide (EtBr)–stained gel is shown as a loading control. (C) Immunoblotting analysis against total lysates with anti–CAP-D3. A CBB-stained part of the gel is shown as a loading control. (D) Immunoprecipitation analysis. Lysates prepared from M4 and D3-1 were subjected to immunoprecipitations using the antibodies indicated at the top. We analyzed 100% of supernatants (S) and 100 and 200% of immunoprecipitates (P) by immunoblotting using the antibodies indicated at the left. Note that CAP-D3 was not detectable at the expected position (indicated by D3*) in D3-1. (E) Growth curves of M4 and two CAP-D3-KO strains in synchronized cultures. Error bars represent the SD. (F) Time course of the level of condensin subunits during the cell cycle in M4 and D3-1. Note that CAP-D3 was not detectable in D3-1 at the expected position (D3*).
We then investigated the growth property of the CAP-D3-KO strains (D3-1 and D3-8) in synchronous cultures using 2× Allen's medium containing uracil. To our surprise, we could not detect any significant differences in their growth rate between D3-1/D3-8 and their parental strain M4 (Figure 5E and Supplemental Figure S9B). Immunoblotting analysis of lysates prepared from the synchronized cultures demonstrated that SMC2, CAP-D2, CAP-G, CAP-G2, and other cell cycle markers appear and disappear in a normal timing in D3-1 (Figure 5F and Supplemental Figure S9C).
To further substantiate the conclusion that condensin II function is not essential for the survival and division of C. merolae at least under the present growth conditions in the laboratory, we generated mutant strains lacking another condensin II subunit, CAP-H2. We found that the CAP-H2-KO strains were also viable despite the fact that the condensin II complex was disrupted more completely than in the CAP-D3-KO strains (Supplemental Figures S9B and S11). On the other hand, our attempts to generate CAP-D2-KO or CAP-G-KO strains have been unsuccessful, implying that, unlike condensin II, condensin I is essential for the survival of C. merolae.
CAP-D3-KO strains display a defect in sister centromere resolution in the absence of functional microtubules
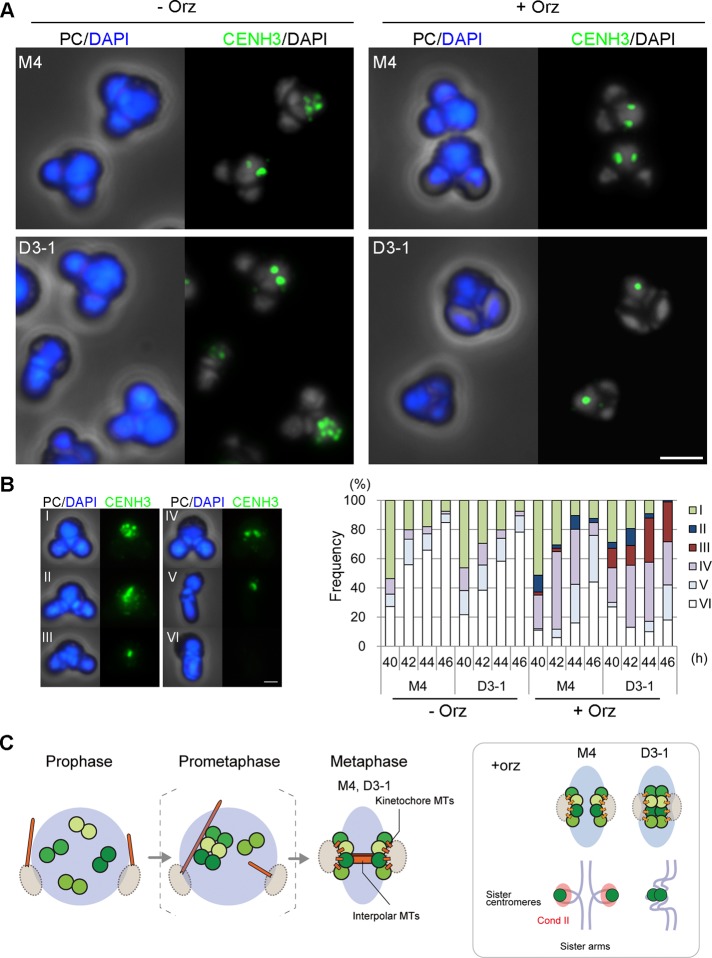
Because our immunofluorescence and ChIP-qPCR analyses showed that condensin II is enriched at centromeres in metaphase, we examined the possibility that condensin II might play a nonessential yet important function in the assembly and/or behavior of centromeres in C. merolae. To this end, we tested for the effect of a microtubule-polymerization inhibitor, oryzalin, in the two strains. In M4, the addition of oryzalin increased the frequency of metaphase-like cells with two centromere clusters (Figure 6A, M4, –/+Orz). Remarkably, however, we found that a substantial fraction of D3-1 cells displayed single (rather than two) centromere clusters in the presence of oryzalin (Figure 6A, D3-1, –/+Orz). To further extend our observations, we classified the observed morphologies into six different types (I–VI) and plotted the time course of their frequencies (Figure 6B). In the absence of oryzalin, virtually no difference was observed between M4 and D3-1 in the frequencies of the type I, IV, V, and IV cells. The type II cells, displaying a bar-like CENH3 signal, were observed in both M4 and D3-1 but only in the presence of oryzalin. On the other hand, the type III cells, displaying single dot-like clusters of centromeres (shown in Figure 6A, D3-1, +Orz), were observed and accumulated over time only in D3-1 and were hardly detectable in M4. Of importance, we found that the type III cells were readily detectable in a CAP-H2-KO strain in the presence of oryzalin but not in its absence (Supplemental Figure S12). Immunofluorescence labeling with antibodies against CENH3 and α-tubulin showed that oryzalin treatment disrupted interpolar microtubules but did not completely remove tubulin-positive signals from spindle poles or their vicinity (Supplemental Figure S9D). The residual signals could represent very short kinetochore microtubules that connect centromeres and spindle poles: it is well known that kinetochore microtubules are more resistant to microtubule depolymerization conditions than are interpolar microtubules (e.g., Rieder, 1981).
FIGURE 6:
CAP-D3-KO cells display a defect in sister centromere resolution in the presence of oryzalin. (A) Immunofluorescence labeling with anti-CENH3 of M4 and D3-1 strains cultured in the absence (–Orz) or presence (+Orz) of oryzalin. Phase-contrast (PC) and DAPI-stained (blue) images are also shown. Bar, 2 μm. (B) Classification of cell types based on DAPI-stained and CENH3 labeled images, and frequencies of the different cell types observed in M4 and D3-1 in the absence or presence of oryzalin (n > 100). Type I, speckles; type II, bar-like shapes; type III, a single cluster; type IV, a pair of clusters; V, telophase cells; VI, G1 cells. Bar, 1 μm. (C) Schematic diagram of predicted centromere–microtubule interactions in C. merolae. The dark/light green circles indicate CENH3-positive sister centromeres, and the light blue circles/ovals indicate arm regions. Inset, observed behaviors of centromeres (top) and hypothesized organization of a pair of sister centromeres/arms (bottom) in M4 and D3-1 strains cultured in the presence of oryzalin.
Our interpretations of the observed phenotypes are summarized in Figure 6C. In the absence of oryzalin, intranuclear microtubules are formed and start searching for kinetochores in the postulated prometaphase stage, eventually establishing bipolar attachments by metaphase in collaboration with interpolar microtubules (Figure 6C; also see Imoto et al., 2010). Sister centromeres are precociously separated from each other on the metaphase spindle, being recognized as a pair of discrete centromere clusters. These processes can occur apparently normally even in cells lacking functional condensin II. On the other hand, disruption of interpolar microtubules by oryzalin helps to uncover a cryptic yet important function of condensin II in resolving sister centromeres (Figure 6C, inset): the pair of sister centromere clusters are no longer discernible from each other when functions of both microtubule and condensin II are compromised at the same time. We suggest that condensin II–dependent centromere resolution and microtubule-dependent (precocious) separation of sister centromeres cooperate to support faithful segregation of chromosome and that the latter process functions dominantly at least under the present growth conditions.
DISCUSSION
Most previous studies on condensins used a subset of standard model organisms and experimental systems, such as vertebrate tissue culture cells, Xenopus egg extracts, Drosophila melanogaster, Caenorhabditis elegans, and yeasts. All of these organisms belong to the supergroup Unikonta, although some genetic studies have been reported in Arabidopsis thaliana, which belongs to the evolutionary distant supergroup Plantae (e.g., Liu et al., 2002; Siddiqui et al., 2006; Sakamoto et al., 2011). In the present study, we used the unicellular red alga C. merolae because it is the smallest and simplest organism predicted to have both condensins I and II (note that yeasts and other fungi lack condensin II). By combining cytology, biochemistry, and genetics, we demonstrate that C. merolae indeed possesses two different biochemically defined condensin complexes, which display differential spatiotemporal dynamics and have nonoverlapping functions. Equally important, the present study establishes a highly reliable and reproducible method for one-step gene replacement by homologous recombination in C. merolae and demonstrates its fruitful use for both gene tagging and targeted gene disruption.
Redefining the cell cycle of C. merolae
We analyzed the nuclear division cycle of C. merolae in great detail by introducing a new set of immunofluorescence markers. Our results show that PCNA and H3S10ph can be used as excellent markers for S and M phase, respectively, in this organism. Although previous work suggested that C. merolae would display “closed” mitosis (Yagisawa et al., 2012), the present data provide evidence that the nuclear envelope dissolves at least partially and the nuclear pore complexes disperse into the cytoplasm in metaphase. The appearance of condensin I subunits on chromosomes is indeed coincident with these large-scale changes in nuclear architecture, implying that condensin I gains access to chromosomes through partially disrupted nuclear envelopes. Moreover, careful observations of DAPI-stained and H3S10ph-positive regions indicate that C. merolae chromosomes undergo substantial structural changes from prophase through metaphase, most likely through the action of condensin I.
During the transition from prophase to metaphase, speckled signals of CENH3 in the prophase nucleus are converted into a pair of two discrete clusters, as described previously (Maruyama et al., 2007). We reason that, although the clusters of sister centromeres are separated for a short distance (∼0.5 μm), sister chromatids are held together along their arm regions at this stage. The clustering and precocious separation of sister centromeres at metaphase observed here in C. merolae are highly reminiscent of those reported in S. cerevisiae (Goshima and Yanagida, 2000; He et al., 2000; Anderson et al., 2009). Moreover, a subfraction of condensin I is found between the two CENH3 clusters (possibly corresponding to the pericentromeric region) at metaphase in C. merolae, again reminiscent of observations in S. cerevisiae (Stephens et al., 2011).
Unlike in S. cerevisiae, however, no centromere-specific sequences common to all chromosomes have been identified in C. merolae (Maruyama et al., 2008). Furthermore, the C. merolae genome encodes a simple set of conserved kinetochore components (Matsuzaki et al., 2004). Future studies of centromere structure and dynamics in this model organism will be of great interest and will undoubtedly provide fundamental insights into the evolution of centromere specification and kinetochore assembly.
Spatiotemporal regulation of condensins I and II is widely conserved among eukaryotes
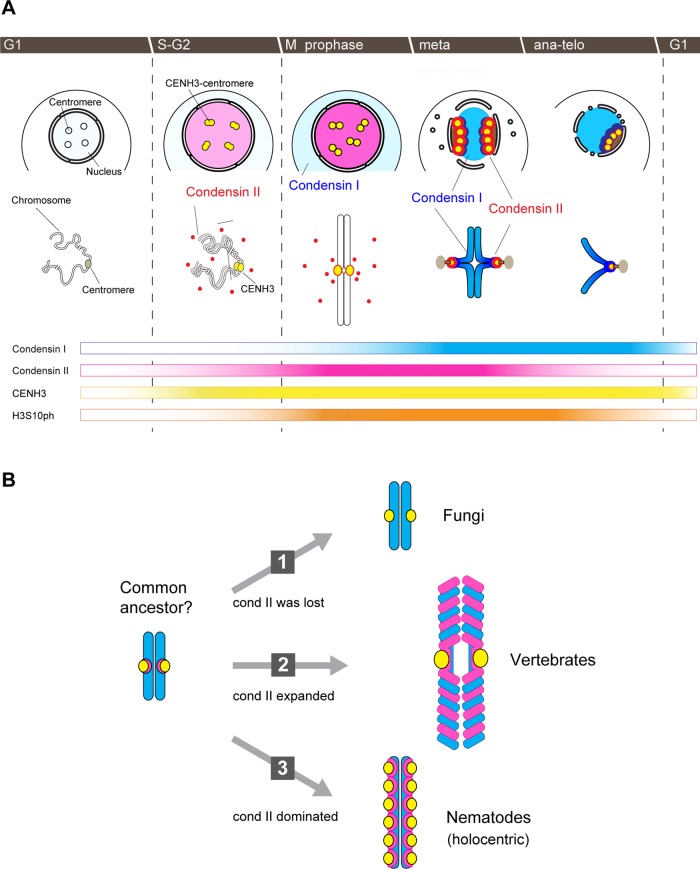
Immunofluorescence microscopy shows that condensin II subunits are detectable as speckles within the nucleus in S phase. Whereas their distribution within the nucleus from S phase through G2 phase does not overlap with CENH3, condensin II becomes enriched at centromeres and colocalized with CENH3 by metaphase. In striking contrast, immunofluorescence signals of condensin I become detectable on chromosomes only after metaphase. Together with immunoblotting data, these results imply that condensin I subunits present during S/G2 phase are dispersed in the cytoplasm (as shown in Figure 7A), although this fraction cannot be visualized under the present fixation and labeling conditions. It should be emphasized that the differential nucleocytoplasmic localization and the order of actions of condensins I and II observed here in C. merolae are surprisingly similar to those reported in animals cells, including human tissue culture cells (Ono et al., 2004, 2013; Gerlich et al., 2006), mouse oocytes (Lee et al., 2011), and Xenopus egg cell-free extracts (Shintomi and Hirano, 2011). Thus these highly characteristic behaviors of condensins are most likely to represent an evolutionarily conserved, most fundamental mechanism of chromosome assembly in both mitosis and meiosis. We propose that the sequestration of condensin I in the cytoplasm until nuclear envelope breakdown helps to specify the order of action of the two condensin complexes (i.e., condensin II first, condensin I later), thereby ensuring the two-step process of chromosome condensation (Marko, 2008; Hirano, 2012).
FIGURE 7:
Dynamics of condensins I and II in the C. merolae cell cycle and evolutionary implications. (A) Spatiotemporal localization of condensins I and II during the cell cycle of C. merolae. (Top) Nuclear and cytoplasmic distributions of CENH3, condensin I, and condensin II. (Middle) Schematic diagrams of individual chromosomes. (Bottom) Changes in the nuclear levels of four components (condensin I, condensin II, CENH3, and H3S10ph), as judged by immunofluorescence. (B) Hypothetical paths during the evolution of chromosome architecture based on the localization and functions of condensins I (blue) and II (magenta). The yellow circles indicate sister centromeres. See the text for details.
Specific contribution of condensin II to sister centromere resolution in C. merolae
The distributions of the two condensin complexes in metaphase chromosomes are clearly distinct from each other in C. merolae, as judged by both immunofluorescence microscopy and ChIP-qPCR analyses: condensin II is highly enriched at centromeres, whereas condensin I distributes broadly along chromosome arms as well as at centromeres. To our surprise, targeted gene disruption analyses show that condensin II–specific subunits (CAP-D3 and CAP-H2) are nonessential for the survival or growth of C. merolae cells under the present laboratory conditions. In the presence of a microtubule drug, however, condensin II mutants fail to resolve sister centromeres. Although this phenotype is perfectly consistent with the specific enrichment of condensin II at centromeres in metaphase, our present data do not exclude the possibility that condensin II participates in chromosomal events during interphase or in proper segregation of chromosome arms during mitosis. It is also possible that C. merolae condensin II becomes essential under diverse nonlaboratory conditions. For example, a recent genetic study demonstrated that condensin II confers tolerance for excess boron and helps to reduce the incidence of DNA double-strand breaks in A. thaliana (Sakamoto et al., 2011).
Evolutionary insights into differential use of condensins I and II
Whereas C. merolae has one of the smallest and simplest nuclear genomes among nonsymbiotic eukaryotes studied so far, its chloroplast genome contains a large number of genes, implying that C. merolae is a very ancient organism (Martin et al., 2002; Nozaki et al., 2003). It is therefore tempting to speculate that the last eukaryotic common ancestor (LECA) might have shared many structural and functional properties of condensins I and II with C. merolae (i.e., condensin I along arms and condensin II at centromeres). This hypothesis, if correct, would enable us to consider at least three different paths of eukaryotic chromosome evolution in terms of the use of the two condensin complexes (Figure 7B). In the first path, condensin II was lost during evolution, and condensin I took over the primary job of condensin II at centromeres, as observed in fungi (Sutani et al., 1999; Freeman et al., 2000). In the second path, a specific fraction of condensin II remained at centromeres, but the bulk of condensin II spread along whole chromosome arms, thereby gaining an ability to participate in their axial condensation, as observed in vertebrates (Ono et al., 2004; Shintomi and Hirano, 2011; Green et al., 2012). The third and arguably most intriguing product of evolution is found in the nematode C. elegans, in which condensin II's function became dominant over that of condensin I (Csankovszki et al., 2009). It is reasonable to hypothesize that this is because C. elegans chromosomes have a unique holocentric configuration in which numerous centromeres assemble along the entire length of chromatids. In fact, condensin II-deficient C. elegans embryos exhibit very severe defects in chromosome rigidity and centromere resolution (Stear and Roth, 2002; Moore et al., 2005). Thus we argue that the LECA's fortuitous possession of two different condensin complexes could have provided great opportunities and plasticity for the evolution of chromosome architecture and dynamics in Eukarya.
It is also of great interest to consider the very deep origin of the chromosome segregation machinery. Primitive forms of condensin complexes are widely conserved among Archaea and Bacteria. Recent studies in Bacillus subtilis show that bacterial condensin is recruited to centromere-like sequences (known as parS) and promotes chromosome segregation by compacting replicated DNA molecules (Gruber and Errington, 2009; Sullivan et al., 2009). It is of interest to note that this function of bacterial condensins at centromere-like sequences is more similar to that of condensin II than that of condensin I. Thus the most ancestral function of condensins could have been to organize and segregate centromere-like structures, a job that is performed by condensin II in many present-day eukaryotic organisms. Future studies to further clarify the similarities and differences between the two condensin complexes not only will facilitate our understanding of chromosome architecture and dynamics in general, but they also will provide a valuable hint for the question of how an early form of life might have devised a strategy to handle ever-increasing lengths of DNA molecules.
MATERIALS AND METHODS
Strains, culture conditions, and drug treatments of C. merolae
The wild-type C. merolae strain 10D-14 (Toda et al., 1998) and the uracil-auxotrophic mutant M4 (Minoda et al., 2004) were used in the present study. They were maintained by gyratory culture (130 rpm) in either 2× Allen's medium (Allen, 1959) or MA2 medium (Ohnuma et al., 2008) containing uracil (0.5 mg/ml) at pH 2.3 and 42°C under continuous light. Cell synchronization was performed as described previously (Suzuki et al., 1994). In brief, 10D cell cultures were diluted to an OD750 of 0.4 and cultured under a 12-h light–dark cycle with vigorous aeration. Alternatively, cultures of M4 and M4-derived transformants were diluted to an OD750 of 0.4 and cultured under a modified light–dark cycle (16 h in the light, 8 h in the dark, 12 h in the light, and 12 h in the dark) with vigorous aeration. Drug treatment of C. merolae was performed as described previously (Nishida et al., 2005) with minor modifications. When necessary, oryzalin was added at a final concentration of 40 μM (at 38 h in the synchronous cultures of M4 and CAP-D3-KO). MG132 was added at a final concentration of 100 μM (at 16 h in the culture of 10D; at 38 h in the cultures of M4, CAP-D3-KO, and CAP-D3-HA). Stock solutions of 100 mM oryzalin (Wako Pure Chemical Industries, Osaka, Japan) and 50 mM MG132 (Peptide Institute, Osaka, Japan) were made in dimethyl sulfoxide.
Generation and purification of antibodies
DNA fragments encoding a full or a partial amino acid sequence of PCNA (CMS101C; gene locus in Cyanidioschyzon merolae Genome Project, http://merolae.biol.s.u-tokyo.ac.jp/), SMC2 (CMG189C), SMC4 (CME029C), CAP-D2 (CMR484C), CAP-G (CMS422C), CAP-D3 (CMQ236C), and CAP-G2 (CMA089C) were PCR amplified using specific primers containing restriction sequences or the adaptor sequence for In-Fusion reaction (Supplemental Table S1). The PCR products were inserted into pQE80 expression vector (Qiagen, Valencia, CA), resulting in constructs with a six-histidine tag at their N-termini. The recombinant polypeptides of PCNA and SMC4 were purified using a HisTrap column (GE Healthcare, Piscataway, NJ), whereas those of SMC2, CAP-D2, CAP-G, CAP-D3, and CAP-G2 were purified by electroelution after SDS-PAGE. Alternatively, a synthetic peptide corresponding to the C-terminal sequence of CAP-D3 (CRRHLDGRYPSPF) was conjugated to KLH. Antisera were raised by immunizing three different species (a mouse and a rat for PCNA; mice and rabbits for SMC2; rabbits for CAP-D2, CAP-G, CAP-D3, and CAP-G2). The antiserum against SMC4 was affinity purified using a HiTrap N-hydroxysuccinimide (NHS)–activated HP column (GE Healthcare), and the rabbit antisera against SMC2, CAP-D2, CAP-G, CAP-D3, and CAP-G2 were affinity purified using Affi-Gel 15 (Bio-Rad, Hercules, CA).
Immunoprecipitation
Cells synchronized at M phase (2 × 107 cells/ml; 120 ml) were collected by centrifugation at 1500 × g for 2 min and washed once with TBS (20 mM Tris-HCl, pH 7.5, and 150 mM NaCl). The cell pellet was frozen in liquid nitrogen and then resuspended and lysed in 1.2 ml of TBS containing a protease inhibitor cocktail (1× Complete [Roche, Indianapolis, IN]). The lysate was centrifuged at 20,000 × g for 5 min, and the supernatant was supplemented with Triton X-100 at a final concentration of 0.1%. For immunoprecipitations, 50 μl of the supernatant was mixed with either 5 μg of affinity-purified antibodies or 1 μg of anti-HA (clone 3F10) on ice for 1 h and then incubated with protein A or protein G Sepharose (GE Healthcare) on ice for 1 h. After washed with TBS containing 0.1% Triton X-100, the immunoprecipitates were eluted with 2× SDS sample buffer at 100°C for 2 min. Rabbit immunoglobulin G (IgG; Sigma-Aldrich, St. Louis, MO) and rat monoclonal IgG (clone 1H5; MBL, Woburn, MA) were used as negative controls.
ChIP-qPCR analysis
Cells were fixed with 1% formaldehyde at 30°C for 10 min and quenched with 250 mM glycine on ice for 5 min. The cells were then pelleted and washed once with TBS. The cell pellet was resuspended in micrococcal nuclease (MNase) digestion buffer (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40, 1× Complete [EDTA-free; Roche], 1 mM dithiothreitol, and 5 mM CaCl2) at concentration of 2 × 106 cells/μl and was digested with 10,000 gels units/ml of MNase (New England BioLabs, Ipswich, MA) at 37°C for 15 min. EDTA was then added at a final concentration of 5 mM on ice to stop the reaction. The samples were centrifuged at 20,000 × g for 5 min, and the pellet was suspended in lysis buffer (LB; 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.1% SDS, and 1× Complete) at a concentration of 1 × 106 cells/μl. The cells were broken in two steps. The first step was performed using glass beads (425–600 μm in diameter; Sigma-Aldrich) and a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan) with the following parameters: vortexed at 2700 rpm for 1 min; chilled at 0°C for 1 min by the cooling circulator MBC-100 (Yasui Kikai); 20 cycles. The second step was performed by sonication (Sonifier 250; Branson, Danbury, CT) with the following parameters: output 15, duty cycle 10%, sonication for 15 s, chilling for 30 s, 10 cycles. The lysate was centrifuged at 20,000 × g for 5 min, and the resultant supernatant was used as the input for ChIP analyses. For ChIP of condensin subunits, the same amounts of antibodies and the same volumes of input materials were used as for the conventional immunoprecipitations described. In addition, 125 ng of an anti-CENH3 antibody (Maruyama et al., 2007) and control rabbit IgG (Sigma-Aldrich) were used. The beads were washed once with 500 μl of LB, five times with RIPA buffer (50 mM Tris-HCl, pH 7.5, 0.5 M LiCl, 1 mM EDTA, 0.5% Na deoxycholate, and 1% NP-40), and finally once with 50TE (50 mM Tris-HCl, pH 7.5, and 10 mM EDTA). To elute the immunoprecipitated materials, the beads were resuspended in 200 μl of 50TE containing 1% SDS at 65°C for 20 min. The eluates were further incubated at 65°C overnight to reverse cross-linking. After treatment with RNaseA (Qiagen) and proteinase K (Sigma-Aldrich), DNA was purified by extracting with phenol:chloroform:isoamylalcohol, 25:24:1, followed by ethanol precipitation with 4 μg of glycogen. In addition, the DNA pellets were dissolved in TE and were purified through QIAquick columns (Qiagen). The DNA was reconstituted with 100 μl of elution buffer (10 mM Tris-HCl, pH 8) and then used for quantitative real-time PCR analysis. Real-time PCR was performed using a CFX96 Real-Time PCR Detection System and C1000 Thermal Cycler (Bio-Rad) in a 20-μl reaction mixture containing 1 μl DNA, 0.5 nM primers (Supplemental Table S1), and 10 μl SsoFast EvaGreen Super Mix (Bio-Rad). Standard curves were constructed using serially diluted solutions of DNA isolated from C. merolae cells and the relevant sets of primers, and the recovery of each DNA fragment relative to the input DNA was estimated.
Acknowledgments
We are grateful to F. Yagisawa and other members of the Kuroiwa lab (Rikkyo University, Tokyo, Japan) for providing us with a calnexin antibody and technical advice on experiments using C. merolae. We are greatly indebted to Y. Kanesaki (Tokyo University of Agriculture, Tokyo, Japan) and S. Imamura (Tokyo Institute of Technology, Tokyo, Japan) for sharing the information on C. merolae centromere sequences identified by ChIP-chip analysis. R. Nakato, Y. Kato, M. Komata, and K. Shirahige (University of Tokyo, Tokyo, Japan) gave us technical advice on ChIP. We also thank A. Matsuura for excellent technical assistance and members of the Hirano lab for critically reading the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research (A) (to K.T.) and by a Grant-in-Aid for Specially Promoted Research (to T.H.) from the Japan Society for the Promotion of Science. T.F. was a RIKEN Special Postdoctoral Researcher.
Note added in proof. Additional information regarding the newly developed technique for gene targeting in C. merolae has been published by Fujiwara et al. (2013).
Abbreviations used:
- CENH3
centromere-specific histone H3 variant
- ChIP
chromatin immunoprecipitation
- H3S10ph
phosphorylated histone H3 at serine 10
- PCNA
proliferating nuclear antigen
- qPCR
quantitative PCR
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-04-0208) on June 19, 2013.
*Present address: Center of Frontier Research, National Institute of Genetics, Mishima, Shizuoka 411-8540, Japan.
REFERENCES
- Allen MB. Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch Microbiol. 1959;32:270–277. doi: 10.1007/BF00409348. [DOI] [PubMed] [Google Scholar]
- Anderson M, Haase J, Yeh E, Bloom K. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris JP, Blobel G. Yeast nuclear envelope proteins cross react with an antibody against mammalian pore complex proteins. J Cell Biol. 1989;108:2059–2067. doi: 10.1083/jcb.108.6.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis JE, Celis A. Cell cycle-dependent variations in the distribution of the nuclear protein cyclin proliferating cell nuclear antigen in cultured cells: subdivision of S phase. Proc Natl Acad Sci USA. 1985;82:3262–3266. doi: 10.1073/pnas.82.10.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, et al. Three distinct condensin complexes control C. elegans chromosome dynamics. Curr Biol. 2009;19:9–19. doi: 10.1016/j.cub.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S, Haering CH. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 2011;21:552–559. doi: 10.1016/j.tcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Freeman L, Aragon-Alcaide L, Strunnikov AV. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Ohnuma M, Yoshida M, Kuroiwa T, Hirano T. Gene targeting in the red alga Cyanidioschyzon merolae: single- and multi-copy insertion using authentic and chimeric selection markers. PLoS ONE. 2013. (in press) [DOI] [PMC free article] [PubMed]
- Fujiwara T, et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16:59–72. doi: 10.1093/dnares/dsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D, Hirota T, Koch B, Peters J-M, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in living cells. Curr Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Green LC, et al. Contrasting roles of condensin I and II in mitotic chromosome formation. J Cell Sci. 2012;125:1591–1604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- He X, Asthana S, Sorger PK. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: organizing and segregating the genome. Curr Biol. 2005;15:R265–R275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659–1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- Imamura S, et al. Nitrate assimilatory genes and their transcriptional regulation in a unicellular red alga Cyanidioschyzon merolae: genetic evidence for nitrite reduction by a sulfite reductase-like enzyme. Plant Cell Physiol. 2010;51:707–717. doi: 10.1093/pcp/pcq043. [DOI] [PubMed] [Google Scholar]
- Imamura S, Kanesaki Y, Ohnuma M, Inouye T, Sekine Y, Fujiwara T, Kuroiwa T, Tanaka K. R2R3-type MYB transcription factor, CmMYB1, is a central nitrogen assimilation regulator in Cyanidioschyzon merolae. Proc Natl Acad Sci USA. 2009;106:12548–12553. doi: 10.1073/pnas.0902790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto Y, Fujiwara T, Yoshida Y, Kuroiwa H, Maruyama S, Kuroiwa T. Division of cell nuclei, mitochondria, plastids, and microbodies mediated by mitotic spindle poles in the primitive red alga Cyanidioschyzon merolae. Protoplasma. 2010;241:63–74. doi: 10.1007/s00709-010-0107-y. [DOI] [PubMed] [Google Scholar]
- Imoto Y, Yoshida Y, Yagisawa F, Kuroiwa H, Kuroiwa T. The cell cycle, including the mitotic cycle and organelle division cycles, as revealed by cytological observations. J Electron Microsc. 2011;60(Suppl 1):S117–S136. doi: 10.1093/jmicro/dfr034. [DOI] [PubMed] [Google Scholar]
- Lee J, Ogushi S, Saitou M, Hirano T. Condensins I and II are essential for construction of bivalent chromosomes in mouse oocytes. Mol Biol Cell. 2011;22:3465–3477. doi: 10.1091/mbc.E11-05-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-M, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AAM, Meinke D. Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 2002;29:405–415. doi: 10.1046/j.1365-313x.2002.01224.x. [DOI] [PubMed] [Google Scholar]
- Marko JF. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008;16:469–497. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99:12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S, Kuroiwa H, Miyagishima S-Y, Tanaka K, Kuroiwa T. Centromere dynamics in the primitive red alga Cyanidioschyzon merolae. Plant J. 2007;49:1122–1129. doi: 10.1111/j.1365-313X.2006.03024.x. [DOI] [PubMed] [Google Scholar]
- Maruyama S, Matsuzaki M, Kuroiwa H, Miyagishima S-Y, Tanaka K, Kuroiwa T, Nozaki H. Centromere structures highlighted by the 100%-complete Cyanidioschyzon merolae genome. Plant Signal Behav. 2008;3:140–141. doi: 10.4161/psb.3.2.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Minoda A, Sakagami R, Yagisawa F, Kuroiwa T, Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004;45:667–671. doi: 10.1093/pcp/pch087. [DOI] [PubMed] [Google Scholar]
- Moore LL, Stanvitch G, Roth MB, Rosen D. HCP-6/CENP-C promotes the prophase timing of centromere resolution by enabling the centromere association of HCP-6 in Caenorhabditis elegans. Mol Cell Biol. 2005;25:2583–2592. doi: 10.1128/MCB.25.7.2583-2592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K, Yagisawa F, Kuroiwa H, Nagata T, Kuroiwa T. Cell cycle-regulated, microtubule-independent organelle division in Cyanidioschyzon merolae. Mol Biol Cell. 2005;16:2493–2502. doi: 10.1091/mbc.E05-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H, Ohta N, Matsuzaki M, Misumi O, Kuroiwa T. Phylogeny of plastids based on cladistic analysis of gene loss inferred from complete plastid genome sequences. J Mol Evol. 2003;57:377–382. doi: 10.1007/s00239-003-2486-6. [DOI] [PubMed] [Google Scholar]
- Ohnuma M, Yokoyama T, Inouye T, Sekine Y, Tanaka K. Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2008;49:117–120. doi: 10.1093/pcp/pcm157. [DOI] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector D, Hirano T. Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Ono T, Yamashita D, Hirano T. Condensin II initiates sister chromatid resolution during S phase. J Cell Biol. 2013;200:429–441. doi: 10.1083/jcb.201208008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1981;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Inui YT, Uraguchi S, Yoshizumi T, Matsunaga S, Mastui M, Umeda M, Fukui K, Fujiwara T. Condensin II alleviates DNA damage and is essential for tolerance of boron overload stress in Arabidopsis. Plant Cell. 2011;23:3533–3546. doi: 10.1105/tpc.111.086314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011;25:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui NU, Rusyniak S, Hasenkampf CA, Riggs CD. Disruption of the Arabidopsis SMC4 gene, AtCAP-C, compromises gametogenesis and embryogenesis. Planta. 2006;223:990–997. doi: 10.1007/s00425-006-0234-z. [DOI] [PubMed] [Google Scholar]
- Stear JH, Roth MB. Characterization of HCP-6, a C. elegans protein required to prevent chromosome twisting and merotelic attachment. Genes Dev. 2002;16:1498–1508. doi: 10.1101/gad.989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Ehara T, Osafune T, Kuroiwa H, Kawano S, Kuroiwa T. Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur J Cell Biol. 1994;63:280–288. [PubMed] [Google Scholar]
- Toda T, Takano H, Miyagishima S, Kuroiwa H, Kuroiwa T. Characterization of a chloroplast isoform of serine acetyltransferase from the thermo-acidiphilic red alga Cyanidioschyzon merolae. Biochim Biophys Acta. 1998;1403:72–84. doi: 10.1016/s0167-4889(98)00031-7. [DOI] [PubMed] [Google Scholar]
- Yagisawa F, Fujiwara T, Kuroiwa H, Nishida K, Imoto Y, Kuroiwa T. Mitotic inheritance of endoplasmic reticulum in the primitive red alga Cyanidioschyzon merolae. Protoplasma. 2012;249:1129–1135. doi: 10.1007/s00709-011-0359-1. [DOI] [PubMed] [Google Scholar]
- Yeong FM, et al. Identification of a subunit of a novel kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr Biol. 2003;13:2058–2064. doi: 10.1016/j.cub.2003.10.032. [DOI] [PubMed] [Google Scholar]