Abstract
Hsp40 proteins of bacterial and human origin are suspected to be involved in the pathogenesis of rheumatoid arthritis (RA). It has been shown that sera of RA patients contain increased levels of antibodies directed to bacterial and human Hsp40s. The aim of this work was to explore immunological similarities between the bacterial (DnaJ) and human (DNAJA1 and DNAJA2) Hsp40 proteins in relation to their possible involvement in the RA. Using polyclonal antibodies directed against a full-length DnaJ or its domains, against DNAJA1 and DNAJA2, as well as monoclonal anti-DnaJ antibodies, we found immunological similarities between the bacterial and human Hsp40s. Both ELISA and Western blotting showed that these similarities were not restricted to the conserved J domains but were also present in the C-terminal variable regions. We also found a positive correlation between the levels of the anti-DnaJ and anti-DNAJA1 antibodies in the sera of RA patients. This finding supports the molecular mimicry hypothesis that human Hsp40 could be the targets of antibodies originally directed against bacterial DnaJ in RA.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-013-0407-1) contains supplementary material, which is available to authorized users.
Keywords: Anti-Hsp40 autoantibodies, Hsp40 in rheumatoid arthritis, Cross-reactivity of anti-Hsp40 antibodies, Anti-DNAJ monoclonal antibodies, Molecular mimicry
Introduction
Heat shock proteins (Hsps) are a family of evolutionarily conserved proteins, which play an important role in cell physiology under the normal and stress conditions. At times of cellular stress, including infection and chronic inflammation, such as present in autoimmune diseases, the expression of Hsps is markedly elevated (reviewed in van Eden et al. 2005; Borges et al. 2012). Hsps are also a group of major bacterial antigens (Albani et al. 1995; reviewed in Borges et al. 2012), and the conservation of their structure from bacteria to man, as well as high immunogenicity, makes them attractive targets for investigation in the area of autoimmunity. In this area, the Hsps of the Hsp60 and Hsp70 families were the ones most extensively studied, especially since the discovery that the T cells isolated from the rats with adjuvant-induced arthritis were responding to mycobacterial Hsp60 (van Eden et al. 1988; reviewed in van Eden et al. 2005). To the contrary, the research on Hsp40 involvement in autoimmune diseases has been less extensive, in spite of the fact that Hsp40 is probably the largest Hsp family in humans, with at least 50 members (reviewed in Kampinga et al. 2009).
The model representative of the Hsp40 family is the Escherichia coli DnaJ protein, composed of 375 amino acids in four domains. The amino-terminal 75 residues of DnaJ constitute an evolutionarily highly conserved motif, the J domain, which together with the adjacent region, rich in glycine and phenylalanine, is essential for DnaJ’s interactions with Hsp70 chaperone. The third domain, rich in cysteine residues, together with the least conserved C-terminal region, functions to bind substrate proteins (Qiu et al. 2006; reviewed in Kampinga et al. 2009). Of human Hsp40, the DNAJB1 (Hdj1), DNAJA1 (Hdj2), and DNAJA2 (Hdj3) proteins are best characterized (Terada and Mori 2000; reviewed by Sterrenberg et al. 2011). DNAJA1 and DNAJA2, belonging to the class A of Hsp40, bear the highest structural similarity to the bacterial DnaJ and possess all the domains characteristic for DnaJ, while DNAJB1 does not have the cysteine-rich domain (Cheetham and Caplan 1998; Kampinga et al. 2009).
Bacterial and human Hsp40 are suspected to participate in the autoimmune response during pathogenesis of rheumatoid arthritis (RA) and juvenile idiopathic arthritis (JIA). The presence of elevated levels of antibodies against the E. coli DnaJ has been shown in RA (Albani et al. 1994, 1995; Chukwuocha et al. 1999; Tukaj et al. 2010a), with especially high response to the conserved J domain of DnaJ (Albani et al. 1995; Tukaj et al. 2010a). Additionally, an overexpression of human Hsp40s and significantly increased levels of the anti-DNAJA1 and anti-DNAJA2 antibodies have been found, respectively, in the synovial tissue and sera of RA patients (Kurzik-Dumke et al. 1999; Tukaj et al. 2010a). Initially, a “molecular mimicry hypothesis” has been proposed, suggesting that the immune response directed against the bacterial DnaJ protein cross-reacts with the human homologous protein(s) and promotes development of RA; an infection with various bacterial species could trigger the response, since DnaJ is highly conserved among bacteria (Albani et al. 1995; Albani and Carson 1996). Indeed, bacterial infection is considered as one of the possible factors promoting development of RA (reviewed in Lundberg et al. 2010). Later on, another aspect of the Hsp40s role in inflammatory diseases emerged, showing an immunomodulatory role of Hsp40s in downregulating immune response. It has been demonstrated that T cells from patients with JIA respond differentially to peptides derived from bacterial and human Hsp40s and that regulatory T cells, induced by a peptide derived from a human Hsp40, downregulate proliferation of synovial fluid mononuclear cells of JIA patients (Massa et al. 2007). Our previous study showed that DnaJ and human Hsp40 proteins inhibited proliferation of T cells of the RA patients and had an immunomodulatory effect on cytokine secretion by the patients’ lymphocytes (Tukaj et al. 2010a). Although the exact role of bacterial and human Hsp40s in the autoimmune response requires further elucidation, it is clear that those proteins have a relevance in the clinical setting, especially since Hsp40s are considered as potential targets for anti-inflammatory therapy in JIA (Massa et al. 2007) and RA (Tukaj et al. 2010a).
Previously, we have shown a significant immunological similarity between bacterial DnaJ and human DNAJB1, not restricted to the conserved J domains of the proteins (Krzewski et al. 2003). The aim of this work was to further investigate the immunological similarities among the bacterial and human Hsp40s and focus on the DNAJA1 and DNAJA2, the human Hsp40s best structurally matched with DnaJ. We tested cross-reactivity of the bacterial and human Hsp40s with the monoclonal antibodies, recognizing different and defined DnaJ epitopes, and with polyclonal antibodies against full-length DnaJ, DnaJ N-terminal domain (N-DnaJ), DnaJ C-terminal domain (C-DnaJ), DNAJA1, and DNAJA2. We also assayed the humoral anti-Hsp40 response in RA patients and analyzed correlation between the bacterial and human anti-Hsp40 antibody levels.
Results and discussion
Polyclonal antibodies against DnaJ, N-DnaJ, and C-DnaJ react with human DNAJA1 and DNAJA2
We purified recombinant bacterial (E. coli DnaJ, N-DnaJ, C-DnaJ) and human (DNAJA1 and DNAJA2) Hsp40 proteins, overproduced in bacterial cells (Fig. 1a), and used them as antigens to obtain rabbit polyclonal antibodies. Cross-reactivity of these antibodies with the Hsp40 proteins was tested by Western blotting technique and ELISA tests.
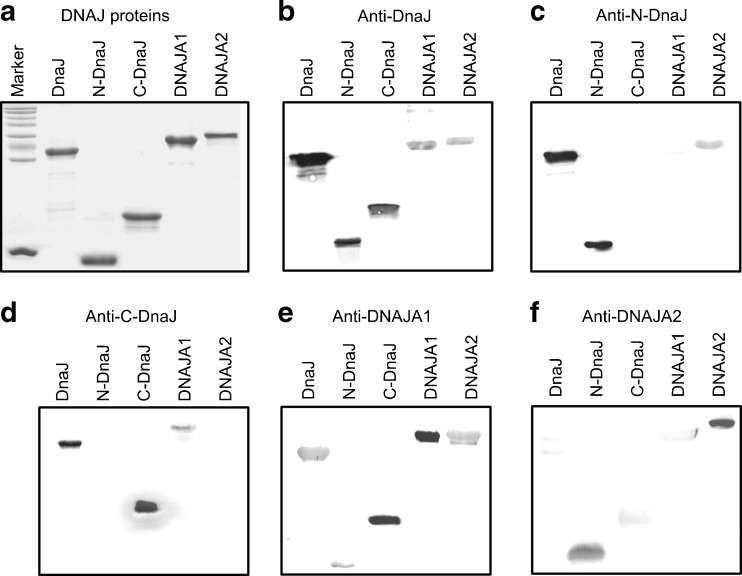
Fig. 1.
Cross-reactions between anti-DNAJ(Hsp40) polyclonal antibodies and DNAJ(Hsp40) proteins tested by Western blotting. The DnaJ, N-DnaJ (DnaJΔ107-375), C-DnaJ (DnaJΔ1-199), DNAJA1, and DNAJA2 recombinant proteins were overproduced in E. coli cells, purified, and resolved by SDS-PAGE. Of each Hsp40 protein, 2.5 μg has been loaded onto the gel (a). The polyclonal antibodies indicated above the panels were used in Western blotting as the primary antibodies (b–f)
The immunoblotting results showed that the anti-DnaJ antibodies were able to cross-react with both DNAJA1 and DNAJA2 (Fig. 1b). The anti-N-DnaJ antibodies reacted with DNAJA2 but not with DNAJA1 (Fig. 1c). Conversely, the anti-C-DnaJ antibodies reacted with DNAJA1 but not with DNAJA2 (Fig. 1d). These results indicate immunological similarity between DnaJ, and human DNAJA1 and DNAJA2, with DNAJA1 bearing a higher similarity to the C-terminal part of DnaJ and DNAJA2 to the N-terminal domain. This conclusion was confirmed further by the fact that the anti-DNAJA1 serum reacted strongly with C-DnaJ protein and weakly with N-DnaJ (Fig. 1e), while the anti-DNAJA2 antibodies reacted well with N-DnaJ but very weakly with C-DnaJ (Fig. 1f). The identity at the amino acid level between DnaJ and DNAJA1 or DNAJA2 is 38 or 39 %, respectively. Surprisingly, we noticed a very weak reaction of anti-DNAJA1 serum with DNAJA2 in spite of 54 % identity at the amino acid level between DNAJA1 and DNAJA2 (Fig. 1e). In parallel, the anti-DNAJA2 antibodies reacted very weakly or not at all with DNAJA1 (Fig. 1f and results not shown). The latter finding is in agreement with the results of Terada and Mori (2000), who showed the lack of cross-reactivity of the anti-Hdj3 (DNAJA2) antibodies with Hdj2 (DNAJA1) in immunoblotting. This also shows that the degrees of sequence identity and immunological similarity may differ. It should be noted that the anti-DNAJA1 serum recognized the full-length DnaJ (Fig. 1e).
The results obtained by the Western blotting technique, in which only linear epitopes were recognized, were confirmed by the ELISA tests in which non-denatured antigens were used. The anti-DnaJ antibodies cross-reacted with both DNAJA1 and DNAJA2 (Fig. 2a). The anti-N-DnaJ antibodies reacted well with both DNAJA1 and DNAJA2 (Fig. 2b), while in the immunoblots, the reaction with DNAJA1 was almost non-detectable. The reason why we could observe the reaction with DNAJA1 here but not in the immunoblots could be due to the presence of the native antigens and thus of conformational epitopes and/or the higher sensitivity of the ELISA test compared to Western blotting. It should be noted that the J domains of DnaJ and DNAJA1 or DNAJA2 bear about 50 % identity (as calculated for the residues 1–77, 3–78, and 5–80 of DnaJ, DNAJA1, and DnaAJA2, respectively). In the case of anti-C-DnaJ antibodies, a cross-reaction with DNAJA1 and a minimal binding to DNAJA2 was observed (Fig. 2c), which was consistent with the immunoblotting results. Like in immunoblotting, anti-DNAJA1 and anti-DNAJA2 showed very weak cross-reactivity with DNAJA2 and DNAJA1, respectively (Fig. 2d, e). In ELISA tests, these antibodies showed poorer reaction with bacterial proteins than in Western blotting. This could be due to inaccessibility of the linear epitopes which could be exposed only after a denaturation process takes place during SDS-PAGE in Western blotting experiments.
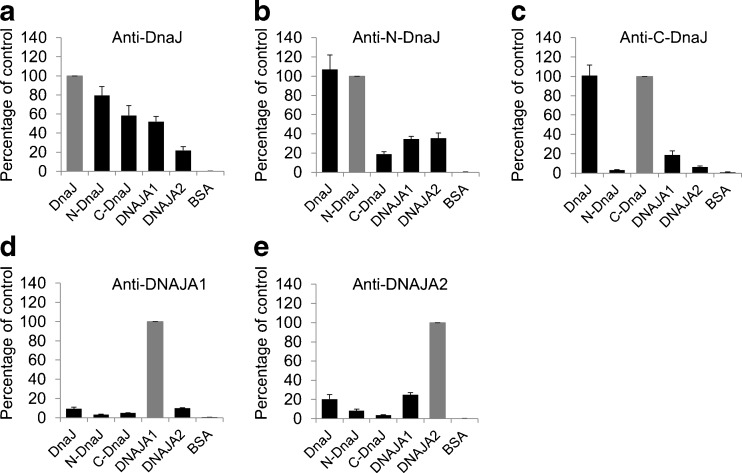
Fig. 2.
Cross-reactions between anti-DNAJ(Hsp40) polyclonal antibodies and DNAJ(Hsp40) proteins tested by ELISA. The reactivity of polyclonal anti-DnaJ (a), anti-N-DnaJ (b), anti-C-DnaJ (c), anti-DNAJA1 (d), and anti-DNAJA2 (e) with purified non-denatured DNAJ antigens was tested as described in “Materials and methods.” Data obtained for the antigens at 40 μg per milliliter are presented as the mean ± SEM of three independent experiments and expressed as a percentage of control (i.e., reactivity with the antigen used to raise a given antibody; indicated as the gray bar)
Taken together, all the above results show a significant immunological similarity between DnaJ and DNAJA1/DNAJA2 proteins, which in the case of DNAJA1 was found not only in the conserved J domain but also in the poorly conserved C-terminal region. Additionally, a very low immunological similarity was found between DNAJA1 and DNAJA2 proteins, suggesting that these antigens induce different immune responses.
The monoclonal antibodies recognizing DnaJ epitopes react with DNAJA1 and DNAJA2
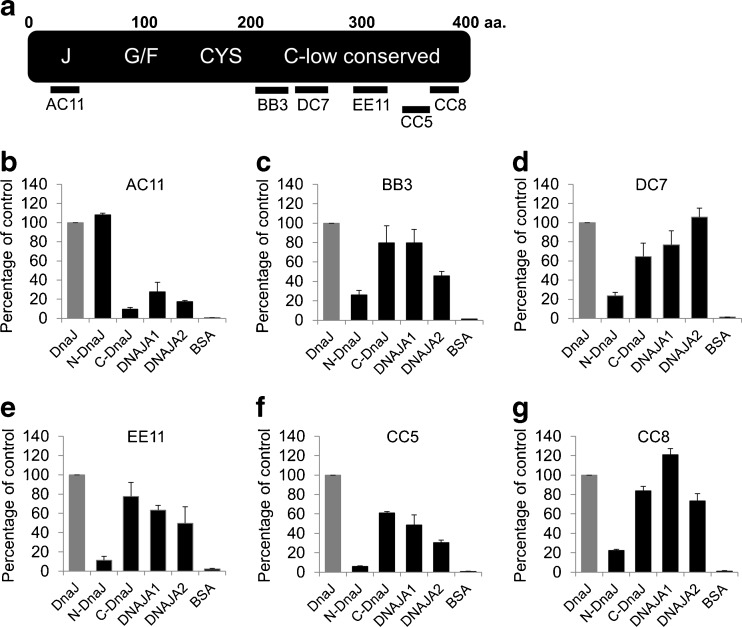
To further characterize response of human Hsp40 proteins to anti-DnaJ antibodies, we used a set of six anti-DnaJ monoclonal antibodies (mAbs), whose epitopes have been characterized previously (Krzewski et al. 2003); localization of these epitopes in DnaJ protein is shown schematically in Fig. 3a. The mAb AC11 epitope is localized in a highly conserved region of the J domain while the other epitopes (BB3, DC7, EE11, CC5, and CC8) are placed in the weakly conserved C-terminal region. It has been previously shown (Krzewski et al. 2003) that all these mAbs bind more efficiently to the native DnaJ compared to the denatured protein. In ELISA tests, the AC11 mAb reacted with both DNAJA1 and DNAJA2 (Fig. 3b). Surprisingly, all the mAbs directed to the C-terminal region of DnaJ (Fig. 3a) recognized both human Hsp40s with high efficiency (Fig. 3c–g). These results indicate that, despite a low identity (not exceeding 20 %) of amino acid sequence among the C-terminal regions of DnaJ and DNAJA1/DNAJA2, they do have some similar epitopes (the amino acid residues used to calculate the identity were 198–373 for DNAJA1 and 206–381 for DNAJA2). These epitopes are most probably of the conformational type, since the mAbs used here preferentially bind to the native DnaJ, and also alignment of the mAbs epitope consensus sequences with the DNAJA1 and DNAJA2 amino acid sequences did not reveal any putative linear epitopes (results not shown). These results further confirm that the anti-DnaJ antibodies can recognize human DNAJA1 and DNAJA2, and the recognition is not restricted to the conserved J domains. They also suggest that the C-terminal domains of DnaJ and DNAJA1/DNAJA2 have a similar three-dimensional structure. To date, the structures of the C-terminal, substrate-binding regions of these proteins have not been solved. Within the Hsp40 family of proteins, the C-terminal domains of the yeast Sis1 (Sha et al. 2000) and human Hdj1 (DNAJAB1) protein (Suzuki et al. 2010) have been crystallized and characterized.
Fig. 3.
Reactivity of anti-DnaJ monoclonal antibodies (mAbs) with human DNAJA proteins. Schematic outline of the DnaJ linear structure and localization of the epitopes recognized by the mAbs are presented in a. The reactivity of six mAbs (b–g) with purified non-denatured DNAJ(Hsp40) antigens was assayed by ELISA test as described in “Materials and methods.” Data obtained for the antigens at 40 μg per milliliter are presented as the mean ± SEM of three independent experiments and expressed as a percentage of the reactivity with DnaJ control (gray bar)
The major conclusion that the anti-DnaJ antibodies can recognize human DNAJA1 and DNAJA2, and the recognition is not restricted to the conserved J domain, supports the “molecular mimicry” hypothesis. This hypothesis states that in RA, human Hsp40 protein(s) may be the targets of immune response induced by a bacterial DnaJ antigen (Albani et al. 1995; Albani and Carson 1996). Our previous observations that sera derived from RA patients contain significantly elevated levels of anti-DnaJ, anti-DNAJA1, and anti-DNAJA2 (but not anti-DNAJB1) antibodies (Tukaj et al. 2010a, b) are also consistent with the mimicry hypothesis. In this study, using a different set of the RA patient sera, we obtained similar results (Table S1, Supplementary data). Furthermore, we found a positive correlation between the levels of the anti-DnaJ and anti-DNAJA1 antibodies in the RA sera (Table 1). Since DNAJA1 and DNAJA2 are modified by farnesylation in humans (Terada and Mori 2000) but not in the bacterial systems, we also purified and tested the overproduction and farnesylation of these proteins in the baculovirus eukaryotic expression system. We did not find any significant differences in the reactivity of the farnesylated and non-farnesylated proteins with the RA sera (Table S1, Supplementary data). In the case of the farnesylated proteins, the positive correlation between the anti-DnaJ and anti-DNAJA1 responses was also found (Table 1). The observed correlations indicate that in RA patients, the anti-DnaJ response may be directed against the DNAJA1 protein.
Table 1.
Correlation of the level of the anti-DnaJ antibodies (IgG) with the levels of the anti-DNAJA1/2 autoantibodies (IgG) in the sera of the rheumatoid arthritis (RA) patients
| DnaJ (2.5) | DnaJ (0.15) | |
|---|---|---|
| DNAJA1 | ||
| (2.5) | 0.631, p < 0.001 | |
| (0.15) | 0.847, p < 0.001 | |
| DNAJA2 | ||
| (2.5) | −0.151, p = 0.333 | |
| (0.15) | −0.150, p = 0.338 | |
| DNAJA1f | ||
| (2.5) | 0.442, p = 0.003 | |
| (0.15) | 0.33, p = 0.029 | |
| DNAJA2f | ||
| (2.5) | 0.176, p = 0.259 | |
| (0.15) | 0.161, p = 0.302 | |
The levels of the antibodies were assayed by ELISA test in the sera of 43 RA patients (presented in Table S1, Supplementary data) as described in “Materials and methods,” using antigens at the concentrations (in micrograms per milliliter) indicated in parentheses. DNAJA1f and DNAJA2f are the farnesylated proteins produced in the baculovirus system. The other proteins were produced in the bacterial system. The correlations between the antibody levels are presented as Pearson’s coefficients (r). The statistically significant values are shown in italics
The question whether the postulated cross-reaction between the anti-DnaJ antibodies and human Hsp40s may lead to pathological consequences remains open. There is increasing evidence that Hsps may function as factors downregulating immune response. It has been shown that the T cells immunoreactive to various types of self-Hsps display regulatory properties (van Eden et al. 2005; Wieten et al. 2007, 2010; reviewed in Borges et al. 2012). The regulatory role of Hsp40s encouraged the idea that they may be potential targets for designing new anti-inflammatory therapies (Massa et al. 2007; Tukaj et al. 2010a). Furthermore, a peptide derived from the J domain of DnaJ has been successfully applied in clinical trials of immunotherapy of RA patients (Koffeman et al. 2009). We believe that the information presented in this work increases our understanding of the anti-Hsp40 immune response and may be useful in designing new anti-inflammatory therapies.
Materials and methods
Protein overproduction and purification
The DnaJ, N-DnaJ (DnaJΔ107-375), C-DnaJ (DnaJΔ1-199), DNAJA1, and DNAJA2 recombinant proteins were overproduced in E. coli cells and purified as described previously (Krzewski et al. 2003; Tukaj et al. 2010a). To obtain the DNAJA1 and DNAJA2 proteins modified by farnesylation, the insect cells (Spodoptera frugiperda Sf9) and baculovirus expression system Bac-To-Bac (Invitrogen) were used, according to the manufacturer’s instruction. We constructed the pFastBac1/DNAJA1 and pFastBac1/DNAJA2 transfer plasmids, encoding the His6-DNAJA1 and His6-DNAJA2 proteins, respectively. The plasmid pFastBac1/DNAJA1 was derived from the pMal-c2x/hdj2 (Kanazawa et al. 1997) by excising the Ecl136-XbaI fragment and inserting it into a pFastBac1 vector. The plasmid pFastBac1/DNAJA2 was constructed by excising the BamHI–PstI fragment of the pVL1393/hdj3 plasmid (Terada et al. 1997) and inserting it into a pFastBac1 vector. Other procedures were as described in the manufacturer’s manual. Farnesylation of His-DNAJA1/2 proteins was enhanced by exogenously adding a mevalonate precursor, mevalonolactone (Sigma), to the Sf-900 II SFM medium, at 5 mM. The His-DNAJA1/2 proteins produced in insect cells were purified from the cytoplasmic fraction using a Ni-NTA agarose (Qiagen), according to the manufacturer’s manual.
Polyclonal and monoclonal antibodies
The polyclonal antisera were raised by immunizing rabbits with the purified recombinant Hsp40 proteins according to the method described by Szewczyk and Harper (1994) and tested by Western blotting with appropriate antigens. The mouse monoclonal antibodies AC11, BB3, CC8, EE11, DC7, and CC8 were generated and characterized as described by Krzewski et al. (2003).
Protein electrophoresis and Western blotting
Hsp40 proteins were resolved in 15 % polyacrylamide gels, in denaturing conditions. Western blotting was performed, using the polyclonal rabbit anti-Hsp40 sera as the primary antibodies and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulins (Roche) as secondary antibodies, as described previously (Krzewski et al. 2003). The primary antibodies were used at the following dilutions: anti-DnaJ (1:5,000), anti-N-DnaJ (1:200), anti-C-DnaJ (1:500), anti-DNAJA1 (1:500), and anti-DNAJA2 (1:500), for 1 h at room temperature. The secondary antibodies were used at 1:200 dilutions.
ELISA assays
ELISA tests were performed as described by Krzewski et al. (2003), with minor modifications. Costar 3590 96-well plates were coated with serial dilutions of DnaJ, N-DnaJ, C-DnaJ, DNAJA1, or DNAJA2 proteins (40–0.02 μg/mL) and incubated overnight at 37 °C. Then, 50 μL of a diluted anti-DnaJ mAb was added to the wells. The mAbs were used at the following dilutions: AC11—1:6,000; BB3, CC8, and EE11—1:2,000; DC7—1:500; and CC8—1:200. After 60 min of incubation, the secondary antibodies coupled with horseradish peroxidase (HRP) (Sigma) were added: the goat anti-mouse IgG antibodies to AC11 and the goat anti-mouse IgM antibodies to the other mAbs, at 1:2,000 dilutions. The bound conjugates were detected colorimetrically after 60 min of incubation, by using a horseradish peroxidase substrate, tetramethylbenzidine (Sigma).
When the reactivity of the Hsp40 proteins with the rabbit polyclonal anti-Hsp40 sera was tested, the sera were used at the following dilutions: anti-DnaJ (1:40,000), anti-N-DnaJ (1:1,000), anti-C-DnaJ (1:4,000), anti-DNAJA1 (1:4,000), and anti-DNAJA2 (1:4,000). HRP-conjugated goat anti-rabbit IgG immunoglobulins (Dako) (1:2,000) were used as secondary antibodies.
The levels of the anti-DnaJ, anti-DNAJ1, and anti-DNAJ2 antibodies (IgG) in the sera of the RA patients (n = 43) and the control individuals (n = 35) were assayed by the ELISA test performed as described before (Tukaj et al. 2010b). The antigens used were the Hsp40 proteins produced in the bacterial cells and, additionally, the DNAJA1 and DNAJA2 produced and farnesylated in the baculovirus system (designated as DNAJA1f and DNAJA2f).
The RA patients
The RA patients’ sera were obtained from 43 (29 females and 14 males) patients (the mean age was 59, range 21–82), with the early RA (the mean disease duration was 9 months). For controls, we used sera of 35 sex- and age-matched healthy volunteers. The study was approved by the Local Committee for Biomedical Research Ethics at the Medical University of Gdansk. All subjects were informed of the details of the experiment prior to the taking of a sample of 10 mL peripheral venous blood.
Statistical analysis
Statistical analyses were performed using the Mann–Whitney U test and STATISTICA 7.1 software. For correlation analyses, Pearson test was used. p values less than 0.05 were considered significant.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 47.5 kb)
Acknowledgments
We would like to express our thanks to Dr. B. Szewczyk for his help with overproducing proteins in the baculovirus system. We are very grateful to Dr. Kazutoyo Terada for his generous gift of the pMal-c2x/hdj2 and pVL1393/hdj3 plasmids. This work was supported by a grant from the Polish Ministry of Science and Higher Education (no. 1610/B/P01/2008/35) to BL.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- Albani S, Carson DA. A multistep molecular mimicry hypothesis for the pathogenesis of rheumatoid arthritis. Immunol Today. 1996;17:466–470. doi: 10.1016/0167-5699(96)20029-G. [DOI] [PubMed] [Google Scholar]
- Albani S, Ravelli A, Massa M, De Benedetti F, Andree G, Roudier J, Martini A, Carson DA. Immune responses to the Escherichia coli dnaJ heat shock protein in juvenile rheumatoid arthritis and their correlation with disease activity. J Pediatr. 1994;124:561–565. doi: 10.1016/S0022-3476(05)83134-8. [DOI] [PubMed] [Google Scholar]
- Albani S, Keystone EC, Nelson JL, Ollier WE, La Cava A, Montemayor AC, Weber DA, Montecucco C, Martini A, Carson DA. Positive selection in autoimmunity: abnormal immune responses to a bacterial dnaJ antigenic determinant in patients with early rheumatoid arthritis. Nat Med. 1995;1:448–452. doi: 10.1038/nm0595-448. [DOI] [PubMed] [Google Scholar]
- Borges TJ, Wieten L, van Herwijnen MJ, Broere F, van der Zee R, Bonorino C, van Eden W. The anti-inflammatory mechanisms of Hsp70. Front Immunol. 2012;3:95. doi: 10.3389/fimmu.2012.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukwuocha RU, Zhang B, Lai CJ, Scavulli JF, Albani S, Carson DA, Chen PP. Isolation of an IgG monoclonal anti-dnaJ antibody from an immunoglobulin combinatorial library from a patient with rheumatoid arthritis. J Rheumatol. 1999;26:1439–1445. [PubMed] [Google Scholar]
- Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, Cheetham ME, Chen B, Hightower LE. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa M, Terada K, Kato S, Mori M. (1997) HSDJ, a human homolog of DnaJ, is farnesylated and is involved in protein import into mitochondria. J Biochem 121:890–895 [DOI] [PubMed]
- Koffeman EC, Genovese M, Amox D, Keogh E, Santana E, Matteson EL, Kavanaugh A, Molitor JA, Schiff MH, Posever JO, Bathon JM, Kivitz AJ, Samodal R, Belardi F, Dennehey C, van den Broek T, van Wijk F, Zhang X, Zieseniss P, Le T, Prakken BA, Cutter GC, Albani S. Epitope-specific immunotherapy of rheumatoid arthritis: clinical responsiveness occurs with immune deviation and relies on the expression of a cluster of molecules associated with T cell tolerance in a double-blind, placebo-controlled, pilot phase II trial. Arthritis Rheum. 2009;60:3207–3216. doi: 10.1002/art.24916. [DOI] [PubMed] [Google Scholar]
- Krzewski K, Kunikowska D, Wysocki J, Kotlarz A, Thompkins P, Ashraf W, Lindsey N, Picksley S, Głośnicka R, Lipińska B. Characterization of the anti-DnaJ monoclonal antibodies and their use to compare immunological properties of DnaJ and its human homologue HDJ-1. Cell Stress Chaperones. 2003;8:8–17. doi: 10.1379/1466-1268(2003)8<8:COTAMA>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzik-Dumke U, Schick C, Rzepka R, Melchers I. Overexpression of human homologs of the bacterial DnaJ chaperone in the synovial tissue of patients with rheumatoid arthritis. Arthritis Rheum. 1999;42:210–220. doi: 10.1002/1529-0131(199902)42:2<210::AID-ANR2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA—the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6:727–730. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- Massa M, Passalia M, Manzoni SM, Campanelli R, Ciardelli L, Yung GP, Kamphuis S, Pistorio A, Meli V, Sette A, Prakken B, Martini A, Albani S. Differential recognition of heat-shock protein dnaJ-derived epitopes by effector and Treg cells leads to modulation of inflammation in juvenile idiopathic arthritis. Arthritis Rheum. 2007;56:1648–1657. doi: 10.1002/art.22567. [DOI] [PubMed] [Google Scholar]
- Qiu X-B, Shao Y-M, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha B, Lee S, Cyr DM. The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure. 2000;8:799–807. doi: 10.1016/S0969-2126(00)00170-2. [DOI] [PubMed] [Google Scholar]
- Sterrenberg JN, Blatch GL, Edkins AL. Human DNAJ in cancer and stem cells. Cancer Lett. 2011;312:129–142. doi: 10.1016/j.canlet.2011.08.019. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Noguchi S, Arakawa H, Tokida T, Hashimoto M, Satow Y. Peptide-binding sites as revealed by the crystal structures of the human Hsp40 Hdj1 C-terminal domain in complex with the octapeptide from human Hsp70. Biochemistry. 2010;49:8577–8584. doi: 10.1021/bi100876n. [DOI] [PubMed] [Google Scholar]
- Szewczyk B, Harper DR. Use of blotted proteins as immunogens. In: Dunbar B, editor. Protein blotting—a practical approach. Oxford: IRL Press; 1994. pp. 189–205. [Google Scholar]
- Terada K, Mori M. Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J Biol Chem. 2000;275:24728–24734. doi: 10.1074/jbc.M002021200. [DOI] [PubMed] [Google Scholar]
- Terada K, Kanazawa M, Bukau B, Mori M (1997) The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J Cell Biol 139:1089–1095 [DOI] [PMC free article] [PubMed]
- Tukaj S, Kotlarz A, Jozwik A, Smolenska Z, Bryl E, Witkowski JM, Lipinska B. Hsp40 proteins modulate humoral and cellular immune response in rheumatoid arthritis patients. Cell Stress Chaperones. 2010;15:555–566. doi: 10.1007/s12192-010-0168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukaj S, Kotlarz A, Jóźwik A, Smoleńska Z, Bryl E, Witkowski JM, Lipińska B. Cytokines of the Th1 and Th2 type in sera of rheumatoid arthritis patients; correlations with anti-Hsp40 immune response and diagnostic markers. Acta Biochim Pol. 2010;57:327–332. [PubMed] [Google Scholar]
- van Eden W, Thole JE, van der Zee R, Noordzij A, van Embden JD, Hensen EJ, Cohen IR. Cloning of the mycobacterial epitope recognized by T lymphocytes in adjuvant arthritis. Nature. 1988;331:171–173. doi: 10.1038/331171a0. [DOI] [PubMed] [Google Scholar]
- van Eden W, van der Zee R, Prakken B. Heat-shock proteins induce T-cell regulation of chronic inflammation. Nat Rev Immunol. 2005;5:318–330. doi: 10.1038/nri1593. [DOI] [PubMed] [Google Scholar]
- Wieten L, Broere F, van der Zee R, Koerkamp EK, Wagenaar J, van Eden W. Cell stress induced HSP are targets of regulatory T cells: a role for HSP inducing compounds as anti-inflammatory immuno-modulators? FEBS Lett. 2007;581:3716–3722. doi: 10.1016/j.febslet.2007.04.082. [DOI] [PubMed] [Google Scholar]
- Wieten L, van der Zee R, Spiering R, Wagenaar-Hilbers J, van Kooten P, Broere F, van Eden W. A novel heat-shock protein coinducer boosts stress protein Hsp70 to activate T cell regulation of inflammation in autoimmune arthritis. Arthritis Rheum. 2010;62:1026–1035. doi: 10.1002/art.27344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 47.5 kb)