Abstract
TGF-β and Notch signaling pathways play important roles in regulating self-renewal of stem cells and gastrointestinal carcinogenesis. Loss of TGF-β signaling components activates Notch signaling in esophageal adenocarcinoma, but the basis for this effect has been unclear. Here we report that loss of TGF-β adapter β2SP (SPNB2) activates Notch signaling and its target SOX9 in primary fibroblasts or esophageal adenocarcinoma cells. Expression of the stem cell marker SOX9 was markedly higher in esophageal adenocarcinoma tumor tissues than normal tissues, and its higher nuclear staining in tumors correlated with poorer survival and lymph node invasion in esophageal adenocarcinoma patients. Downregulation of β2SP by lentivirus short hairpin RNA increased SOX9 transcription and expression, enhancing nuclear localization for both active Notch1 (intracellular Notch1, ICN1) and SOX9. In contrast, reintroduction into esophageal adenocarcinoma cells of β2SP and a dominant-negative mutant of the Notch coactivator mastermind-like (dnMAN) decreased SOX9 promoter activity. Tumor sphere formation and invasive capacity in vitro and tumor growth in vivo were increased in β2SP-silenced esophageal adenocarcinoma cells. Conversely, SOX9 silencing rescued the phenotype of esophageal adenocarcinoma cells with loss of β2SP. Interaction between Smad3 and ICN1 via Smad3 MH1 domain was also observed, with loss of β2SP increasing the binding between these proteins, inducing expression of Notch targets SOX9 and C-MYC, and decreasing expression of TGF-β targets p21(CDKN1A), p27 (CDKN1B), and E-cadherin. Taken together, our findings suggest that loss of β2SP switches TGF-β signaling from tumor suppression to tumor promotion by engaging Notch signaling and activating SOX9.
Introduction
Despite a decrease in the overall cancer incidence in the united States, the incidence of esophageal cancer continues to increase with an estimated 17,460 new cases reported in 2012 (1) and a 5-year survival rate for patients with advanced disease of only 0.9% (2). Eesophageal adenocarcinoma, the predominant form in western world, typically arises from Barrett's esophagus, a metaplastic transformation of the native esophageal squamous epithelium into columnar epithelium in response to gastroesophageal reflux (3). The risk of malignant progression among patients with Barrett's esophagus is 0.22% to 0.5% per year (4). A lack of molecular predictors and clear mechanisms for Barrett's esophagus progression are the critical barriers for developing clinical useful strategies for Barrett's esophagus managements. The TGF-β and Notch signaling pathways play important roles in regulating stem cell self-renewal, cell fate determination and frequently implicated in gastrointestinal carcinogenesis including esophageal adenocarcinoma (5–8). Deregulation of these pathways along with improper interactions between them may represent key events for esophageal adenocarcinoma carcinogenesis.
TGF-β is a pleiotropic cytokine that plays a central role in maintaining epithelial homeostasis. Dysfunction of TGF-β signaling is widely associated with many tumors (9–10). TGF-β signals are conveyed from type I and type II transmembrane serine/threonine kinase receptors to the intracellular mediators—Smad2 and Smad3, which further complex with Smad4, translocate to the nucleus and bind to Smad-binding elements (SBE; GTCTAGAC) in target gene promoters and activates its targets such as p21, p15, p16, p27 (11–13). Proper control of TGF-β signaling tumor suppressor function requires an additional adaptor protein β-2 spectrin (β2SP; ref. 14). β2SP plays an essential role in cell–cell interactions and maintenance of epithelial cell polarity. Interestingly, 40% of mice with heterozygous deletion of β2SP developed hepatocellular carcinoma and 90% of β2SP+/–/Smad4+/– mice developed gastric cancer and other gastrointestinal cancers (15). Dysfunctional TGF-β signaling has been reported in esophageal adenocarcinoma. Smad4 mRNA expression progressively decreases in the metaplasia–dysplasia–adenocarcinoma sequence and smad4 promoter methylation was found in 70% of primary Barrett’ adenocarcinoma samples (16). However, the functionality of TGF-β signaling loss and the mechanisms of its action, especially the role of β2SP has not yet to be established in esophageal adenocarcinoma.
Notch signaling is another key pathway in the stem cell signaling network. Aberrant Notch signaling has been implicated in a variety of tumors including colon cancer, glioma, and T-cell leukemia, etc (17–20). Binding of Notch ligands to receptors leads to a release of the intracellular region of Notch1 [the active form of Notch1, intracellular Notch1 (ICN1)] cleaved by the γ-secretase protease complex, which is composed of 4 integral membrane proteins, presenilin1 and presenilin2, nicastrin, and PEN2; ICN1 translocate to the nucleus, forms a complex with RBP-Jk and the transcriptional coactivator mastermind-like (MAML) in the RBP-Jk–binding site (TGTGGGAA) and activates transcription of its target genes (21, 22). SOX9, a high-mobility group box transcription factor, is required for development and lineage commitment. SOX9 has been reported to be a direct target for Notch signaling (23) and documented a stem cell marker (24). Recently, it was reported that SOX9 is highly upregulated in many premaligmant processes and tumor tissues and plays a functional role in tumor progression and invasion. Patients with higher SOX9 mRNA level had shorter overall survival in human breast tumors (25). In prostate tissue, SOX9 as part of a developmental pathway is reactivated in prostate neoplasia where it promotes tumor cell proliferation (26). Gene knockdown studies have showed a role for SOX9 in cell migration and invasion in UroCa cancers (27). Notch signaling and SOX9 have been implicated in cancer development; however, they have not been well studied in esophageal adenocarcinoma.
Our previous study (28) showed an inverse expression pattern between TGF-β and Notch signaling components in esophageal adenocarcinoma tissues. Here, we provide evidence that loss of β2SP directly activates Notch signaling and increased expression of its target SOX9 at the level of transcription through interactions between Smad3 and ICN1 at Smad3 MH1 domain. Our study for the first time provides evidence that loss of β2SP in TGF-β signaling may switch Smad3 function from tumor suppression to tumor promotion by engaging Notch signaling and increasing expression of SOX9.
Materials and Methods
Cells and reagents
The human esophageal adenocarcinoma cell lines FLO-1, SKGT-4, and BE3 have been previously described (29, 30) and these cell lines were authorized and recharacterized in the characterized cell line core facility of The University of Texas MD Anderson Cancer Center (Houston, TX) every 6 months. γ-Secretase inhibitor (GSI) XXI was from Calbiochem. Antibodies p21, p27, and C-MYC were obtained from Santa Cruz Biotechnology. Antibodies Smad3, Cleaved Notch1 (ICN1, VAL1744), E-cadherin, Twist, were purchased from Cell Signaling. SOX-9 and Hes-1 were from Chemicon. The antibody against β2SP has been previously described (14). Dominant-negative MAML (MigR-dnMAN) was provided by Dr. Pingyu Zhang (The University of Texas MD Anderson Cancer Center) and previously described (31).
Cell proliferation assay
Cell proliferation assays were previously described (28). β2SP+/+ and β2SP–/– MEFs cells morphology was illustrated by EVOS digital inverted microscope at ×10 magnification.
Protein extraction and Western blot analysis
Protein isolation and Western blot analyses were done as previously described (32).
Transient transfection and luciferase reporter assays
SOX9 luciferase reporter and their deletions (33) were kindly provided by Dr. Sonsoles Piera-Velazquez (Thomas Jefferson University, Philadelphia, PA). Jagged1 promoter (pJPF2) and Hes-1 promoter were kindly provided by Dr. Randy L Johnson (the University of Texas MD Anderson Cancer Center). Transient cotransfection with luciferase reporters and Renilla vector were done as previously described (32).
Immunohistochemistry
Immunohistochemical staining for SOX9 was done on tissue microarray slides consisting of 113 esophageal adenocarcinoma and nonneoplastic esophageal tissue samples from patients who underwent esophagogastrectomy without neoadjuvant therapy using antibody against SOX9 (1:2,000 dilution) as described previously (28). The staining results were evaluated by a pathologist (D.M.) on the basis of the percentage of tumor cell nuclei stained (0, no staining; 1, ≤10%; 2, 10–50% and 3, >50%) and the staining intensity (0, negative; 1, weak; 2, moderate; and 3, strong).
Matrigel invasion assay
The invasive capability of cells was determined by using Matrigel-coated invasion chambers with 0.8-μm pore size according to the protocol (BD Biosciences) as previously described (34).
Indirect immunofluorescence and flow cytometry
Indirect immunofluorescence staining was done as described (34). Putative cancer stem cells was labeled by indirect Alex-anti-OCT4 antibody at 1:1,000 and analyzed by flow cytometry using BD FACS Caliburs (BD Biosciences).
Quantitative real-time PCR
To quantify the changes in SOX9 mRNA level, real-time reverse transcription (RT)-PCR was done on the ABI Prism 7900 (Applied Biosystems) using the commercially available gene expression assay for both SOX9 (Hs00165814-ml; Applied Biosystems) and cyclophilin A (4326316E; Applied Biosystems) as described previously (34). The C1000 Thermal Cycler-CFX96TM Real-Time System (Bio-Rad) automatically determined the amount of change for SOX9 in each sample using the δδCt method with 95% confidence interval.
GST–pull-down assay and immunoprecipitation
Immunoprecipitation from total cell lysate were done as previously described (34). Full-length and various domains of GST-tagged Smad3 pGEX-4T-1 constructs and Flag-Smad3 were from Dr. Lopa Mishra's laboratory. GST fusion proteins were purified as formerly described (35). For pull-down studies, aliquots (800 μg) of cell total protein from esophageal adenocarcinoma cell lines SKGT-4 and BE3 were mixed with either pGEX-4T (vector only), full-length GST-Smad3, or GST-Smad3 MH1, MH2 or link domains immobilized on glutathione-agarose beads for overnight at 4°C. Bound proteins were collected by centrifugation and resolved by 12% SDS-PAGE. Western blotted using an antibody against active intracellular Notch domain1 (val1744) antibody (Cell Signaling Inc.).
Mobility shift assays
Nondenatured nuclear extracts were prepared using the NE-PER extraction kit (Thermo Scientific). Mobility shift assays were done using Gel-shift kit (Panomics) according to the protocol as described (36).
Tumor sphere formation assay
Single cells (2,500/well) were seeded in triplicate onto a 6-well ultra-low attachment plate (Corning) in serum-free DMEM/F-12 supplemented with 10 ng/mL epidermal growth factor, 5 μg/mL insulin, 0.5 μg/mL hydrocortisonum, and bovine pituitary extract (Invitrogen). After 10 to 14 days of culture, the number of tumor spheres formed (diameter >100 μm) was counted under microscope.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was done in accordance with the manufacturer's instructions (Upstate) as described (36). The DNA–protein complex was sheared by sonication. A 1% portion of the sheared DNA–protein complex was kept as an input DNA sample. Anti-Smad3 and normal IgG were used for immunoprecipitation. Enrichment of promoter-binding levels was analyzed by real-time PCR. Specific SOX9 promoter primers flanking the SBE and RBP-Jκ sites of the SOX9 proximal promoter were as follows: (forward) 5′-CGA ATA CTG CAA ACT CCA GCT-3′ and (reverse) 5′-CGA ATC TTG TGT GTG TGT GTG-3′.
In vivo xenograft mouse model
Esophageal adenocarcinoma cells with genetically knockdown β2SP or/and shSOX9 and control cells were subcutaneously injected with 2 × 106 cells in nude mice. When tumors reached a size of approximately 50 mm2, shβ2SP group was divided by 2, one group was treated by intraperitoneal (i.p.) injections with buffer alone (control), the other group treated by i.p. injections with GSI (200 μg/kg) for 2 weeks; n = 6 each group. The tumor size was measured by using a digital caliper (VWR International), and the tumor volume was determined with the formula: tumor volume (mm3) = [length (mm)] × [width (mm)]2 × 0.52. All the measurements were compared using unpaired Student t test.
Statistical analysis
Data were analyzed using the Student t test. Tissue sample sets of immunohistochemical data were assessed the significance using the Kaplan–Meier estimate. A P value of <0.05 was required for statistical significance, and all tests were 2-sided. All tests were done with SPSS 10.1 software (SPSS Inc.).
Results
Nuclear SOX9 expression in esophageal adenocarcinoma tumor tissues was correlated with a poorer clinical outcome
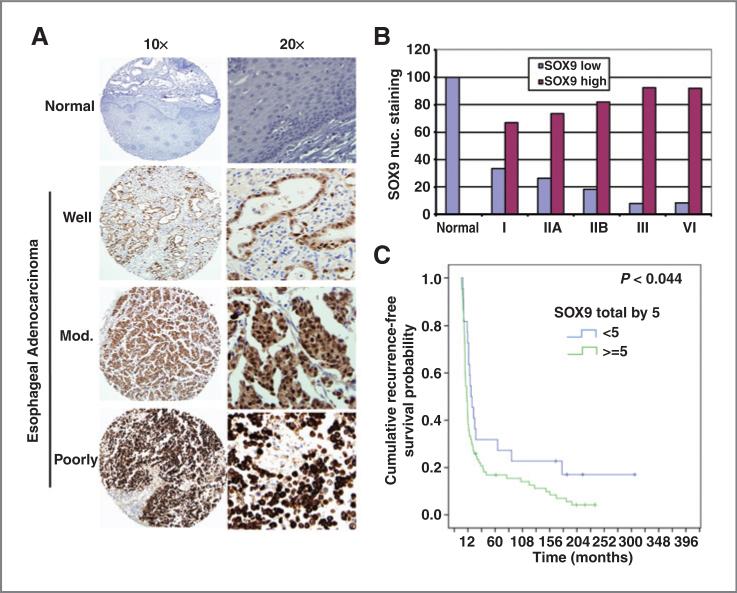
SOX9 is a direct Notch signaling target that maintains stem or progenitor (transit-amplifying) cells (37). To determine SOX9 expression in human esophageal adenocarcinoma tissues, immunohistochemistry was done in 113 cases tumor tissue microarray using specific SOX9 antibody. As shown in Fig. 1A, nuclear SOX9 expression was progressively increased along with the disease stages and differentiation status. The nuclear staining of SOX9 was absent or weakly expressed in normal squamous epithelium; however, SOX9 was positive in majority of esophageal adenocarcinoma tumor cells nuclei in esophageal adenocarcinoma tumor tissues. The tumors were categorized into SOX9-low (combined scores <5) and SOX9-high (combined score ≥ 5) groups. The Fig. 1B shows that high nuclear SOX9 expression increases along with raised tumor stage. Further study of SOX9 nuclear expression by Kaplan–Meier analysis (Fig. 1C) indicated that high levels of nuclear SOX9 expression was associated with poor survival in esophageal adenocarcinoma patients in univariate analysis (P < 0.05). Furthermore, high SOX9 nuclear expression was significantly correlated with lymph node metastasis as shown in Supplementary Table S1 (P = 0.03). These data suggest that nuclear expression of SOX9 is an adverse prognostic factor for survival in esophageal adenocarcinoma patients.
Figure 1.
Nuclear SOX9 expression in esophageal adenocarcinoma tumor tissues correlates with poorer clinical outcome. A, tissue microarray slides were immunohistochemically stained using SOX9 antibody as described in Materials and Methods. A, representative staining of SOX9 was shown in normal esophagus and esophageal adenocarcinoma with well-, moderate-, and poorly differentiated tumors. B, nuclear SOX9 staining in different stages of esophageal adenocarcinoma tumor tissues. The tumors were categorized into SOX9 low and SOX9 high based on the scoring of nuclear staining in tumor cells and staining intensity. C, Kaplan–Meier survival analysis indicated that high levels of nuclear SOX9 expression are associated with poor survival in esophageal adenocarcinoma patients (P < 0.05).
Deletion of β2SP in MEFs leads to upregulation of Notch signaling and increased expression of SOX9
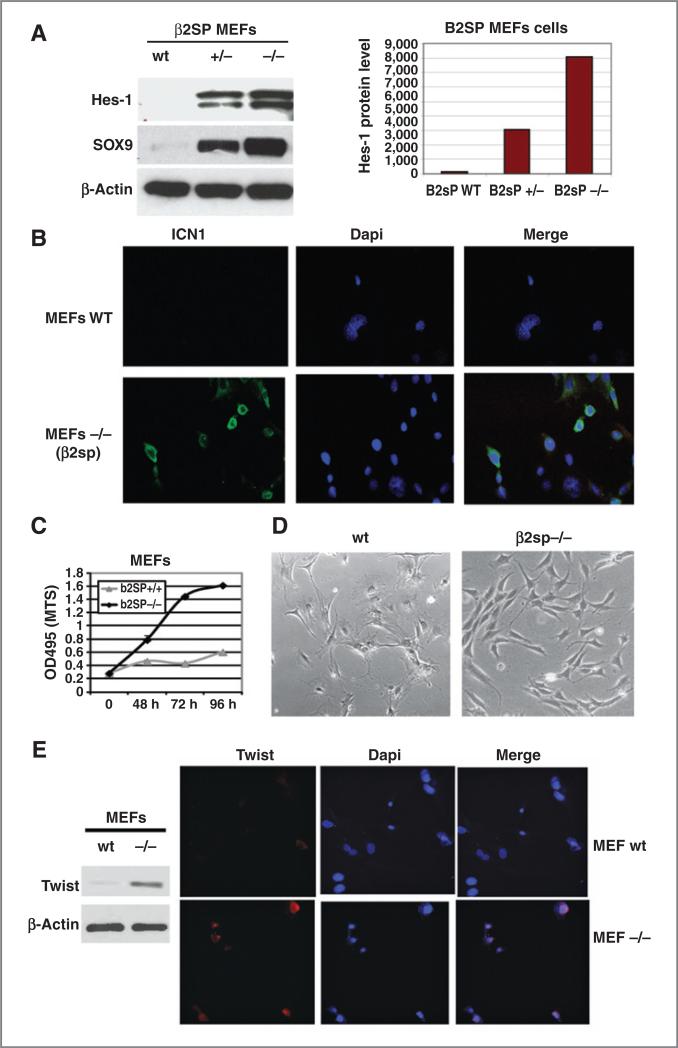
To determine if SOX9 upregulation is because of loss of TGF-β signaling, we found that Hes1 and SOX9 expression increased by 15- and 40-fold, respectively, in MEFs–/– (Fig. 2A). The quantification of Hes-1 expression was determined by densitometry (Fig. 2A, right). Upregulation of SOX9 and Hes1 was accompanied by increased nuclear localization of ICN1 by immunofluorescence in MEFs with loss of β2SP (Fig. 2B). Notch1 receptor and Hes-1 promoter activity were increased in MEFs cells and esophageal adenocarcinoma tumor cells with loss of β2SP (Supplementary Fig. S1). Furthermore, β2SP–/– MEFs grew faster, showed mesenchymal phenotype, and acted like immortalized cells, whereas wt MEFs grew slowly had an epithelial phenotype (Fig. 2C and D). Correspondingly, expression of EMT transcription factor Twist as well as Slug and Snail was increased in β2SP–/– MEFs compared with β2SP wt MEFs cells (Fig. 2E and Supplementary Fig. S3). These data suggest that Notch signaling activation because of loss of β2SP in MEFs may activate an epithelial–mesenchymal transition (EMT)-like phenotype or proliferative potential of the cells.
Figure 2.
Deletion of β2SP in MEFs leads to upregulation of Notch signaling and increased expression of SOX9. A, Hes-1 and SOX9 expression was detected by Western blotting (left), and expression of Hes-1 was quantified by densitometry (right). B, expression and localization of ICN1 in MEFs was determined by indirect immunofluorescence. C, cell growth of β2SP+/+ and β2SP–/– MEFs was done to determine the rate of proliferation; *, P < 0.05. D, β2SP+/+ and β2SP–/– MEFs cells morphology was illustrated by EVOS digital inverted microscope at ×10 magnification (right). E, twist expression was detected by Western blotting (left) and by immunofluorescence (right).
Downregulation of β2SP in esophageal adenocarcinoma tumor cells increases SOX9 expression and nuclear localization
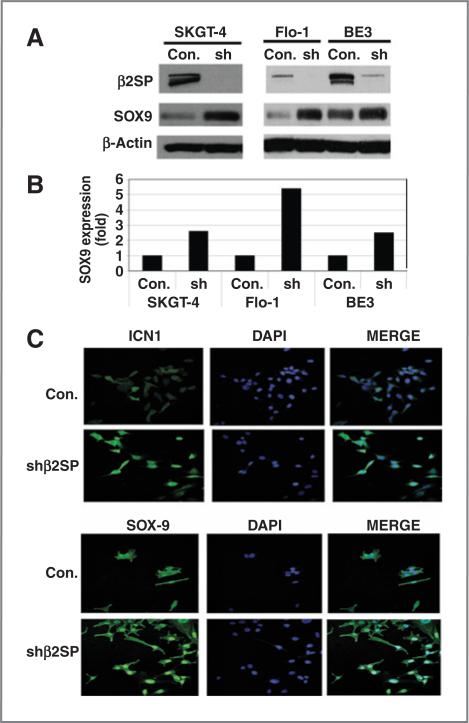
To determine the direct control of Notch and SOX9 by TGF-β signaling in esophageal adenocarcinoma tumor cells, we knocked down β2SP by lentivirus short hairpin RNA (shRNA) in 3 esophageal adenocarcinoma cancer cell lines: SKGT-4, FLO-1, and BE3 (Fig. 3A). As expected, downregulation of β2SP increased SOX9 expression by 2.6-, 5.4-, and 2.5-fold in SKGT-4, FLO-1, and BE3 cells, respectively (Fig. 3A and B). ICN1 and SOX9 levels were greatly increased in the nucleii of β2SP knockdown cells compared to vector control cells (Fig. 3C). Notch target Hes-1 and its ligand Jagged1 promoter activities were increased because of knockdown β2SP (Supplementary Fig. S1B and S1C). These data suggest that TGF-β signaling directly activate Notch signaling and increases SOX9 in esophageal adenocarcinoma tumor cells.
Figure 3.
Downregulation of β2SP in esophageal adenocarcinoma tumor cells increases SOX9 expression and its nuclear localization. A, expression of SOX9 was analyzed in esophageal adenocarcinoma cells with β2SP shRNA knockdown by immunoblotting. B, quantification of SOX9 was determined by densitometry. C, SOX9 and ICN1 expression and localization were analyzed by immunofluorescence.
Loss of β2SP increases transcription of SOX9 via Notch signaling
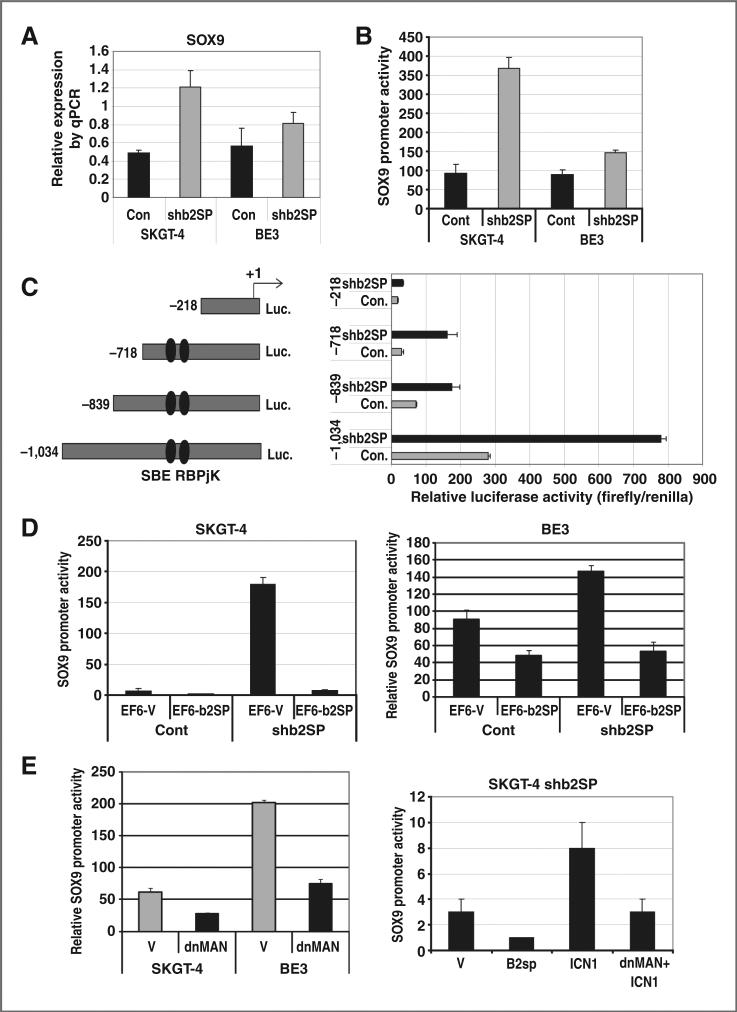
We detected the mRNA level of SOX9 in SKGT-4 and BE3 using quantitative RT-PCR. Results in Fig. 4A show that the mRNA level of SOX9 was increased by β2SP knockdown in both SKGT-4 and BE3 cells. Using transient transfection of a SOX9 luciferase promoter into esophageal adenocarcinoma cells, we found that loss of β2SP increased SOX9 promoter luciferase activities in both SKGT-4 and BE3 esophageal adenocarcinoma cells (Fig. 4B).
Figure 4.
Loss of β2SP increases transcription of SOX9. A, mRNA level of SOX9 was determined by quantitative RT-PCR. B, SOX9 promoter activity was determined by transient transfection of SOX9 luciferase promoter reporter. C, deletion analysis of SOX9 promoters was done by transient transfection with different lengths of SOX9 promoters in SKGT-4 cells with shβ2SP compared with control cells. D, cotransfection of the SOX9 promoter (–1,034) with a β2SP expression vector (pEF6β2SP) or its control vector (pEF6EV) into esophageal adenocarcinoma cells. E, cotransfection of dominant negative MAML(MigR-dnMAML) with SOX9 promoter luciferase into esophageal adenocarcinoma cells (left); cotransfection of β2SP expression vector (pEF6β2SP) or active Notch1 (MigR-ICN1) and/or MigR-dnMAML into SKGT4 shβ2SP cells (right); luciferase reporter activity was measured after 48 hours. For all experiments, values shown represent the mean and SD of at least triplicate assays (*, P < 0.001; **, P < 0.05).
To further analyze SOX9 promoter that is responsive for loss of β2SP, we transfected luciferase reporters containing various lengths of the SOX9 proximal promoter (Fig. 4C, left) into SKGT-4 cells with or without β2SP knockdown. SOX9 promoter activity was reduced dramatically with deletion of both the Notch-RBP-Jκ and Smad3 SBE-binding sites in the promoter and the responsiveness to loss of β2SP on SOX9 promoter activity was greatly reduced as well (–218 construct). Conversely, 3 other SOX9 promoter constructs (–718, –839, and –1,034) kept the high responsiveness to loss of β2SP because they maintained intact binding sites for both RBP-Jκ and Smad3. These findings indicate that cooperation of the Notch RBP-Jκ–binding site and SBE in the SOX9 promoter maycontrol the upregulation of SOX9 transcription. Reintroducing β2SP (pEF6-β2SP) into β2SP knockdown cells blocked the induction of SOX9 promoter activity by loss of β2SP (Fig. 4D). Cotransfection of dominant-negative MAML (MigR-dnMAN) with SOX9 promoter luciferase into esophageal adenocarcinoma cells greatly reduced SOX9 promoter activity (Fig. 4E, left). Moreover, cotransfection of β2SP cDNA decreased SOX9 transcriptional activity and cotransfection active Notch1 (MigR-ICN1) expression vector increased SOX9 promoter activity, whereas dominant-negative MAML blocked MigR-ICN1–induced SOX9 activation (Fig. 4E, right). This further indicated that SOX9 upregulation induced by loss of β2SP through Notch signaling.
Interaction between Smad3 and ICN1 via Smad3 MH1 domain
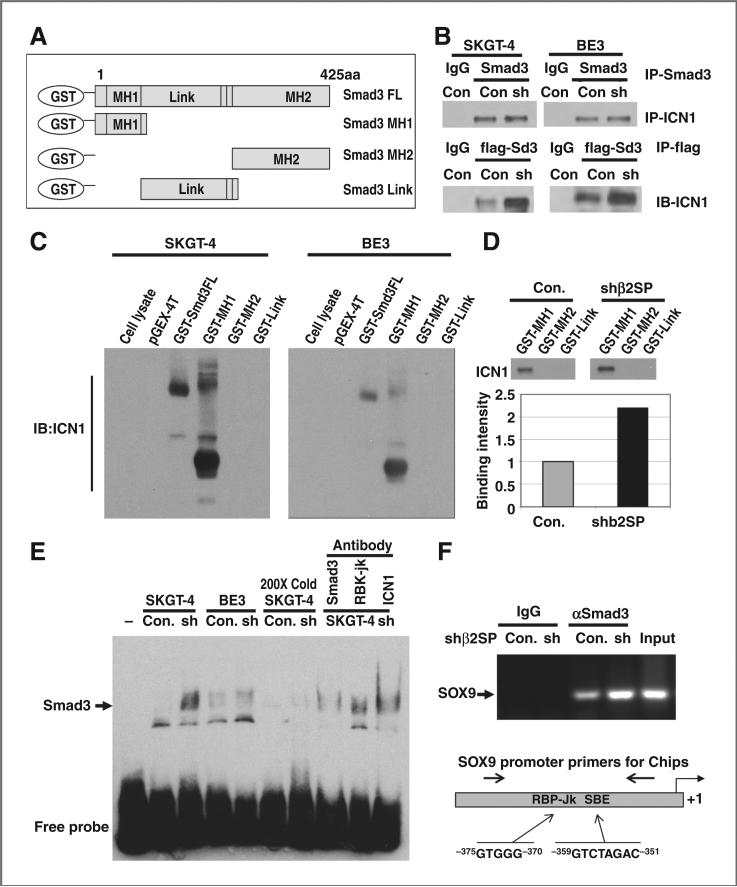
To determine the exact mechanisms by which loss of β2SP increased SOX9, we proposed that interaction between Smad3 and active Notch intracellular domain (ICN1) may mediate induction of SOX9. We incubated lysates from bacteria expressing different lengths of GST-Smad3 including full length and the MH1, MH2, and Link domains (Fig. 5A) with total protein lysate from SKGT-4 and BE3 esophageal adenocarcinoma cells and pulled down with GST-sepharose-4B beads and detected with ICN1 antibody, which recognize only cleaved active Notch1. Normally, active Notch1 is barely detected in the total cell lysate (Fig. 5C, lane 1), but we detected strong interactions between Smad3 and ICN1 in both SKGT-4 and BE3 esophageal adenocarcinoma cells after full-length Smad3 expressed in the pGEX-4T vector (Fig. 5C, lane 3), but not in control vector pGEX-4T (Fig. 5C, lane 2). Furthermore, the interactions between Smad3 and ICN1 were limited to the MH1 domain of Smad3 as showed in Fig. 5C, lane 4 and downregulation of β2SP increased this binding by 2-fold (Fig. 5D). The direct binding between constitutive Smad3 and ICN1 further confirmed by co-IP with anti-Smad3 antibody and detected with ICN1 antibody showed in Fig. 5B (top). Interestingly, when transfection with Flag-Smad3 and pull-down with anti-Flag antibody in these cell lysate, stronger bindings were found in esophageal adenocarcinoma cells with knockdown of β2SP (Fig. 5B, bottom). These further indicated that there is direct interaction between Smad3 and ICN1 especially when loss of β2SP.
Figure 5.
Smad3 and ICN1 interact via Smad3 MH1 domain. A, diagram of GST expressed Smad3 for GST-pull-down experiments. B, constitutive interaction between Smad3 and intracellular notch domain (ICN1) was detected by Co-IP (top). Esophageal adenocarcinoma cells were transfected with Flag-Smad3 and pull-down with anti-Flag antibody detected ICN1 (bottom). C, interaction between full-length GST-smad3 and MH1 domain with ICN1 by GST-pull-down in SKGT4 and BE3 cells. D, reduction of β2SP expression by shRNA increased the binding of Smad3 and ICN1 in SKGT4 cells. E, a gel shift assay was done using lysate from SKGT-4 and BE3 cells. A 200-fold excess of unlabeled SBE probe (cold probe) was used for competitive binding. Antibodies against Smad3, RBP-Jκ, and INC1 were used for the protein–DNA complexes. F, ChIP assay was done in SKGT-4 cells, and SOX9 promoter primers were designed flanking the binding sites of RBP-Jκ and Smad3 (SBE) and used for SOX9 amplification by PCR.
In addition, Smad3 DNA-binding ability was increased in shRNA β2SP knockdown esophageal adenocarcinoma cells compared with control cells as assessed by gel shift assay (Fig. 5E). The specific binding of Smad3 on the SBE DNA-binding site was confirmed using 200-fold excess unlabeled probes to block this binding and the Smad3 DNA-binding ability was disrupted when Smad3, RBP-Jκ, or ICN1 antibody was incubated with SKGT-4 cell lysate before adding to the SBE probe (Fig. 5E). This indicates that the DNA-binding protein complex may contain Smad3, RBP-Jκ, and ICN1. ChIP assay (Fig. 5F) showed that Smad3 binds in the proximal promoter of SOX9, and this binding was enhanced by loss of β2SP in esophageal adenocarcinoma cells. The design of SOX9 promoter primers used for ChIP assay depicted in Fig. 5F (bottom), which flanks the binding sites of RBP-Jκ and Smad3 (SBE).
Functional consequence of loss of β2SP and activation of Notch signaling in esophageal adenocarcinoma cells
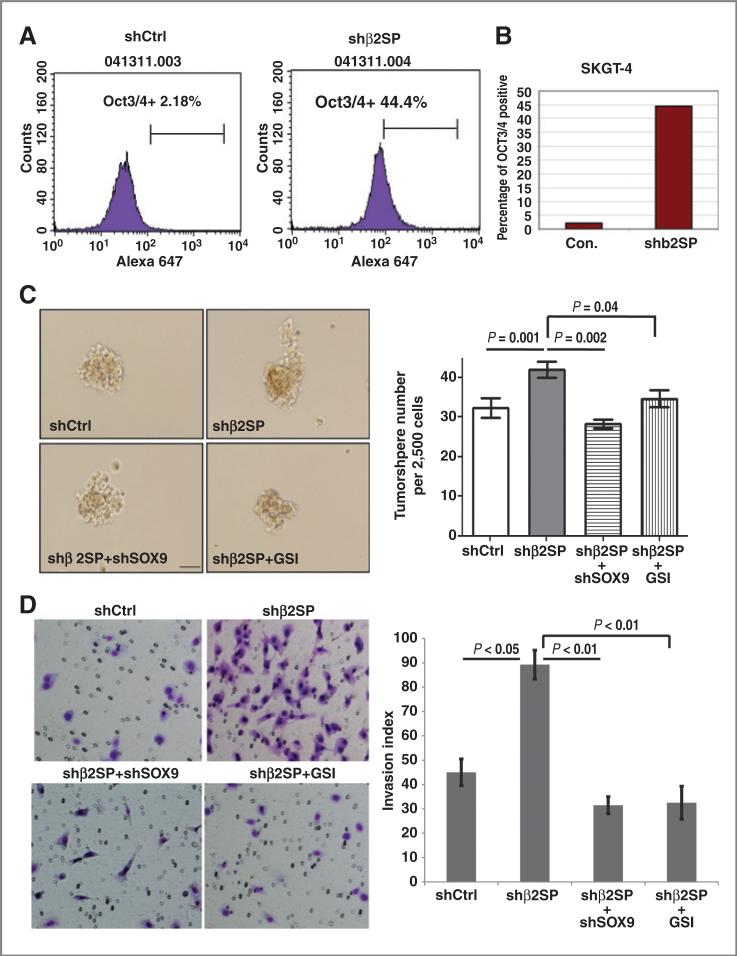
We proposed that increased expression of SOX9 and Notch signaling because of loss of β2SP may expand the putative cancer stem cell proportion. By flow cytometry analysis with stem cell marker OCT3/4, we found that knockdown of β2SP expanded putative cancer stem cells labeled with OCT3/4 by more than 20-fold in esophageal adenocarcinoma cells compared with control cells (Fig. 6A and B). Expression of OCT3/4 and Lgr5 was increased in shβ2SP SKTG4 cells compared to its control (Supplementary Fig. S4).
Figure 6.
Loss of β2SP increases oncogenic potential in esophageal adenocarcinoma cells. A, flow cytometry analysis using Oct3/4 antibody in esophageal adenocarcinoma cells with control or shβ2SP was conducted. B, bar graph showed the difference of OCT3/4 positive cells in control and shβ2SP cells. C, tumor sphere assays were done in SKGT-4 cells with modified β2SP and or SOX9 level as described in Materials and Methods. Representative fields are shown in C (left) and the bar graph in C (right). D, Matrigel-invasion assays were done in SKGT-4 cells (1 × 105) with modified β2SP or SOX9 as indicated in Materials and Methods. Representative fields are shown in D (left) and the bar graph in D (right). Experiments conducted in triplicate; bars, standard errors, *, P < 0.05 or lower.
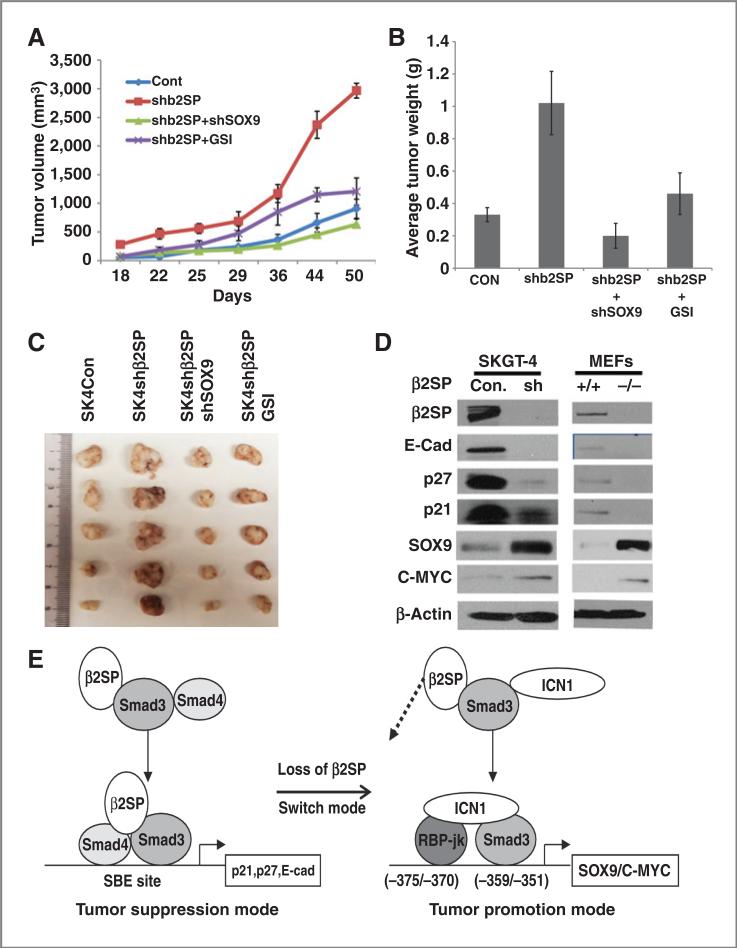
Downregulation of β2SP significantly increased tumor sphere numbers in esophageal adenocarcinoma cells (Fig. 6C) and enhance cell invasion (Fig. 6D). Furthermore, knockdown SOX9 in the background of depleted β2SP or Notch inhibitor GSI decreased the capacities of tumor sphere formation and invasion (Fig. 6C and D). Results from in vivo xenograft model further confirmed that mice with knockdown β2SP group significantly increased tumor growth compared with control group (P < 0.001), whereas double knockdown SOX9 and β2SP or GSI reduced tumor growth compared to shβ2SP alone group (P < 0.001; Fig. 7A–C). The expression of stem cell markers as well as active form of Notch1 (ICN1) were increased in mice tumors with knockdown β2SP (Supplementary Fig. S2B). This indicates loss of β2SP in tumor cells enhances oncogenic properties of esophageal adenocarcinoma cells, which may be because of activating Notch signaling and its target SOX9, whereas knockdown SOX9 or blocking Notch signaling by GSI rescued the phenotype of loss of β2SP.
Figure 7.
Loss of β2SP switches TGFβ function from tumor suppression to tumor promotion by engaging Notch signaling. A–C, effects of knockdown β2SP on esophageal adenocarcinoma cancer growth in the nude mouse of xenograft. SKGT-4 cells with shβ2SP alone, double shβ2SP and shSOX9, and shControl were inoculated into nude mice (n = 6 per group) Tumor volume and weight were calculated as indicated in Materials and Methods. D, immunoblots were conducted as described in Materials and Methods. E, proposed model by which loss of β2SP engages Notch signaling and SOX9. Loss of β2SP disrupts the canonical Smad3 complex with β2SP and Smad4 and leads to reduced expression of p27, p21, and E-cadherin. Instead, a tumor promotion complex forms by recruiting ICN1 and RBP-Jκ and subsequently activating the Notch signaling targets SOX9 and C-MYC.
The functionality of β2SP loss was illustrated by reduced canonical TGF-β signaling targets and increased expression of Notch signaling targets. As showed in Fig. 7D, loss of β2SP in esophageal adenocarcinoma tumor cells or in MEFs led to decrease in canonical TGF-β signaling targets—p27, p21, and E-cadherin, whereas increased expression of Notch targets—SOX9 and C-MYC. Diagram in Fig. 7E illustrates the proposed model by which loss of TGF-β signaling adaptor β2SP engages and activates Notch signaling and increases the expression of SOX9. Loss of β2SP disrupts canonical Smad3 complex with β2SP, Smad4 and leads to reduced expression of p27, p21, and E-cadherin. Instead, a tumor promotion complex forms by recruiting ICN1 and RBP-Jκ and subsequently activating Notch signaling targets SOX9 and C-MYC.
Discussion
TGF-β pathway plays a dual role in the development and progression of epithelial cancers (7). In normal and premalignant cells, TGF-β signaling enforces homeostasis and suppresses tumor progression, whereas in established malignancy, TGF-β signaling abused by cancer cells plays a role in tumor promotion and invasion. The mechanism of this signaling function switch during carcinogenesis and cancer progression is unclear. In this study, we show that loss of an important TGF-β adaptor β2SP in esophageal adenocarcinoma and MEF cells leads to activation of Notch signaling and increased expression of SOX9; high levels of nuclear SOX9 expression are associated with poor survival and adverse disease status (lymph node metastasis) in esophageal adenocarcinoma patients. A direct interaction between Smad3 and ICN1 via Smad3 MH1 domain was observed and loss of β2SP increases the binding of Smad3 with ICN1 and induces Notch targets SOX9. Our findings suggest that loss of β2SP may switch TGF-β signaling function from tumor suppression to tumor promotion by engaging Notch signaling and increasing the expression of SOX9 (Fig. 7E).
TGF-β signaling restricts carcinogenesis in earlier stage of tumors. Proper control of TGF-β signaling tumor suppressive function requires β2SP (14, 38). Previously, we reported that loss of TGF-β signaling components—β2SP, Smad4, and TBRII—correlates with increased expression of Notch signaling components—Hes1 and Jagged1—in esophageal adenocarcinoma tissues (28). In this study, we show that loss of β2SP in TGF-β signaling directly gives rise to activation in Notch signaling exemplified by increased Notch1 receptor and nuclear ICN1 expression in both MEFs and esophageal adenocarcinoma cells. But other Notch receptors (Notch 2–4) and γ-secretase components such as presenilin1/2, PEN2, and nicastrin were not affected (Supplementary Fig. S1A). This suggests that loss of β2SP mainly increased Notch1 activation in esophageal adenocarcinoma cells; and this may occur downstream of the cleavage of Notch1 receptor by γ-secretase complex through increased binding between Smad3 and ICN1 and prevent its degradation and facilitate its translocation to the nucleus.
SOX9 plays a pivotal role in embryonic development and its expression marks a subset of CD24-expressing adult epithelial stem cells (37) and links to progenitor status in gastrointestinal tract, liver, and pancreas (24, 39). The proximal promoter of SOX9 contains both SBE and RBP-Jκ–binding sites in close proximity to each other. We found that nuclear SOX9 expression in tumor tissue increases along with tumor stage and indicate an adverse clinical outcome. For the first time, we showed that loss of β2SP leads to an increase of SOX9 expression at the transcriptional level by the coordination among Smad3, ICN1, and RBP-Jκ in the promotion of SOX9 upon loss of β2SP. The cross-talk between Notch and TGF-β signaling has been reported previously and showed activation of TGF-β signaling upregulation of Hes-1 through cooperation between Smad3 and ICN1 (40). Our findings suggest loss of β2SP may permit Smad3 to recruit ICN1 as its coactivator and binds more avidly to its SBE and RBP-Jκ sites in the form of a complex with ICN1 and activate Notch signaling target SOX9.
Dysfunctional TGF-β signaling via loss of β2SP has the potential to increase the putative cancer stem cell proportion and significantly enhance tumor sphere formation and invasive capacities, which are critical for their malignancy. This may be because of a TGF-β tumor suppression complex including Smad3, Smad4, and β2SP disrupted upon loss of β2SP and leads to reduced expression of p27, p21, and E-cadherin. Instead, a tumor promotion complex may form by recruiting ICN1 and RBP-Jκ and activating Notch signaling targets SOX9 and C-MYC (Fig. 7E). Our findings introduce an important mechanism for how TGF-β signaling function switches from tumor suppression to tumor promotion during esophageal carcinogenesis by loss of β2SP and subsequent activation of Notch and SOX9 oncogenic signaling.
In conclusion, this study provides evidence for the first time loss of β2SP directly activates Notch signaling and increased the expression of SOX9 by the interplay between Smad3 MH1 domain and ICN1. This interplay seems critical for switching TGF-β signaling function from tumor suppression to tumor promotion. Results from this study begin to shed some light on the complex role of TGF-β signaling in esophageal tumor biology and promise to elucidate the mechanisms and predictors of Barrett's progression and yield novel target therapeutics for esophageal adenocarcinoma.
Supplementary Material
Acknowledgments
We thank Dr. Sonsoles Piera-Velazquez for the kind gifts of the SOX9 promoter luciferase constructs. We also thank Mr. and Mrs. Parker Quillen for their contribution to UTMDACC, Department of Gastroenterology/Hepatology and Nutrition, which supports this publication and color printing of the article.
Grant Support
American Gastroenterological Association Research Scholar Award (S. Song), and Public Health Service Grant DF56338, which supports the Texas Medical Center Digestive Diseases Center (S. Song), UTMDACC IRG (3-0026317; S. Song), and PO1-CA130821 (L. Mishra).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Authors’ Contributions
Conception and design: S. Song
Development of methodology: S. Song, D.M. Maru, C.-H. Chan, A.M. Correa
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): S. Song, D.M. Maru, S. Honjo, H.-K. Lin, W.L. Hofstetter, M. Davila
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. Song, D.M. Maru, C.-H. Chan, A.M. Correa, W.L. Hofstetter
Writing, review, and/or revision of the manuscript: S. Song, D.M. Maru, J.A. Ajani, W.L. Hofstetter, M. Davila, J.R. Stroehlein, L. Mishra
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): S. Song, D.M. Maru, J.A. Ajani, S. Honjo, H.-K. Lin, J.R. Stroehlein
Study supervision: S. Song, H.-K. Lin, J.R. Stroehlein, L. Mishra
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Paulson TG, Reid BJ. Focus on Barrett's esophagus and esophageal adenocarcinoma. Cancer Cell. 2004;6:11–6. doi: 10.1016/j.ccr.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 3.DeMeester SR. Adenocarcinoma of the esophagus and cardia: a review of the disease and its treatment. Ann Surg Oncol. 2006;13:12–30. doi: 10.1245/ASO.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Bhat S, Coleman HG, Yousef F, Johnston BT, McManus DT, Gavin AT, et al. Risk of malignant progression in Barrett's esophagus patients: results from a large population-based study. J Natl Cancer Inst. 2011;103:1049–57. doi: 10.1093/jnci/djr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mishra L, Derynck R, Mishra B. Transforming growth factor-beta signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Li Y, Banerjee S, Sarkar FH. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009;279:8–12. doi: 10.1016/j.canlet.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massague J. TGFbeta in cancer. Cell. 2008;134:215–30. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin L, Velazquez OC, Liu ZJ. Notch signaling: emerging molecular targets for cancer therapy. Biochem Pharmacol. 2010;80:690–701. doi: 10.1016/j.bcp.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 9.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–21. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 10.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–84. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 11.Sporn MB, Roberts AB. The transforming growth factor-betas: past, present, and future. Ann N Y Acad Sci. 1990;593:1–6. doi: 10.1111/j.1749-6632.1990.tb16095.x. [DOI] [PubMed] [Google Scholar]
- 12.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–71. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 13.Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–43. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 14.Katuri V, Tang Y, Marshall B, Rashid A, Jogunoori W, Volpe EA, et al. Inactivation of ELF/TGF-beta signaling in human gastrointestinal cancer. Oncogene. 2005;24:8012–24. doi: 10.1038/sj.onc.1208946. [DOI] [PubMed] [Google Scholar]
- 15.Katuri V, Tang Y, Li C, Jogunoori W, Deng CX, Rashid A, et al. Critical interactions between TGF-beta signaling/ELF, and E-cadherin/beta-catenin mediated tumor suppression. Oncogene. 2006;25:1871–86. doi: 10.1038/sj.onc.1209211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onwuegbusi BA, Aitchison A, Chin SF, Kranjac T, Mills I, Huang Y, et al. Impaired transforming growth factor beta signalling in Barrett's carcinogenesis due to frequent SMAD4 inactivation. Gut. 2006;55:764–74. doi: 10.1136/gut.2005.076430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abstracts of the BNOS (British Neuro-Oncology Society) Meeting. June 23-25, 2010. Glasgow, Scotland, United Kingdom. Neuro Oncol. 2010;12(Suppl 1):i1–12. doi: 10.1093/neuonc/noq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto S, Rosenberg DW. Role of Notch signaling in colon homeostasis and carcinogenesis. Cancer Sci. 2011;102:1938–42. doi: 10.1111/j.1349-7006.2011.02049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirandola L, Comi P, Cobos E, Kast WM, Chiriva-Internati M, Chiaramonte R. Notching from T-cell to B-cell lymphoid malignancies. Cancer Lett. 2011;308:1–13. doi: 10.1016/j.canlet.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Sikandar SS, Pate KT, Anderson S, Dizon D, Edwards RA, Waterman ML, et al. NOTCH signaling is required for formation and self-renewal of tumor-initiating cells and for repression of secretory cell differentiation in colon cancer. Cancer Res. 2010;70:1469–78. doi: 10.1158/0008-5472.CAN-09-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hijioka H, Setoguchi T, Miyawaki A, Gao H, Ishida T, Komiya S, Nakamura N. Upregulation of Notch pathway molecules in oral squamous cell carcinoma. Int J Oncol. 2010;36:817–22. doi: 10.3892/ijo_00000558. [DOI] [PubMed] [Google Scholar]
- 22.Katoh M. Notch signaling in gastrointestinal tract (review). Int J Oncol. 2007;30:247–51. [PubMed] [Google Scholar]
- 23.Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, et al. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–39. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huch M, Clevers H. Sox9 marks adult organ progenitors. Nat Genet. 2011;43:9–10. doi: 10.1038/ng0111-9. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarty G, Moroz K, Makridakis NM, Lloyd SA, Galvez SE, Canavello PR, et al. Prognostic significance of cytoplasmic SOX9 in invasive ductal carcinoma and metastatic breast cancer. Exp Biol Med (May-wood) 2011;236:145–55. doi: 10.1258/ebm.2010.010086. [DOI] [PubMed] [Google Scholar]
- 26.Thomsen MK, Ambroisine L, Wynn S, Cheah KS, Foster CS, Fisher G, et al. SOX9 elevation in the prostate promotes proliferation and cooperates with PTEN loss to drive tumor formation. Cancer Res. 2010;70:979–87. doi: 10.1158/0008-5472.CAN-09-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling S, Chang X, Schultz L, Lee TK, Chaux A, Marchionni L, et al. An EGFR-ERK-SOX9 signaling cascade links urothelial development and regeneration to cancer. Cancer Res. 2011;71:3812–21. doi: 10.1158/0008-5472.CAN-10-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendelson J, Song S, Li Y, Maru DM, Mishra B, Davila M, et al. Dysfunctional transforming growth factor-beta signaling with constitutively active Notch signaling in Barrett's esophageal adenocarcinoma. Cancer. 2011;117:3691–702. doi: 10.1002/cncr.25861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raju U, Ariga H, Koto M, Lu X, Pickett J, Valdecanas D, et al. Improvement of esophageal adenocarcinoma cell and xenograft responses to radiation by targeting cyclin-dependent kinases. Radiother Oncol. 2006;80:185–91. doi: 10.1016/j.radonc.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 30.Soldes OS, Kuick RD, Thompson IA, II, Hughes SJ, Orringer MB, Iannettoni MD, et al. Differential expression of Hsp27 in normal oesophagus, Barrett's metaplasia and oesophageal adenocarcinomas. Br J Cancer. 1999;79:595–603. doi: 10.1038/sj.bjc.6690094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DP. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene. 2010;29:2916–26. doi: 10.1038/onc.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song S, Mazurek N, Liu C, Sun Y, Ding QQ, Liu K, et al. Galectin-3 mediates nuclear beta-catenin accumulation and Wnt signaling in human colon cancer cells by regulation of glycogen synthase kinase-3beta activity. Cancer Res. 2009;69:1343–49. doi: 10.1158/0008-5472.CAN-08-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piera-Velazquez S, Hawkins DF, Whitecavage MK, Colter DC, Stokes DG, Jimenez SA. Regulation of the human SOX9 promoter by Sp1 and CREB. Exp Cell Res. 2007;313:1069–79. doi: 10.1016/j.yexcr.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song S, Ji B, Ramachandran V, Wang H, Hafley M, Logsdon C, et al. Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PLoS One. 2012;7:e42699. doi: 10.1371/journal.pone.0042699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper S, Speicher DW. Purification of proteins fused to glutathione S-transferase. Methods Mol Biol. 2011;681:259–80. doi: 10.1007/978-1-60761-913-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song S, Byrd JC, Mazurek N, Liu K, Koo JS, Bresalier RS. Galectin-3 modulates MUC2 mucin expression in human colon cancer cells at the level of transcription via AP-1 activation. Gastroenterology. 2005;129:1581–91. doi: 10.1053/j.gastro.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298:G590–600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–77. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- 39.Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–48. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, et al. Cross-talk between the Notch and TGF-beta signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003;163:723–8. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.