RanGTP positions the mitotic spindle by controlling cortical LGN and NuMA localization and enhancing astral microtubule elongation. Spindle miscentering caused by the Ran-pathway inhibitor importazole is rescued by overexpression of CLASP1, which functions independently to stabilize astral microtubule interactions with the cell cortex.
Abstract
Accurate positioning of the mitotic spindle is critical to ensure proper distribution of chromosomes during cell division. The small GTPase Ran, which regulates a variety of processes throughout the cell cycle, including interphase nucleocytoplasmic transport and mitotic spindle assembly, was recently shown to also control spindle alignment. Ran is required for the correct cortical localization of LGN and nuclear-mitotic apparatus protein (NuMA), proteins that generate pulling forces on astral microtubules (MTs) through cytoplasmic dynein. Here we use importazole, a small-molecule inhibitor of RanGTP/importin-β function, to study the role of Ran in spindle positioning in human cells. We find that importazole treatment results in defects in astral MT dynamics, as well as in mislocalization of LGN and NuMA, leading to misoriented spindles. Of interest, importazole-induced spindle-centering defects can be rescued by nocodazole treatment, which depolymerizes astral MTs, or by overexpression of CLASP1, which does not restore proper LGN and NuMA localization but stabilizes astral MT interactions with the cortex. Together our data suggest a model for mitotic spindle positioning in which RanGTP and CLASP1 cooperate to align the spindle along the long axis of the dividing cell.
INTRODUCTION
All organisms require proper regulation of cell division to maintain the integrity of their genetic information. In most eukaryotic cells, the position of the cleavage plane is predicted by the location of the metaphase plate (Rappaport, 1971; Albertson, 1984; Strome, 1993; Glotzer, 1997; Grill and Hyman, 2005), and failure to properly position the mitotic spindle can have deleterious consequences, including developmental defects, cell death, aneuploidy, and cancer (O'Connell and Khodjakov, 2007; Gonczy, 2008). Control of spindle positioning is achieved through interactions between the cell cortex and the astral microtubules (MTs), which can either exert pushing forces on the mitotic spindle through MT polymerization or apply pulling forces through MT depolymerization or the activity of motor proteins (Pearson and Bloom, 2004; Siller and Doe, 2009).
Control of mitotic spindle positioning has been studied primarily in organisms that undergo asymmetric cell divisions, such as the Caenorhabditis elegans zygote and Drosophila melanogaster neuroblasts. In these systems, the mitotic spindle is oriented by pulling forces exerted on the astral MTs by dynein/dynactin complexes that are linked to the cell cortex by an evolutionarily conserved tripartite protein complex (Gα/GPR-1/2/Lin-5 in worms and Gα-Pins-Mud in flies; reviewed in Gonczy, 2008; Siller and Doe, 2009; Stevermann and Liakopoulos, 2012; McNally, 2013). A similar mechanism operates to position the spindle in symmetrically dividing mammalian cells, where the membrane-bound, receptor-independent Gαi protein links the dynein/dynactin complex to the cortex through LGN and nuclear-mitotic apparatus protein (NuMA; Du and Macara, 2004).
Whereas key players that position the mammalian mitotic spindle have been identified, less is known about their regulation. Extrinsic cues from the extracellular matrix are known to contribute to spindle orientation (Thery et al., 2005; Toyoshima and Nishida, 2007), but recent work has also identified roles for the RanGTP gradient and CLASP1 in the regulation of spindle positioning (Samora et al., 2011; Kiyomitsu and Cheeseman, 2012). Ran is a Ras-related small GTPase responsible for regulating a variety of processes throughout the cell cycle, including nucleocytoplasmic transport, mitotic spindle assembly, postmitotic nuclear envelope formation, nuclear pore complex biogenesis, and protein ubiquitylation (Harel et al., 2003; Walther et al., 2003; Clarke and Zhang, 2004; Goodman and Zheng, 2006; Terry et al., 2007; Dishinger et al., 2010). Throughout the cell cycle, Ran's guanine nucleotide exchange factor, RCC1, is bound to chromatin, leading to a highly localized production of RanGTP. In mitosis, this generates a gradient of RanGTP centered around chromosomes, which in turn leads to the release and activation of spindle assembly factors from nuclear transport factors, such as importin-β, inducing spindle assembly with high spatial specificity (Kalab et al., 2002, 2006). One Ran/importin-β–regulated factor is NuMA, a large coiled-coil protein that organizes the spindle poles during mitosis and functions in spindle positioning (Gaglio et al., 1995; Gordon et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Joukov et al., 2006; Wong et al., 2006).
Members of the CLASP family of proteins are involved in multiple MT-dependent processes, including Golgi complex organization and trafficking, kinetochore fiber dynamics and flux, mitotic spindle assembly, and the stabilization of MT plus ends near the cortex (Maiato et al., 2005; Mimori-Kiyosue et al., 2005; Miller et al., 2009; Reis et al., 2009; Patel et al., 2012). CLASP proteins regulate MT dynamics by promoting MT rescue and suppressing MT catastrophe through the activity of conserved TOG domains, which recruit tubulin dimers to MT plus ends (reviewed in Al-Bassam and Chang, 2011). Recent work showed that CLASP1 activity is required for proper positioning of the mitotic spindle in both the C. elegans embryo and mammalian cells, but the relationship between the CLASP1 and RanGTP regulated spindle-positioning pathways is unclear (Samora et al., 2011; Espiritu et al., 2012).
Studying the function of Ran in mitosis has been complicated by its multiple essential roles throughout the cell cycle, limiting the usefulness of traditional techniques such as genetics and RNA interference. To overcome this problem, we made use of importazole, a small-molecule inhibitor of RanGTP/importin-β activity, which allowed us to investigate Ran pathway control of spindle positioning with high temporal precision and minimal manipulation of cellular conditions (Soderholm et al., 2011). Here we provide evidence that in addition to regulating the cortical localization of NuMA and LGN, RanGTP enhances astral MT dynamics and functions in conjunction with CLASP1 to promote proper spindle positioning by increasing astral MT interactions with the cell cortex.
RESULTS
Importazole disrupts importin-β–mediated spindle positioning
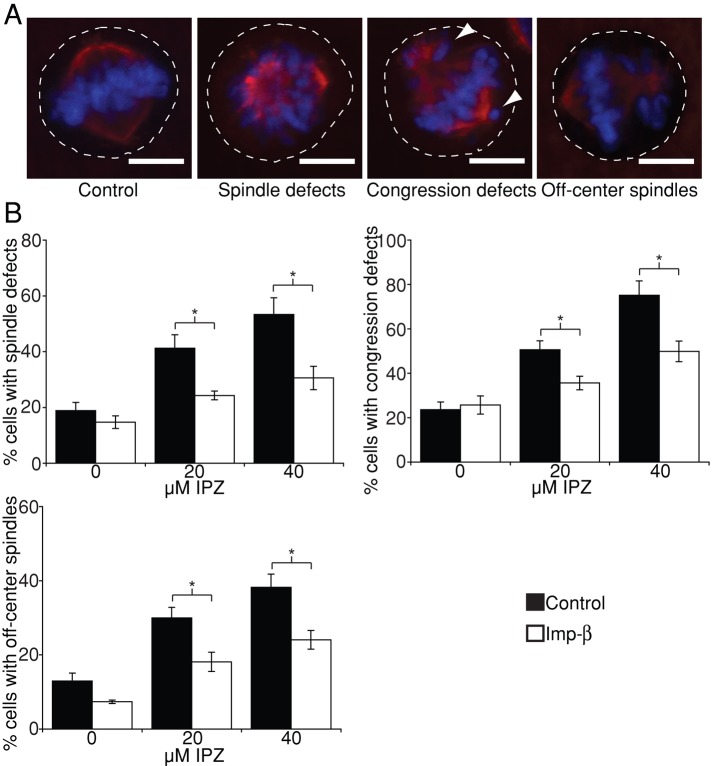
We previously identified importazole as a specific inhibitor of RanGTP/importin-β function (Soderholm et al., 2011). Importazole treatment induces several mitotic phenotypes, often leading to multiple mitotic defects within the same cell (Figure 1A). These include defects in spindle assembly, chromosome congression, and spindle centering. Whereas in control cells >85% of the mitotic spindles were accurately centered within the cell, upon importazole treatment, up to 38% of the cells displayed a spindle-centering phenotype in which the center of the mitotic spindle was visually observed to be closer to the cortex than the center of the cell. To test whether these defects were due to a specific inhibition of the RanGTP/importin-β pathway, we examined whether importin-β overexpression could rescue the importazole-induced mitotic defects. HeLa cells were transiently transfected with either importin-β–yellow fluorescent protein (YFP) or YFP alone and subsequently treated with the solvent control dimethyl sulfoxide (DMSO) or 20 μM or 40 μM importazole and then fixed 1 h later. Mitotic cells displaying visible YFP fluorescence were then scored for the presence of mitotic defects (Figure 1B). Under both control and importin-β overexpression conditions, importazole treatment resulted in a dose-dependent increase in mitotic spindle assembly, chromosome congression, and spindle-centering defects. The number of mitotic cells displaying each class of mitotic defect, however, was significantly reduced in importazole-treated cells expressing importin-β–YFP (Figure 1B), indicating that these defects result specifically from importazole inhibition of the RanGTP/importin-β pathway.
FIGURE 1:
Importazole specifically disrupts importin-β–mediated spindle positioning. (A) Representative images of DMSO-treated (control) and 40 μM importazole-treated cells displaying importazole-associated mitotic phenotypes in spindle assembly, chromosome congression, and spindle centering. Dashed white lines indicate the cell cortex, and arrowheads point to lagging chromosomes. Note that multiple mitotic phenotypes often occur in the same cell. (B) Asynchronously growing HeLa cells expressing YFP–importin-β or YFP alone (control) were treated with DMSO, 20 μM, or 40 μM importazole for 1 h, and their mitotic defects were quantified as a percentage of total mitotic cells. Note that importin-β overexpression partially rescues importazole-induced defects. Scale bars, 10 μm. In B, n = 5, and 100 metaphase cells were counted per condition. Bars, SE. Asterisks denote statistical significance (p < 0.05).
We next asked whether importazole could disrupt spindle positioning in cells with preformed metaphase spindles. HeLa cells were treated with 10 μM MG132 for 3 h to arrest cells in metaphase. DMSO or 40 μM importazole was added during the last 30 min of MG132 treatment, after which cells were washed twice with clean media before an additional 30 min of DMSO or importazole treatment before fixation. Of interest, MG132 metaphase arrest resulted in a higher percentage of cells displaying spindle defects upon importazole treatment, as well as the appearance of an additional importazole phenotype in which two or more spindle structures were observed within the same cell (Supplemental Figure S1, A and B). By contrast, analysis of mitotic defects in MG132-treated cells revealed a similar percentage of mitotic cells displaying a defect in spindle centering compared with nonarrested cells, indicating that Ran pathway control of spindle position is not dependent on assembly of the spindle (Supplemental Figure S1A).
Importazole impairs localization of cortical factors LGN and NuMA
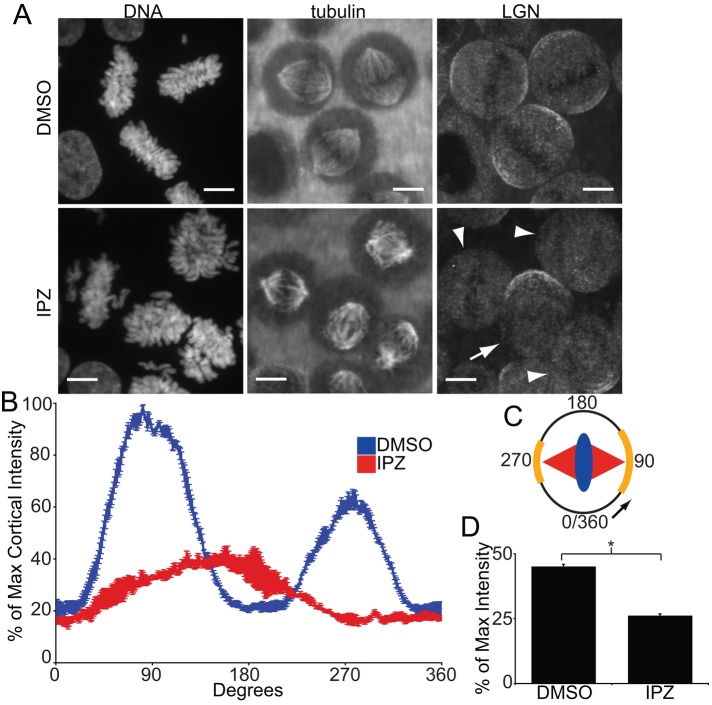
In mammalian cells, the position of the mitotic spindle is determined by pulling forces on the astral MTs exerted by dynein/dynactin complexes (Pearson and Bloom, 2004; Siller and Doe, 2009). These complexes are linked to Gαi at the cortical membrane by LGN and NuMA (Du and Macara, 2004). Previous work established that deactivation of the Ran pathway via transfection of the dominant-negative RanT24N mutant results in a mislocalization of green fluorescent protein (GFP)–LGN along the cortex (Kiyomitsu and Cheeseman, 2012). To test how the Ran/importin-β pathway regulates the localization of cortical positioning factors under endogenous protein conditions, we first observed mitotic localization of LGN in response to importazole treatment. Because the localization of LGN changes during mitosis (Kiyomitsu and Cheeseman, 2012), we synchronized HeLa cells using a double thymidine block and monitored LGN staining specifically at metaphase, 9 h after release from thymidine treatment. Synchronized cells were treated with DMSO or 40 μM importazole 1 h before fixation. In DMSO-treated cells, LGN localized to the cell cortex in a pattern of two arcs adjacent to the two poles of the mitotic spindle, but this cortical staining pattern of LGN was disrupted in importazole-treated cells (Figure 2A). The degree to which cortical LGN staining was disrupted in individual cells corresponded to the severity of importazole defects observed in the cell, as cells that displayed relatively minor defects (Figure 2A, arrow) had minor disruptions in LGN staining, whereas cells with more severe mitotic defects (Figure 2A, arrowheads) showed an almost complete lack of cortical LGN. In addition, in many importazole-treated cells with an observable spindle-miscentering phenotype, LGN was still present at the cortex but localized to a single arc in line with the metaphase plate (Supplemental Figure S2).
FIGURE 2:
Importazole impairs localization of cortical LGN. (A) DNA, tubulin, and LGN localization in synchronized mitotic HeLa cells treated with DMSO or 40 μM importazole for 1 h. LGN in DMSO-treated cells localized to two arcs along the cortex in line with the axis of the mitotic spindle. The cortical staining pattern of LGN was disrupted in importazole-treated cells, with individual cells displaying more severe importazole phenotypes showing a more complete disruption of LGN staining (arrowheads) than cells displaying relatively minor importazole phenotypes (arrow). (B) Quantification of cortical LGN staining in synchronized metaphase HeLa cells treated with DMSO or 40 μM importazole for 1 h. LGN fluorescence intensity was measured as percentage of maximum intensity starting at the cortex in line with the metaphase plate and proceeding in the direction of the more prominent arc of cortical staining, as diagrammed (C). DMSO-treated cells displayed two arcs of cortical LGN staining at 90 and 270°, but importazole-treated cells displayed a single, smaller arc of LGN staining at ∼180°. (D) Mean cortical LGN fluorescence as a percentage of maximum intensity in DMSO- and importazole-treated metaphase cells. Scale bars, 10 μm. For B and D, n = 3, and 40 cells were measured per condition. Bars, SE. Asterisk denotes statistical significance (p < 0.001).
To quantify the role of the Ran pathway in LGN localization, we measured the cortical intensity of LGN in DMSO- and importazole-treated mitotic cells starting at the position closest to the metaphase plate and moving in the direction of the wider arc of LGN staining (Figure 2C). Quantification of DMSO-treated cells revealed two large peaks of LGN localization at ∼90 and 270º, corresponding to the long axis of the mitotic spindle (Figure 2B). Quantification of importazole treated cells, however, revealed a single, smaller LGN peak at ∼180º, corresponding to the position of the metaphase plate. These results suggest that the Ran/importin-β pathway regulates the correct cortical localization pattern of LGN during mitosis. Quantitative analysis of LGN staining revealed a second importazole-induced phenotype, in which the total mean fluorescence intensity of LGN across the entire cortex was reduced in importazole-treated cells compared with the DMSO control (Figure 2D). Taken together, these results show that Ran/importin-β activity is required both for delivery of LGN to the cortex, as well as for its proper pattern of localization at the cortex.
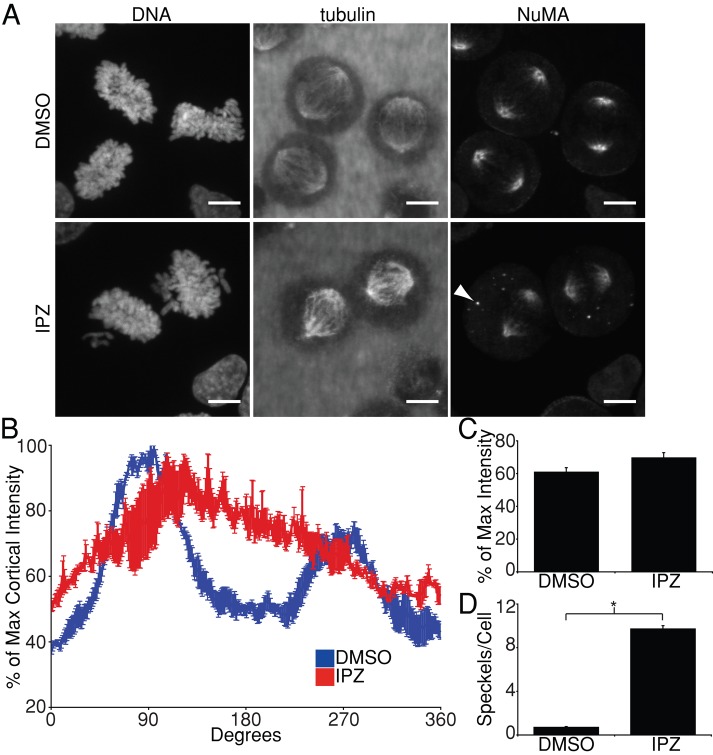
The cortical positioning factor NuMA was a good candidate as a mediator of Ran pathway–regulated control of spindle positioning due to its previously characterized role as an RanGTP-regulated mitotic importin-β cargo (Nachury et al., 2001; Wiese et al., 2001; Joukov et al., 2006; Wong et al., 2006). To test whether the RanGTP gradient regulates spindle positioning through NuMA, we first observed the endogenous localization of NuMA in synchronized mitotic HeLa cells treated with DMSO or 40 μM importazole. In DMSO-treated cells, NuMA localized to the spindle poles as previously described (Lydersen and Pettijohn, 1980; Kalab and Heald, 2008; Haren et al., 2009), but it also localized to the cell cortex in a pattern of two arcs in line with the spindle long axis, similar to what was observed for LGN (Figure 3A). In importazole-treated cells, NuMA still localized to the spindle poles, but its cortical localization was disrupted in a manner similar to LGN (Figure 3A). Similar to what was observed for LGN, quantification of cortical NuMA in DMSO-treated cells revealed two large peaks of NuMA staining in line with the long axis of the mitotic spindle, but this pattern of cortical NuMA localization was disrupted by importazole treatment (Figure 3B). In contrast to LGN, however, the mean fluorescence intensity of NuMA across the entire cortex was not reduced in importazole-treated cells, suggesting that Ran/importin-β function is not essential to bring NuMA to the cortex but is critical for its correct cortical localization (Figure 3C). In addition, we observed that NuMA formed cytoplasmic foci in response to importazole treatment (Figure 3, A, arrowhead, and D). Although the significance of the importazole-induced cytoplasmic NuMA foci is unclear, these data further support a role for the Ran pathway in the regulation of NuMA localization.
FIGURE 3:
Importazole impairs localization of cortical NuMA. (A) DNA, tubulin, and NuMA localization in synchronized mitotic HeLa cells treated with DMSO or 40 μM importazole for 1 h. NuMA in DMSO-treated cells localized to spindle poles, as well as to two arcs along the cortex in line with the long axis of the mitotic spindle, similar to the cortical staining pattern of LGN. NuMA's cortical localization was disrupted in importazole-treated cells. Cytoplasmic NuMA foci also appeared in importazole-treated cells (arrowhead). (B) Quantification of cortical NuMA staining in synchronized metaphase HeLa cells treated with DMSO or 40 μM importazole was performed as described in Figure 2B. (C) Mean cortical NuMA fluorescence as percentage of maximum intensity in DMSO and importazole-treated metaphase cells. (D) Quantification of the mean number of cytoplasmic NuMA foci observed per cell in DMSO- and importazole-treated cells. Scale bars, 10 μm. For B and C, n = 3, and 40 cells were measured per condition; for D, n = 3, and 20 cells were measured per condition. Bars, SE. Asterisk denotes statistical significance (p < 0.001).
It was recently shown that the RanGTP/importin-β pathway regulates the stability of some of its mitotic targets (Song and Rape, 2010). To determine whether importazole treatment affects the stability of LGN and NuMA, we measured the relative abundance of LGN and NuMA upon DMSO and importazole treatment in mitotic HeLa cells by quantitative Western blot. Importazole treatment did not result in a detectable decrease in the stability of either protein, suggesting that Ran and importin-β do not regulate spindle positioning through a mechanism of selective protein degradation (Supplemental Figure S3).
Importazole causes astral MT-dependent spindle movement
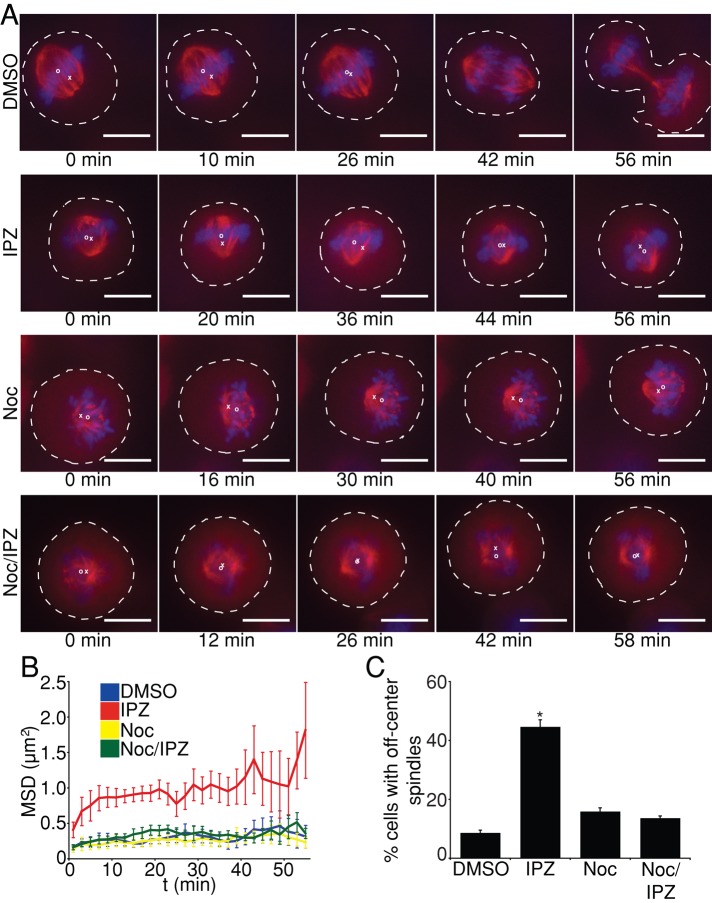
To better characterize the importazole-induced spindle-miscentering phenotype, we performed live-cell time-lapse fluorescence microscopy on mitotic HeLa cells stably expressing GFP-tubulin and mCherry-H2B. Cells were treated with a DMSO solvent control or 40 μM importazole, and early metaphase cells were imaged every 2 min for a total of 60 min starting 10 min after treatment. Spindles in DMSO-treated cells aligned with the long axis of the cell and remained close to this position before progressing through anaphase (Figure 4A and Supplemental Movie S1). In contrast, spindles in importazole-treated cells did not properly maintain their position near the middle of the cell but instead appeared to move throughout the cytoplasm in a random manner (Figure 4A and Supplemental Movie S2). Of importance, importazole-treated cells with miscentered spindles were sometimes capable of progressing through anaphase, suggesting that the spindle-movement phenotype produced by importazole did not result from a general disruption of cell function (Supplemental Movie S3). We measured the movement of the center of spindles corrected for the position of the center of the cell and calculated the average mean square displacement (MSD) of spindles in DMSO- and importazole-treated cells. MSD analysis revealed that spindles in importazole-treated cells traveled throughout a much larger area of the cytoplasm and were much less constrained compared with DMSO-treated cells (Figure 4B). In addition, importazole treatment resulted in an apparent increase in the mean speed of spindle movement, although the statistical significance of this increase could not be established due to the small number of cells measured (Supplemental Figure S4A).
FIGURE 4:
Importazole causes astral MT-dependent spindle movement. (A) Selected frames from 60-min movies of HeLa cells stably expressing GFP-tubulin and mCherry-H2B treated with DMSO, 40 μM importazole, 20 nM nocodazole, or 20 nM nocodazole and 40 μM importazole. Spindles in DMSO-treated cells move to the cell center before anaphase, whereas spindles in importazole-treated cells move about the cytoplasm. Spindles in nocodazole- and nocodazole/importazole-treated cells remained close to the center of the cell. (B) Mean square displacement as a function of time measured for mitotic spindles from GFP-tubulin– and mCherry-H2B–expressing HeLa cells under the indicated treatment conditions. (C) Asynchronously growing HeLa cells were treated with DMSO, 40 μM importazole, 20 nM nocodazole, or 20 nM nocodazole and 40 μM importazole for 1 h, and cells displaying spindle-centering defects were quantified as a percentage of total mitotic cells. Dashed white lines indicate the cell cortex, white Xs indicate the cell center, and white Os indicate the spindle center. Scale bars, 10 μm. Spindles from six cells were analyzed for each condition in B. For C, n = 3, and 100 cells were counted per condition. Bars, SE. Asterisk denotes statistical significance from all other conditions (p < 0.001).
To test the possibility that importazole-induced spindle movement resulted from aberrant astral MTs, we treated HeLa cells with a low dose (20 nM) of nocodazole, which depolymerizes astral MTs (Thery et al., 2005). Disruption of astral MTs upon nocodazole treatment was confirmed via live-cell fluorescence microscopy of HeLa cells stably expressing the MT plus end–binding protein EB3 (Supplemental Movie S4). Strikingly, spindles in GFP-tubulin– and mCherry-H2B–expressing HeLa cells treated with either a low dose of nocodazole alone or nocodazole in combination with importazole remained close to the center of the cell and did not move about the cytoplasm (Figure 4A and Supplemental Movies S5 and S6). The calculated MSD and spindle movement speed of cells treated with low nocodazole alone or both nocodazole and importazole was comparable to that of DMSO-treated cells (Figure 4B and Supplemental Figure S4A). In addition, when treated with both importazole and nocodazole, the percentage of mitotic HeLa cells displaying spindle-centering defects was rescued to near-wild-type levels (Figure 4C). Nocodazole treatment did not rescue importazole-induced defects, however, in either spindle assembly or chromosome congression (Supplemental Figure S4B). Thus destabilization of astral MTs itself does not cause aberrant mitotic spindle movement or positioning, suggesting that disruption of Ran pathway function leads to the misregulation of the connection between astral MTs and the cell cortex, which in turn results in actively moving mitotic spindles.
RanGTP and CLASP1 independently promote spindle positioning through astral MTs
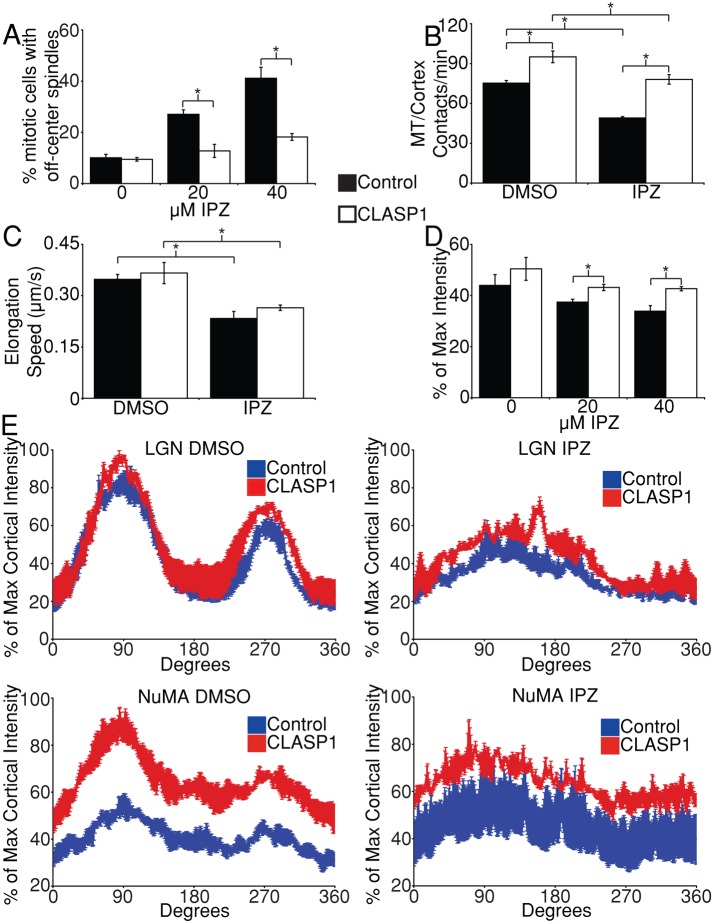
Recently, a role for CLASP1 in mitotic spindle positioning has been demonstrated in which CLASP1 helps to initiate and stabilize end-on astral MT attachments with the cell cortex (Samora et al., 2011). We therefore sought to elucidate the interplay between CLASP1 and the Ran pathway in spindle positioning. We overexpressed GFP-CLASP1 in HeLa cells and quantified the frequency of mitotic defects in DMSO- and importazole-treated cells. CLASP1 overexpression had little to no effect on importazole-induced spindle assembly and chromosome congression defects (Supplemental Figure S5A). CLASP1 overexpression, however, significantly reduced the number of mitotic cells displaying defects in spindle centering, demonstrating that increased CLASP1 activity is sufficient to rescue spindle positioning in the absence of proper Ran pathway function (Figure 5A).
FIGURE 5:
RanGTP and CLASP1 independently promote spindle positioning through astral MTs. (A) Quantification of spindle-centering defects in asynchronously growing HeLa cells expressing either GFP-CLASP1 or YFP alone (control) treated with DMSO, 20 μM, or 40 μM importazole for 1 h. (B) The mean number of astral MT/cortex contacts per minute measured in asynchronous HeLa cells stably expressing GFP-EB3 transfected with BFP-CLASP1 or BFP alone (control) and treated with DMSO or 40 μM importazole 10 min before live-cell imaging for 2 min. (C) Mean astral MT elongation speed determined in EB3 HeLa cells transfected with BFP-CLASP1 and treated with DMSO or 40 μM importazole as in B. (D) Mean cortical LGN fluorescence as a percentage of maximum intensity in synchronized metaphase HeLa cells expressing GFP-CLASP1 or YFP alone (control) treated with DMSO, 20 μM, or 40 μM importazole for 1 h before fixation. (E) Quantification of cortical LGN and NuMA staining in synchronized metaphase HeLa cells expressing GFP-CLASP1 or YFP treated with DMSO or 40 μM importazole for 1 h before fixation. For A–E, n = 3, with 100 metaphase cells counted/condition in A, 5 cells analyzed/condition in B, 50 MTs from 5 cells measured/condition in C, and 20 cells measured/condition in D and E. Bars, SE. Asterisks denote statistical significance (p < 0.05).
To clarify the contribution of CLASP1 to spindle positioning, we compared the effects of CLASP1 and the Ran pathway on astral MTs. To do so, we used HeLa cells stably expressing the MT plus-tip protein GFP-EB3, in which growing astral MTs are clearly visible in both DMSO- and importazole-treated cells (Supplemental Movies S7 and S8). GFP-EB3 HeLa cells were transiently transfected with blue fluorescent protein (BFP)–CLASP1 or BFP alone and treated with DMSO or 40 μM importazole before imaging via live-cell time-lapse fluorescence microscopy. Metaphase cells were imaged every 5 s in a single z-plane that bisected both spindle poles for a total of 2 min starting 10 min after DMSO or importazole treatment. We then quantified the number of astral MTs observed to make contact with the cell cortex. In agreement with the literature (Mimori-Kiyosue et al., 2005; Espiritu et al., 2012), we found that overexpression of CLASP1 resulted in a significant increase in the number of astral MTs contacting the cortex (Figure 5B). In contrast, disruption of the Ran pathway with importazole caused a significant decrease in astral MT/cortex contacts. This observation was supported by data from fixed mitotic HeLa cells, in which importazole treatment resulted in significantly fewer and shorter astral microtubules (Supplemental Figure S5, B and C). Overexpressing CLASP1 in importazole-treated cells restored astral MT contacts with the cortex to wild-type numbers. To determine how RanGTP and CLASP1 affect astral MTs, we measured the elongation speed of astral MTs that remained in the same z-plane between subsequent image acquisitions. We observed a significant reduction in astral MT elongation speed from 0.348 ± 0.014 in DMSO- to 0.233 ± 0.021 μm/s in importazole-treated cells, demonstrating that Ran pathway activity promotes astral MT growth (Figure 5C). In contrast, overexpression of CLASP1 had no significant effect on astral MT elongation speed in either DMSO- or importazole-treated cells. Together these results show that both RanGTP and CLASP1 affect spindle positioning by regulating contact of astral MTs with the cortex, but that they do so through independent mechanisms.
To further dissect the roles of CLASP1 and Ran in spindle positioning, we asked whether CLASP1 overexpression affected LGN and NuMA localization. Quantification of the cortical localization patterns of LGN and NuMA in DMSO- and importazole-treated cells revealed that CLASP1 overexpression did not rescue the arc-like localization pattern of either LGN or NuMA (Figure 5E). This demonstrates that CLASP1 promotes spindle positioning independently of the Ran-regulated localization of LGN and NuMA. However, mean LGN intensity at the cortex was higher in importazole-treated cells expressing GFP-CLASP1 compared with control cells, suggesting that CLASP1 stabilization of astral MT/cortex interactions can promote LGN localization at the cortex in general (Figure 5D).
DISCUSSION
Our work, in combination with recently published data (Kiyomitsu and Cheeseman, 2012), clearly demonstrates a role for Ran and importin-β in the regulation of mitotic spindle positioning in mammalian cells via the cortical positioning factors LGN and NuMA. When Ran activity was inhibited either by transfection of RanT24N or by use of a RCC1ts mutant, GFP-LGN ceased to localize to two cortical arcs in line with the long spindle axis but instead localized uniformly to the entire cortex (Kiyomitsu and Cheeseman, 2012). Similarly, importazole treatment disrupted the localization pattern of both LGN and NuMA. Importazole inhibition of the Ran/importin-β pathway, however, also resulted in lower levels of LGN localizing to the cortex. The difference in the observed localization of LGN between these two studies could be due to differences in experimental technique. Rather than completely disrupting Ran activity like RanT24N or RCC1ts, importazole treatment likely causes a conformational change in importin-β that diminishes the gradient of importin-β cargo release without completely inhibiting Ran activity (Soderholm et al., 2011). Because RanT24N and RCC1ts affect the Ran pathway upstream of importin-β, however, we cannot formally exclude the possibility that cortical LGN localization is regulated by a Ran pathway gradient independent of importin-β. In either case, our data are consistent with the conclusion that cortical LGN and NuMA localization is inhibited in the proximity of the Ran gradient. Indeed, we often observed disrupted cortical LGN and NuMA staining in the vicinity of lagging chromosomes, in agreement with this model (data not shown). The molecular mechanism by which RanGTP regulates cortical NuMA and LGN is unclear. NuMA's failure to localize to the cortex in the proximity of the RanGTP gradient, where it should be released by importin-β, as well as its formation of cytoplasmic speckles in response to importazole treatment, suggests that additional factors may be involved in Ran regulation of cortical NuMA function, although further work will be required to investigate this possibility.
Our time-lapse imaging of mitotic HeLa cells reveals that spindles in importazole-treated cells move about a significantly larger area of the cytoplasm than spindles in DMSO-treated cells, resulting in the spindle-miscentering phenotype observed with importazole treatment. In addition, both this increase in spindle movement and the resulting defects in spindle centering depend on the presence of astral MTs, whereas importazole-induced spindle assembly and chromosome congression defects do not. Together these results imply that misregulation of astral MTs under importazole treatment induces spindles to actively move about the cell. How might this active spindle movement be generated? Several clues exist that help to explain this phenomenon. First, our observations show that the cortical localization patterns of LGN and NuMA are disrupted in importazole-treated cells. Improper localization of LGN and NuMA is likely to cause abnormal associations between dynein/dynactin and astral MTs capable of pulling the spindle away from the center of the cell. In addition, the effect of importazole on astral MT dynamics may influence the movement of mitotic spindles. Importazole treatment results in a decrease in the number of astral MTs contacting the cortex and lowers the number of these contacts that occur in line with the long axis of the spindle, which may make spindles more vulnerable to improper pulling forces generated by the mislocalization of LGN and NuMA.
Our study reveals that importazole-induced spindle miscentering can be rescued by overexpression of CLASP1. CLASP1 was shown to be important for spindle positioning in multiple organisms, likely due to stabilization of end-on astral MT attachments with the cell cortex (Mimori-Kiyosue et al., 2005; Samora et al., 2011; Espiritu et al., 2012). Our work demonstrates that CLASP1 overexpression can rescue astral MT/cortex contacts in importazole-treated cells without affecting MT elongation speed, showing that RanGTP and CLASP1 affect astral MT dynamics through different mechanisms. In addition, the failure of CLASP1 to rescue the cortical localization pattern of NuMA and LGN in importazole-treated cells suggests that it does not directly regulate these proteins but instead contributes to positioning through a general mechanism of stabilizing MT attachments with the cortex. It appears, however, that some level of cross-talk exists between the CLASP1- and RanGTP-mediated spindle-positioning pathways, as CLASP1 overexpression increases the mean fluorescence intensity of LGN at the cortex in importazole-treated cells. This suggests that astral MT/cortical contact contributes to LGN localization to the cortex but that the action of RanGTP and importin-β is needed for proper cortical localization. This assertion is consistent with recent work the showing astral MTs are required for establishment of LGN's cortical localization pattern (Zheng et al., 2013).
Taken together, our data suggest a model in which Ran controls mitotic spindle positioning through two processes. First, the RanGTP gradient surrounding chromosomes locally inhibits the cortical localization of LGN and NuMA through an unidentified mechanism. In addition, our data support the existence of a second process, in which RanGTP activity stimulates growth of astral MTs to promote MT/cortex contacts. Simultaneously, the activity of CLASP1 independently stabilizes astral MTs in proximity to the cortex. Association of astral MTs with the cortex promotes cortical LGN and NuMA function, which operates in conjunction with dynein/dynactin to generate pulling forces on the spindle (Figure 6). In summary, our data support the presence of a positive feedback loop for spindle positioning in which Ran pathway activity promotes delivery of positioning information from the spindle to the cortex via astral MTs. In turn, the formation of cortical LGN/NuMA complexes provides positioning information to the spindle by generating pulling forces on astral MTs in conjunction with dynein, maintaining the proper position of the spindle within the cell.
FIGURE 6:
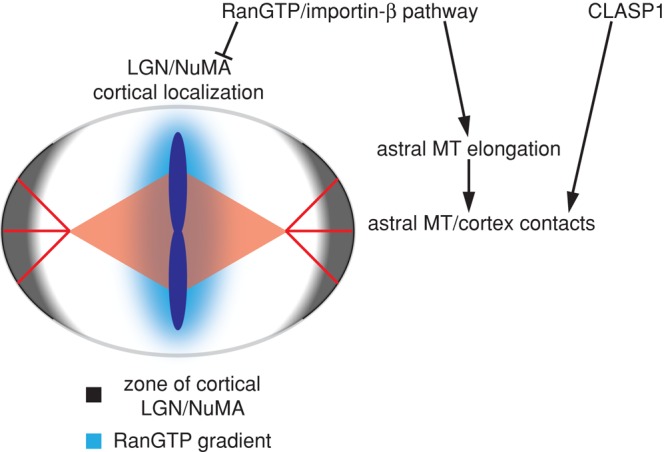
Model for RanGTP and CLASP1 regulation of mitotic spindle positioning. The RanGTP gradient negatively regulates the cortical localization of LGN and NuMA in proximity to the chromosomes along the metaphase plate axis. At the same time, the RanGTP gradient elicits release of importin-β cargo factors that promote astral MT elongation along the long axis of the spindle. Growing astral MTs contact the cortex and are stabilized by CLASP1. The cortical activity of NuMA and LGN is promoted in the vicinity of astral MTs, creating a zone of cortex-bound dynein/dynactin complexes that elicit pulling forces on the astral MTs in line with the long axis of the spindle.
Many questions remain about the role of the Ran pathway in mitotic spindle positioning. Future work will address how the cortical localization of LGN and NuMA is negatively regulated in proximity to the Ran gradient, which should lead to an increase in free NuMA. In addition, future studies should clarify the mechanism by which RanGTP promotes astral MT dynamics, as well as the role these MTs play in directing NuMA and LGN to the cortex.
MATERIALS AND METHODS
Cell lines and tissue culture
HeLa cells stably expressing GFP-tubulin and mCherry-H2B were a gift of J. Ellenberg and were maintained in DMEM plus 10% fetal bovine serum, 1% penicillin/streptomycin, 500 μg/ml G418, and 0.5 μg/ml puromycin. HeLa cells stably expressing GFP-EB3 were a gift of J. Ellenberg and were maintained in DMEM plus 10% fetal bovine serum, 1% penicillin/streptomycin, and 500 μg/ml G418. HeLa cells were grown and maintained according to standard protocols.
Protein overexpression
For importin-β overexpression experiments, 1.6 μg/well of importin-β-YFP (pKW1735) or YFP (pKW1258) plasmid DNA was transfected into six-well dishes containing 30–40% confluent HeLa cells using Lipofectamine LTX with PLUS reagent (Invitrogen, Carlsbad, CA). Cells were allowed to incubate for 45 h posttransfection before fixation. Only cells displaying visible YFP fluorescence were counted for quantification experiments. For CLASP1 overexpression experiments, 536 ng/well of GFP-CLASP1 (pKW3059; a gift of A. Akhmanova), BFP-CLASP1 (pKW3125), control YFP (pKW1258), or BFP (pKW1259) plasmid DNA was transfected into six-well dishes (for fixed samples) or 35-mm glass-bottom Petri dishes (for live-cell movies) containing 30–40% confluent HeLa cells using Lipofectamine LTX with PLUS reagent. Cells were allowed to incubate for 25 h posttransfection before fixation or imaging. Only cells displaying visible GFP, YFP, or BFP fluorescence were counted for quantification experiments.
Cell synchronization
Cells were plated in dishes containing media with 2 mM thymidine and incubated at 37ºC and 5% CO2 for 18 h. Cells were washed twice with clean media and incubated for 8.5 h. Cells were changed into media plus 2 mM thymidine and incubated for 17 h. Cells were washed twice with clean media to release from thymidine treatment and incubated for 9 h before fixation in mitosis.
Live-cell microscopy
GFP-tubulin and mCherry-H2B HeLa cells were imaged at 37ºC and 5% CO2 in Ringer's buffer (155 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 2 mM NaH2PO4, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.2, 10 mM glucose) plus 10% fetal bovine serum and 1% OxyFluor. Images were created from projections of five z-slices (0.5 μm/z-slice) captured every 2 min for a total duration of 60 min with a Nikon TE2000 inverted spinning disk confocal microscope (Nikon, Melville, NY). GFP-EB3 HeLa cells were imaged at 37ºC and 5% CO2 in Ringer's buffer, and single z-slice images were captured every 5 s for a total duration of 2 min with a Nikon TE2000 inverted spinning disk confocal microscope.
Immunofluorescence microscopy
Cells were fixed in 4% formaldehyde with 0.1% Triton X-100 in phosphate-buffered saline (PBS) at 25ºC for 5 min. Cells were then washed and blocked (PBS + 4% bovine serum albumin) and stained by standard techniques using anti-LGN antibody (a gift of Q. Du) diluted at 1:200 or anti-NuMA antibody (a gift of D. Compton) diluted at 1:500 in addition to the E7-A anti–β-tubulin antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) diluted 1:1000 and Hoechst dye. Images were created from projections of 10 z-slices (0.5 μm/z-slice) captured with a Nikon TE2000 inverted spinning disk confocal microscope. We used 100-ms exposures for all LGN and NuMA quantification. Fluorescence intensity was measured in ImageJ (National Institutes of Health, Bethesda, MD).
Image quantification
All image quantification was performed in ImageJ. Astral MT dynamics was determined by measuring the change in position of individual GFP-EB3 speckles that remained within the same z-slice between subsequent movie frames. The contact frequency of astral MT with the cell cortex was determined from the total number of contacts observed in single z-slice 2-min movies of HeLa cells stably expressing GFP-EB3 with exposures taken every 5 s. Spindle movement was quantified in metaphase cells by measuring the change in position of the center of the spindle corrected for movement of the center of the cell. The center of the spindle was determined by the midpoint of a line drawn between the spindle poles, and the center of the cell was determined by the center of a circular shape traced along the position of the cell cortex. Mean square displacement data were then calculated for the corrected movement of mitotic spindles using Matlab (MathWorks, Natick, MA). Cortical LGN and NuMA localization was quantified in synchronized metaphase cells by measuring the fluorescence intensity of a seven-pixel-wide line traced along the cell cortex. The line was always started at the position of the cortex closest to the metaphase plate (as determined by Hoechst staining) and continued in the direction of the more prominent cortical staining. The number of fluorescence intensity measurements along this line was normalized to 360, corresponding to the roughly circular shape of mitotic HeLa cells. Background fluorescence intensity was subtracted from the cortical fluorescence intensities of each individual cell, and the resulting cortical intensities were normalized as the percentage of the maximum observed cortical intensity for each replicate. Mean cortical intensities were determined by averaging the measured intensity along the entire cortex.
Supplementary Material
Acknowledgments
We thank Ryan Joyner for help with the mean square displacement analysis, Jan Ellenberg (EMBL Heidelberg) for the GFP-EB3 and GFP-tubulin/mCherry-H2B HeLa cell lines, Anna Akhmanova (Utrecht University) for the GFP-CLASP1 construct, Quansheng Du (Georgia Regents University) for the LGN antibody, and Duane Compton (Dartmouth College) for the NuMA antibody. This work was supported by the National Institutes of Health (R01 GM065232 and R21 NS53592) and the University of California, Cancer Research Coordinating Committee.
Abbreviations used:
- MSD
mean square displacement
- MT
microtubule
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0150) on June 19, 2013.
REFERENCES
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal control of nuclear envelope assembly by Ran GTPase. Symp Soc Exp Biol. 2004;56:193–204. [PubMed] [Google Scholar]
- Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Espiritu EB, Krueger LE, Ye A, Rose LS. CLASPs function redundantly to regulate astral microtubules in the C. elegans embryo. Dev Biol. 2012;368:242–254. doi: 10.1016/j.ydbio.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Compton DA. NuMA is required for the organization of microtubules into aster-like mitotic arrays. J Cell Biol. 1995;131:693–708. doi: 10.1083/jcb.131.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The mechanism and control of cytokinesis. Curr Opin Cell Biol. 1997;9:815–823. doi: 10.1016/s0955-0674(97)80082-8. [DOI] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Goodman B, Zheng Y. Mitotic spindle morphogenesis: Ran on the microtubule cytoskeleton and beyond. Biochem Soc Trans. 2006;34:716–721. doi: 10.1042/BST0340716. [DOI] [PubMed] [Google Scholar]
- Gordon MB, Howard L, Compton DA. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J Cell Biol. 2001;152:425–434. doi: 10.1083/jcb.152.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill SW, Hyman AA. Spindle positioning by cortical pulling forces. Dev Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Harel A, Chan RC, Lachish-Zalait A, Zimmerman E, Elbaum M, Forbes DJ. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol Biol Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Gnadt N, Wright M, Merdes A. NuMA is required for proper spindle assembly and chromosome alignment in prometaphase. BMC Res. 2009;64 doi: 10.1186/1756-0500-2-64. Notes 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V, Groen AC, Prokhorova T, Gerson R, White E, Rodriguez A, Walter JC, Livingston DM. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–52. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydersen BK, Pettijohn DE. Human-specific nuclear protein that associates with the polar region of the mitotic apparatus: distribution in a human/hamster hybrid cell. Cell. 1980;22:489–499. doi: 10.1016/0092-8674(80)90359-1. [DOI] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller PM, Folkmann AW, Maia AR, Efimova N, Efimov A, Kaverina I. Golgi-derived CLASP-dependent microtubules control Golgi organization and polarized trafficking in motile cells. Nat Cell Biol. 2009;11:1069–1080. doi: 10.1038/ncb1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J Cell Biol. 2005;168:141–153. doi: 10.1083/jcb.200405094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- O'Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- Patel K, Nogales E, Heald R. Multiple domains of human CLASP contribute to microtubule dynamics and organization in vitro and in Xenopus egg extracts. Cytoskeleton (Hoboken) 2012;69:155–165. doi: 10.1002/cm.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Rappaport R. Cytokinesis in animal cells. Int Rev Cytol. 1971;31:169–213. doi: 10.1016/s0074-7696(08)60059-5. [DOI] [PubMed] [Google Scholar]
- Reis R, Feijao T, Gouveia S, Pereira AJ, Matos I, Sampaio P, Maiato H, Sunkel CE. Dynein and mast/orbit/CLASP have antagonistic roles in regulating kinetochore-microtubule plus-end dynamics. J Cell Sci. 2009;(122):2543–2553. doi: 10.1242/jcs.044818. [DOI] [PubMed] [Google Scholar]
- Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M, Weis K, Heald R. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700–708. doi: 10.1021/cb2000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevermann L, Liakopoulos D. Molecular mechanisms in spindle positioning: structures and new concepts. Curr Opin Cell Biol. 2012;24:816–824. doi: 10.1016/j.ceb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Strome S. Determination of cleavage planes. Cell. 1993;72:3–6. doi: 10.1016/0092-8674(93)90041-n. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- Thery M, Racine V, Pepin A, Piel M, Chen Y, Sibarita JB, Bornens M. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–953. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Askjaer P, Gentzel M, Habermann A, Griffiths G, Wilm M, Mattaj IW, Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wong RW, Blobel G, Coutavas E. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc Natl Acad Sci USA. 2006;103:19783–19787. doi: 10.1073/pnas.0609582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Wan Q, Liu J, Zhu H, Chu X, Du Q. Evidence for dynein and astral microtubule-mediated cortical release and transport of Galphai/LGN/NuMA complex in mitotic cells. Mol Biol Cell. 2013;24:901–913. doi: 10.1091/mbc.E12-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.