Abstract
Using simultaneous measurements from multiple channels of a radio-frequency coil array, magnetic resonance inverse imaging (InI) can achieve ultra-fast dynamic functional imaging of the human with whole-brain coverage and a good spatial resolution. Mathematically, the InI reconstruction is a generalization of parallel MRI (pMRI), which includes image space and k-space reconstructions. Because of the auto-calibration technique, the pMRI k-space reconstruction offers more robust and adaptive reconstructions compared to the image space algorithm. Here we present the k-space InI (K-InI) reconstructions to reconstruct the highly accelerated BOLD-contrast fMRI data of the human brain to achieve 100 ms temporal resolution. Simulations show that K-InI reconstructions can offer 3D image reconstructions at each time frame with reasonable spatial resolution, which cannot be obtained using the previously proposed image space minimum-norm estimates (MNE) or linear constraint minimum variance (LCMV) spatial filtering reconstructions. The InI reconstructions of in vivo BOLD-contrast fMRI data during a visuomotor task show that K-InI offer 3 to 5 fold more sensitive detection of the brain activation than MNE and a comparable detection sensitivity to the LCMV reconstructions. The group average of the high temporal resolution K-InI reconstructions of the hemodynamic response also shows a relative onset timing difference between the visual (first) and somatomotor (second) cortices by 400 ms (600 ms time-to-peak timing difference). This robust and sensitive K-InI reconstruction can be applied to dynamic MRI acquisitions using a large-n coil array to improve the spatiotemporal resolution.
Keywords: Event-related, fMRI, InI, Visual, MRI, Neuroimaging, K-InI, Inverse solution, MEG, EEG, Electroencephalography, Magnetoencephalography, Latency, Timing, Rapid imaging, Auto-calibration
Introduction
A radio-frequency (RF) coil array with multiple receivers has been first invented to improve the signal-to-noise ratio (SNR) of MRI (Hyde et al., 1986; Roemer et al., 1990). However, other than SNR improvement, the RF coil array leads to the development of parallel MRI (pMRI) using simultaneous acquisitions from all channels of an array to achieve accelerated data acquisition and/or spatial resolution enhancement (Pruessmann et al., 1999; Sodickson and Manning, 1997). The reconstruction of pMRI data can be categorized into either image space methods, such as SENSE (Pruessmann et al., 1999), or k-space methods, such as SMASH (Sodickson and Manning, 1997) and GRAPPA (Griswold et al., 2002). In functional brain imaging, pMRI has been dominantly combined with the gradient-echo echo-planar imaging (EPI) (Mansfield, 1977) to achieve a higher sampling rate, a higher spatial resolution (Preibisch et al., 2003), reduced geometrical distortion (Schmidt et al., 2005), or a reduced acoustic noise level (de Zwart et al., 2002).
The image reconstruction of pMRI may need explicit coil sensitivity to complete the image reconstruction. Examples include SMASH, SENSE, SPACE-RIP (Kyriakos et al., 2000). However, accurate coil sensitivity map estimation is practically difficult. Error in the estimated coil sensitivity maps can propagate to the reconstructed images. To mitigate this challenge, auto-calibration scans (ACS) have been suggested by AUTO-SMASH (Jakob et al., 1998), PILS (Griswold et al., 2000), and GRAPPA (Griswold et al., 2002). These methods empirically measure a small portion of the fully gradient encoded data, which satisfy the Nyquist sampling theorem to give the desired spatial resolution and FOV, in order to estimate the necessary coefficients to interpolate missing data in the accelerated scans. Since the coil sensitivity embedded in the ACS is implicitly used in the image reconstruction, the reconstructed images are immune from any errors arising from explicit coil sensitivity map estimation. Furthermore, GRAPPA and PILS can achieve coil-by-coil reconstructions. Individual coil images can be later combined in different ways to achieve the optimal performance.
Without considering the finite wavelength effect governed by the electromagnetic theory, increasing the number of channels in an RF coil array can further improve the spatiotemporal resolution of pMRI (Ohliger et al., 2003; Wiesinger et al., 2004). Previously, we have introduced the magnetic resonance inverse imaging (InI) to achieve the ultra-fast functional MRI (fMRI) of the human brain in tasks (Lin et al., 2006; Lin et al., 2008b; Lin et al., 2008c). Mathematically, InI generalizes the pMRI reconstructions from an over-determined linear system to an under-determined linear system in order to reduce the time in k-space traversal and therefore to achieve an unprecedented temporal resolution. InI is closely related to the MR-encephalography (Hennig et al., 2007). Our previous InI reconstructions using either the minimum-norm estimates (MNE) or the linear-constraint minimum variance (LCMV) beamformer spatial filtering are both implemented in the image space. To reveal only the relative changes in the task fMRI, we can reconstruct images by the “in vivo sensitivity” approach (Sodickson, 2000), which uses the fully gradient encoded data themselves as the coil sensitivity maps. However, we cannot reconstruct individual images at each time instant since the pure coil sensitivity maps cannot be measured.
In this study, we investigate the k-space InI (K-InI) reconstruction, which is based on the GRAPPA reconstruction of the pMRI data. The goal of K-InI is to offer robust and sensitive coil-by-coil reconstruction of the highly accelerated fMRI acquisitions using a large-N coil array. Different from MNE or LCMV InI reconstructions, we expect that K-InI can offer 3D volumetric reconstruction at each time instant in addition to the estimates of dynamic changes and the associated statistical inferences. Mathematically, K-InI is a generalization of GRAPPA reconstruction to allow reconstruction of data from an underdetermined system in k-space. In the following sections, we introduce the K-InI reconstruction and quantitatively characterize the spatial resolution and localization accuracy of the K-InI using synthetic simulation data. We then demonstrate the acquisitions and K-InI reconstructions in an event-related visuomotor fMRI experiment for high temporal resolution (100 ms) 4D dynamic statistical characterization of task-related BOLD-contrast responses.
Methods
Participants
Ten healthy participants with normal or corrected-to-normal vision were recruited for the study. Informed consent for these experiments was obtained from each participant approved by the Institutional Review Board of our institutes.
Task
Our visuomotor task required the participants to flex right hand fingers upon perceiving a high-contrast hemifield (right field) visual checkerboard reversing at 8 Hz. The motor task was sequential finger flexion between D1–D3, D1–D5, D1–D2, and D1–D4 (D1: thumb, D2: index finger, D3: middle finger, D4: ring finger, D5: little finger). The purpose of this rather complicated motor task is to elicit a stronger hemodynamic response. The checkerboard subtended 20° of visual angle and was generated from 24 evenly distributed radial wedges (15° each) and eight concentric rings of equal width. The stimuli were generated using the Psychtoolbox (Brainard, 1997; Pelli, 1997). The reversing checkerboard stimuli were presented in 500 ms epochs and the onset of each presentation epoch was randomized with a uniform distribution of inter-stimulus intervals varying from3 to 16 s (average inter-stimulus-interval: 10 s). For event-related fMRI data analyzed using a general linear model (GLM), it is useful to jitter the event onsets in order to optimize the estimates of the HRF by reducing its variance (Dale, 1999). Twenty-four stimulation epochs were presented during four 240 s runs, resulting in a total of 96 stimulation epochs per participant. The choice for 3 to 16 s inter-stimulus intervals was made by consideration of the duration of the HRF and practical concerns on accommodating 24 stimulus events within a 240 s run.
Image data acquisition
MRI data were collected with a 3 T MRI scanner with a 32-channel coil array (Tim Trio, Siemens Medical Solutions, Erlangen, Germany). The InI reference scan was collected using a single-slice echo-planar imaging (EPI) readout, exciting one thick coronal slab covering the entire brain (FOV 256 ×256 ×256 mm; 64×64×64 image matrix) with the flip angle set to the Ernst angle of 30° for the gray matter (considering the T1 of the gray matter is 1 s at 3 T). Partition phase encoding was used to obtain the spatial information along the anterior– posterior axis. The EPI readout had frequency and phase encoding along the superior–inferior and left–right axes respectively. We used TR=100 ms, TE=30 ms, bandwidth=2604 Hz and a 12.8 s total acquisition time for the reference scan, consisting of 64 TRs and two repetitions allowing the coverage of a volume comprising 64 partitions.
For the InI functional scans, we used the same volume prescription, TR, TE, flip angle, and bandwidth as for the InI reference scan. The principal difference was that the partition phase encoding was removed so that the full volume was excited, and the spins were spatially encoded by a single-slice EPI trajectory, resulting in a coronal X/Z projection image with spatially collapsed projection along the anterior–posterior direction. The K-space InI reconstruction algorithm, described in the next section, was then used to estimate the spatial information along the anterior–posterior axis. In each run, we collected 2400 measurements after collecting 32 measurements in order to reach the longitudinal magnetization steady state. A total of four runs of data were acquired from each participant.
In addition to the InI reference and functional scans, structural MRI data for each participant were obtained in the same session using a high-resolution T1-weighted 3D sequence (MPRAGE, TR/TE/flip=2530 ms/3.49 ms/7°, partition thickness=1.33 mm, matrix=256×256, 128 partitions, FOV=21 ×21 cm). Using these data, the location of the gray–white matter boundary for each participant was estimated with an automatic segmentation algorithm to yield a triangulated mesh model with approximately 340,000 vertices (Dale et al., 1999; Fischl et al., 2001; Fischl et al., 1999). This mesh model was then used to facilitate mapping of the structural image from native anatomical space to a standard cortical surface space (Dale et al., 1999; Fischl et al., 1999). To transform the functional results into this cortical surface space, the spatial registration between the InI reference and the native space anatomical data was calculated by FSL (http://www.fmrib.ox.ac.uk/fsl), estimating a 12-parameter affine transformation between the volumetric InI reference and the MPRAGE anatomical space. The resulting spatial transformation was subsequently applied to each time point of the reconstructed K-InI hemodynamic estimates to spatially transform the signal estimates to a standard cortical surface space (Dale et al., 1999; Fischl et al., 1999). Before the spatial transformation, the reconstructed K-InI data were spatially smoothed with a 10 mm full-width-half-maximum (FWHM) 3D Gaussian kernel. This smoothing kernel was chosen to be 2.5 times the native image resolution (4 mm in our reference scan).
Spatiotemporal reconstruction of the InI data
The collection of the InI data set across all channels in an RF coil array and across all time points can be considered as two separate processes in both spatial and temporal domains. Physically, the unknown spin density distribution was spatially modulated by the sensitivity profiles (B1 maps) of the individual channels of the RF coil array; The time series measurements at each channel of the coil array recorded the MR signals including stimulus-induced response as well as other confounds over time, such as system drift and motion artifacts. In this section, we describe how to process the spatiotemporal InI data in order to ultimately yield 3D whole brain images at a high temporal resolution. First, we first processed the data in the time domain in order to derive the coefficients of the hemodynamic response functions (HRF) basis function for each channel of the coil array separately. This was implemented by using Finite-Impulse-Response (FIR) basis function and General Linear Model. FIR basis functions assume no particular shapes of the HRF and they are particularly appropriate for InI, which provides a high degree of freedom in the time domain to characterize dynamic responses. Second, we reconstructed the distribution of the HRF basis function coefficient in the spatial domain using our proposed k-space InI reconstruction (K-InI). Volumetric reconstruction was calculated from the collection of the basis function coefficients across all channels of the RF coil array at each individual particular time instant. Using the FIR basis, we can get the 3D distribution of the HRF covering the whole brain at each TR (100 ms in this study) independently. This first temporal then spatial reconstruction of the InI data is mathematically valid because these two processes are linear. One practical benefits of this order of reconstructing the InI data is that the temporal reconstruction can first reduce the dataset size by eight fold (2400 samples per run to 300 samples, the number of our FIR basis function over the duration of our chosen HRF).
Temporal InI reconstruction: estimation of the coefficients of the HRF basis function in the projection image
The accelerated InI acquisition and the reference scans were processed from k-space to the image domain, using 2D and 3D fast Fourier transformations, respectively. The reference scan in each channel of the coil array was synthetically averaged across partitions in order to simulate the InI acquisitions, whose phase are subsequently used in the accelerated InI acquisition in order to estimate the magnitude changes in the BOLD-contrast fMRI (Lin et al., 2008b; Lin et al., 2008c). After phase-correction of the projection image in the original time series from each coil array channel, we calculated the HRF for each image voxel in the projection images. Specifically, we used a general linear model (GLM) and a basis set of finite impulse response (FIR) to estimate HRF elicited by the stimulus in each channel of the coil array. The basis set was temporally synchronized to the onset of the visual stimulus, spanning a 30 s period that included a 6 s pre-stimulus baseline and a 24 s post-stimulus interval. Given TR=100 ms and a chosen 30 s as the duration for HRF, we had 300 unknown coefficients for the FIR basis functions. HRFs were estimated from the coefficients for each FIR basis in the GLM using a least squares minimization procedure. Additionally, we estimated the noise covariance matrix across different channels of an RF coil array based on the GLM residuals. After model estimation, the coefficients of the FIR basis set across the 30 s epoch for each of the 32 receiver channels were used for the subsequent K-InI reconstruction. At each time instant, the FIR coefficient is a complex number including phase and magnitude information and it represents the instantaneous BOLD-contrast in one particular channel.
Spatial InI reconstruction: estimation of the spatial distribution of the HRF basis function using k-space InI (K-InI) reconstruction
Review of parallel MRI reconstruction in k-space
The parallel imaging reconstruction in the k-space has been proposed by several approaches, such as SMASH (Sodickson and Manning, 1997) and GRAPPA (Griswold et al., 2002). Essentially, these methods estimate those k-space samples not acquired in the accelerated scan by linearly interpolating from other k-space samples acquired in the accelerated scan. Without the loss of generality, we take the GRAPPA formulation here to express the not-acquired k-space data with phase encoding ky = k + m·Δk from coil channel j as a linear combination of the acquired k-space data with ky = k from all channels in the accelerated acquisition:
| (1) |
where nc is the total number of channels in the coil array; βc,m is the unknown fitting coefficients for coil channel c to estimate the data in channel j with phase encoding m·Δk; and Δk is the minimal separation in the phase encoding direction (ky) defined by the FOV, spatial resolution and the Nyquist sampling theorem. Importantly, Eq. (1) describes a linear relationship between data with different phase encodings within a vicinity, which consists of a “block” in GRAPPA (Griswold et al., 2002). For example, a block of four describes the relationship between any four consecutive phase encoding lines using Eq. (1) with m ranging between 1 and 3. One feature of GRAPPA and auto-SMASH (Jakob et al., 1998) is that if some data at the RHS of Eq. (1) are actually acquired for some blocks, βc,m can be first estimated via optimizing the residuals of Eq. (1). And subsequently βc,m can be used to interpolate the not-acquired k-space data in other blocks where the RHS of Eq. (1) are not acquired in the accelerated scan. The data at the RHS of Eq. (1) used for estimating fitting coefficients are called “auto-calibrating scans (ACS)”.
| (2) |
Collecting all data in different frequency encoding steps and all channels in the coil array, we can write Eqs. (1) and (2) using matrices and vectors.
| (3) |
| (4) |
In practice, and AACS are first empirically measured to estimate βj,m, which are then multiplied with AACC constructed from the collected data in the accelerated scan to reconstruct the missing data . As the number of channels in a RF coil array increases, the matrices AACS and AACC will grow horizontally. However, data with different frequency encodings can be included as independent measurements in Eq. (4) and usually a few blocks are used in the auto-calibrating scans. Thus and AACS will also grow vertically as the results of data concatenation. Overall, Eq. (4) usually represents an over-determined linear system as the results of a “long” AACS (more rows than columns). Subject to a constraint, such as the minimal residual power, the fitting coefficients βj,m, can be estimated uniquely.
InI reconstruction in k-space (K-InI)
The InI acquisition achieves massive acceleration by collecting minimal k-space data (one phase encoding line) using a large-N coil array. Therefore the matrix AACS in Eq. (4) is no longer a “long” matrix but a “wide” matrix, since InI usually uses a coil array with many channels and totally 1 block including all partition encodings is collected in the auto-calibrating scan. Practically, AACS includes all data collected with zero partition encoding across all channels of the coil array during the auto-calibrating scan. The auto-calibrating scan also provides describing the measurement at channel j and at m partition encoding steps. Given the size of AACS and , the estimation of βj,m is translated from an over-determined linear system to an under-determined linear system. We cannot uniquely determine the values of βj,m using the constraint of minimizing the residual power. However, we can impose another constraint of, for example, minimizing the power of the fitting coefficients, in order to estimate βj,m. This gives the following optimization formulation:
| (5) |
which can be solved analytically
| (6) |
where ‖•‖2 is the ℓ2-norm, the superscript H denotes the complex conjugate and transpose, C is the noise covariance matrix among different channels in the coil array, and λ is a regularization parameter. From our previous experience, λ can be reasonably estimated from the pre-specified signal-to-noise ratio (SNR) of the measurement (Lin et al., 2006; Lin et al., 2008b):
| (7) |
Here Tr(•) represents the trace of the matrix. This is similar to the minimum-norm estimate (MNE) of the InI data (Lin et al., 2006; Lin et al., 2008b). However, the reconstruction is now done in the k-space directly without involving explicit coil sensitivity estimates.
Provided with the estimated β̂j,m and empirically collected AACC in the accelerated scan, the missing data can be reconstructed sequentially for individual channels and individual m·Δk in the phase encoding direction using Eq. (3). Note that the same set of β̂j,m was used to reconstruct all HRF coefficients for the same channel (channel j) in the coil array at different time instants. And different sets of the β̂j,m were used to reconstruct the time series at different channels. The combined images can thus be calculated using the sum-of-squares:
| (8) |
where FT{•} denotes the discrete Fourier transform.
Statistical inference
In fMRI, we are interested in estimating the statistical significant activity spatiotemporally. Eq. (6) can be used to reconstruct InI data in 3D time point by time point. To facilitate statistical inference, the noise levels in the reconstructed images were estimated from the baseline data after K-InI reconstruction. A baseline noise volume can be calculated after defining a baseline interval, which was chosen as 4 to 0 s before the onset of the visual stimulus in our study. Using these noise estimates, dynamic statistical parametric maps (dSPMs) can be derived as the time-point by time-point ratio between the K-InI reconstruction values and the baseline noise estimates. dSPMs should be t distributed under the null hypothesis of no hemodynamic response (Dale et al., 2000). When the number of time samples used to calculate the noise covariance matrix C exceeds 100, the t distribution approaches the unit normal distribution and the individual t-statistics approximate z-scores.
Spatial resolution analysis
We performed numerical simulations to evaluate the spatial resolution and localization accuracy of the K-InI reconstructions. The reference data and the noise covariance matrix were obtained from empirical data. The simulation procedure began by creating a source ROIs. These ROIs were manually selected at the primary visual (V1), auditory (A1), and somatomotor (SM1) cortices in the left hemisphere. We then estimated the ideal measurements from all coil array channels by computing the product of the reference data and individual ROI. We created 100 realizations of synthetic noise with spatial coloring according to the noise covariance matrix. At a specified SNR, the noise was scaled and subsequently added to the idealized measurements to generate the synthetic measurements. K-InI reconstructions were subsequently calculated using Eqs. (3) and (4).
The performance of the reconstructions was quantified using a similar procedures in MEG/EEG source analysis (Dale et al., 2000; Liu et al., 1998; Liu et al., 2002) and our previous InI analysis (Lin et al., 2008b; Lin et al., 2008c). We estimated the average point spread function (aPSF) at each location to quantify the spatial distribution of the reconstruction:
| (9) |
where |di| indicates the distance between the reconstructed source at location i and the center of mass of a simulation ROI ρ⃑. χK-InI(i) represents vector entries in the K-InI reconstruction with values exceeding 50% of the maximum, and l is the number of voxels to be spatially resolved by the K-InI reconstructions. This procedure allows the estimation of the full-width-half-maximum (FWHM) of the point spread function.
Similar to other InI reconstructions, since K-InI is an ill-posed inverse problem, the reconstructed image may not accurately reflect the original spatial distribution of spins contributing to the actual measurements. Thus, we next explored the localization accuracy by estimating discrepancies between the reconstructed and original sources. Quantification of localization accuracy was done by calculating the distance between the center of mass of the K-InI reconstruction and the simulated sources:
| (10) |
where r⃑(i) is the 3D coordinate of the K-InI reconstruction. Since the K-InI inverse depends on both the SNR and the measurement data, the SNRs were parametrically varied from 0.1 to 100. For comparison, we also calculated the reconstructions using the minimum-norm estimates (MNE), the dynamic statistical parametric maps of the MNE (MNE-dSPM) (Lin et al., 2008b), and the spatial filtering using the linear constraint minimum variance (LCMV) beamformer (Lin et al., 2008c).
The image reconstruction and statistical analysis procedures were implemented in Matlab (The Mathworks, Natick, MA).
Results
Spatial resolution analysis of simulated data
Spatial resolution
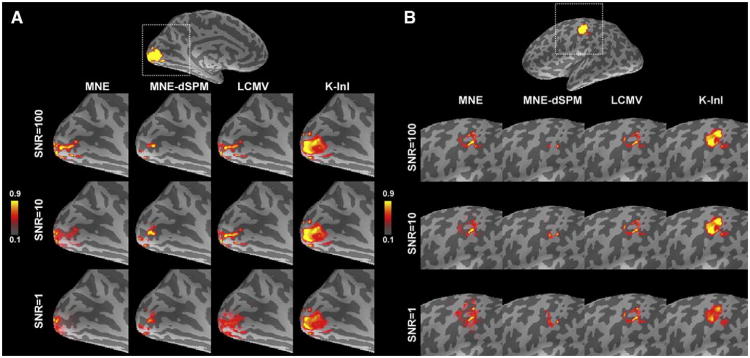
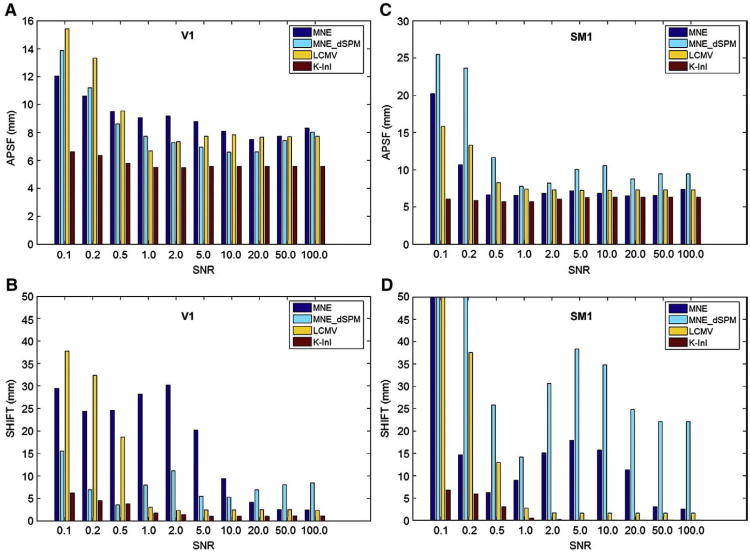
Fig. 1A shows the MNE, MNE-dSPM, LCMV, and K-InI reconstructions of the simulation source at the primary visual cortex (V1). To illustrate the spatial distribution, each reconstruction was linearly scaled between 1 and 0. As SNR varies between 1 and 100, all four reconstructions show high values around the occipital pole. However, compared to MNE, MNE-dSPM and LCMV, only K-InI gives the reconstruction with a comparable size to that of the simulation source. This feature of preserving the size of the simulated source was found stable at low (SNR= 1) and high (SNR= 100) SNRs. To further quantify the reconstruction performance, we calculated the aPSF and SHIFT metrics shown in Figs. 2A and B. As SNRs increase from 0.1 to 100, generally the aPSF metrics decrease for all reconstruction methods. We found that K-InI reconstructions have the aPSF smaller than 6 mm across SNRs. And K-InI has the smallest aPSF compared to MNE, MNE-dSPM, and LCMV reconstructions at individual SNRs. This suggests that the K-InI reconstruction has the least spatial dispersion. Fig. 2B shows the SHIFT metrics. The localization accuracy of the MNE and LCMV reconstructions closely correlates with the SNRs: a smaller SHIFT metric is found at a higher SNR. K-InI does not have such a strong dependency on SNR and it has the smallest SHIFT metric compared to MNE, MNE-dSPM, and LCMV reconstructions at individual SNR. The localization precision for K-InI was found to be approximately 2 mm as SNR higher than 1. MNE-dSPM generally has a smaller SHIFT value than MNE at low SNRs as the result of noise normalization. However, at higher SNR (SNR>20), MNE can outperform MNE-dSPM. Overall, K-InI has the highest localization accuracy (SHIFT<2 mm).
Fig. 1.
Simulation sources and reconstructions at visual (A) and somatomotor (B) cortices in the left hemisphere. The brain was rendered in an inflated surface model where dark and light gray represent sulci and gyri respectively. The MNE, MNE-dSPM, LCMV, and K-InI reconstructions of visual and somatomotor cortices at different SNRs are linearly scaled between 0 and 1 to illustrate the spatial distribution.
Fig. 2.
The aPSF (A and C) and SHIFT (B and D) metrics for MNE, MNE-dSPM, LCMV, and K-InI reconstructions at visual (A and B) and somatomotor (C and D) cortices at different SNRs.
In addition to the visual cortex, we also investigate the spatial dispersion and localization accuracy of the primary somatosensory cortex (SM1). Fig. 1B shows the location of the simulated SM1 source and MNE, MNE-dSPM, LCMV, and K-InI reconstructions. Similar to the results in the V1 simulations, all reconstructions are reasonably localized at the pre-central gyrus, central sulcus, and post-central gyrus. Notably, K-InI reconstructions have a similar size compared to the simulated source, while MNE, MNE-dSPM, and LCMV reconstructions have smaller and distorted size. Again, the ability of K-InI reconstruction to preserve the size of the simulation source is constant over SNRs (SNR>1). Figs. 2C and D summarizes the aPSF and SHIFT metrics for the simulation over the SM1. For MNE, MNE-dSPM and LCMV the spatial dispersion quantified by aPSF shows a strong SNR dependency. K-InI reconstructions have stable aPSF across SNRs, presumably due to the auto-calibrating process in the reconstruction (Eqs. (3) and (4)). As SNR is higher than 1, all methods have aPSF less than 10 mm. Still, K-InI outperforms all other reconstructions by giving aSPF approximately 5 mm at all SNRs. The localization accuracy of MNE and MNE-dSPM are worse than that of LCMV and K-InI reconstructions. Both LCMV and K-InI have mislocalization of less than 3 mm as SNR is higher than 1. K-InI reconstructions give smaller SHIFT metrics at all SNR than other reconstructions. The difference in aPSF and SHIFT metrics between V1 and SM1 is likely attributed to the relative position between the simulated source and the coil array. A shorter distance between the simulated source and the coil array, such as SM1, can provide more disparate spatial information from all channels of the coil array and therefore helpful to correctly reconstruct the image.
Reconstruction of individual time frame
Fig. 3 shows the reconstruction of one time point of the K-InI reconstruction at different axial slices. For comparison, the MNE reconstruction and the sum-of-squares image in the reference scan are also shown. LCMV cannot reconstruct the individual volumes of the time series acquisition, since the LCMV reconstructions require a data covariance matrix, which can only be estimated from an interval of the time series containing the dynamic changes (Lin et al., 2008c). Such a data covariance requirement imposes a constraint of time integration and thus it is contradictory to the goal of obtaining instantaneous reconstructed volumes here. As expected, due to the auto-calibrating nature and the coil-to-coil reconstruction, K-InI reconstructions shows much improved quality in terms of anatomical features and constrict compared to the MNE reconstruction. Note that the InI acquisition generated projection images along the left–right direction. Therefore the K-InI reconstructions still demonstrate spatial smearing as the result of resolving the spatial information solely from the RF coils.
Fig. 3.
Reconstructed instantaneous images using MNE and K-InI. LCMV does not offer instantaneous image reconstruction because of no data covariance. For comparison, the sum-of-squares (SoS) images from the reference are also shown.
Functional activation
Strong task-related activity effects can also be easily seen in group average time series. Fig. 4 shows the medial view of 100 ms duration frames of InI MNE-dSPM, LCMV, and K-InI t-values at the occipital lobe averaged over 10 participants. The individual frames of this group average show progressively increasing activity starting at 1.0 s after the stimulus onset (critical threshold t>2; uncorrected p-value<10−2). The signal returns to baseline approximately 6.5 s after stimulus onset. Both MNE-dSPM and K-InI reconstructions show similar spatial distribution of the visual cortex activity. But MNE-dSPM reconstructions are relatively weaker than K-InI reconstructions. Note that both LCMV and K-InI reconstructions offer very similar estimates of visual cortex activity. Particularly, some deactivation (negative values of the statistical maps) was found around the occipito-parietal junction in K-InI reconstructions. This deactivation location was found to be consistent with the default mode of the brain during the resting state (Raichle et al., 2001).
Fig. 4.
The snapshots of the left hemispheric visual cortex activation in MNE-dSPM, LCMV, and K-InI reconstructions. At top of the figure, we illustrate the cropped ROI as a gray rectangle over the medial aspect of the inflated left hemisphere surface.
The visual cortex time course of the MNE-dSPM, LCMV, and K-InI t-values from the group average are shown in Fig. 5. The shape of this average time series resembles between each other. We observed a reduced variability in the group time course as compared with the individual time courses. Peak task-related activity was found 3.5 s after stimulus onset. Both LCMV and K-InI reconstructions show an approximately three-fold increase in detection sensitivity compared to the MNE-dSPM reconstructions (LCMV: 5.8, K-InI: 5.6; MNE-dSPM: 1.8).
Fig. 5.
The average time courses of the MNE-dSPM, LCMV, spatially smoothed K-InI, and spatially unsmoothed K-InI reconstructions at the visual cortex. The visual cortex ROI is shown in light blue at the medial aspect of the inflated left hemisphere surface. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also concerned that the spatial smoothing procedure can affect the reconstructions. The smoothing kernel we used is about 2.5× larger than the inherent resolution (4 mm). Such a smoothing kernel is common to fMRI analysis with EPI data, where the nominal in-plane spatial resolution was about 3–4 mm and commonly a Gaussian kernel with full-width-half-maximum of 6 to 12 mm was used to spatially smooth the data in order to increase the signal-to-noise ratio. The smoothing was done after the K-InI reconstruction. And the purpose of the spatial smoothing is for visualization only. To demonstrate the reconstruction without smoothing, we also calculated the K-InI reconstruction without smoothing (Fig. 5). The results show similar time courses of the peak t statistics (smoothed K-InI: 5.6; unsmoothed K-InI: 5.5). Thus we conclude that the spatial smoothing only changes the results marginally.
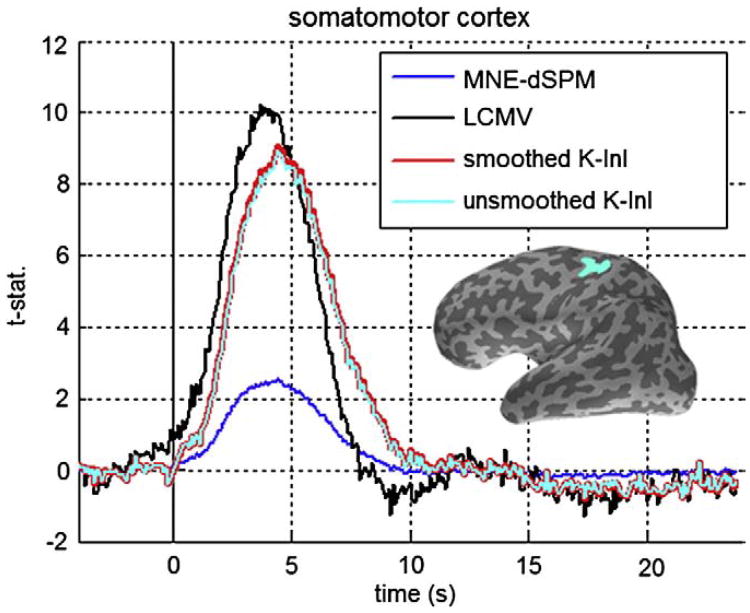
We also studied the hemodynamic responses at the motor cortex. Fig. 6 shows the lateral view of 100 ms duration frames of InI MNE-dSPM, LCMV, and K-InI t-values around the central sulcus averaged over 10 participants. Two clusters of positive responses at the premotor cortex and primary somatomotor cortex were found in both MNE-dSPM and K-InI reconstructions. Comparing between these reconstructions, we found that K-InI reconstructions show much stronger significant responses than MNE-dSPM: between the interval of 3.0 and 5.0 s after the onset of the visual stimulus, MNE-dSPM values are around 2.0, while K-InI values are around 7.0. LCMV reconstructions offer the highest values of statistical significance of somotomotor cortex activation. But the spatial distribution of the activated somatomotor cortexis very similar between LCMV and K-InI reconstruction. All three reconstructions also detected the deactivation in the secondary somatomotor cortex around the inferior part of the central sulcus.
Fig. 6.
The snapshots of the left hemispheric somatomotor cortex activation in MNE-dSPM, LCMV, and K-InI reconstructions. At top of the figure, we illustrate the cropped ROI as a gray rectangle over the lateral aspect of the inflated left hemisphere surface.
Fig. 7 shows the time courses of MNE-dSPM, LCMV, spatially smoothed and spatially un-smoothed K-InI reconstructions over the somatomotor cortex. Qualitatively, all time courses are similar, while minor differences exist still: MNE-dSPM peaks at 3.5 s after the onset of the visual stimulus, and K-InI peaks at 4.0 s after the onset of the visual stimulus. LCMV time courses offer the highest peak t statistics estimate (10.2). Consistent with the visual cortex time courses, we found that K-InI reconstructions give 3.6-fold higher statistical significance than MNE-dSPM: the peak value of smoothed K-InI is 9.1, the peak value of unsmoothed K-InI is 8.9, and MNE-dSPM peak value is 2.5.
Fig. 7.
The average time courses of the MNE-dSPM, LCMV, spatially smoothed K-InI, and spatially unsmoothed K-InI reconstructions at the somatomotor cortex. The somatomotor cortex ROI is shown in light blue at the lateral aspect of the inflated left hemisphere surface. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
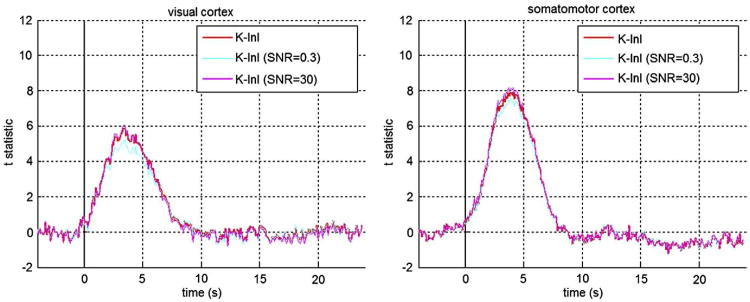
Since K-InI reconstructions depend on a regularization parameter to estimate the interpolation coefficient (Eqs. (3) and (4)), we studied the sensitivity of the K-InI reconstructions by scaling the regularization parameter by 10 or 0.1 fold. Fig. 8 shows the respective time courses at visual and somatomotor cortices. We found that the time courses at either visual or somatomotor cortices are not sensitivity to the regularization parameter: the peak values of the time courses at the visual cortex changes by +5% ((peak t statistics=5.1) or -12% (peak t statistics=6.1) at SNR=0.3 and SNR=30 respectively. Similarly, the peak values of the time courses at the somatomotor cortex changes by +4% ((peak t-statistics=8.1) or -5% (peak t statistics=7.4) at SNR=0.3 and SNR=30 respectively. The shapes of the time courses are similar to each other when we increased or decreased the regularization parameter by 10 folds.
Fig. 8.
The time courses of K-InI reconstruction with 10-fold bigger (light blue) or smaller (pink) regularization parameter at visual and somatomotor cortices. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 9 shows the scaled group average time courses at the visual and somatomotor cortices. To further quantify the timing information of these two waveforms, we first linearly scaled the time courses between 0 and 1. Subsequently, we fitted a canonical hemodynamic response function using two shifted and scaled exponential functions (Glover, 1999). Taking the time arriving the 50% of the peak value as the onset timing, we found that the visual cortex time course leads the somatomor cortex time course by 400 ms (visual cortex: 1.4 s; motor cortex 1.8 s). The timings of reaching the peak value for these two time courses are 3.7 and 4.3 s for visual and motor cortex respectively. Interestingly, both ways of quantifying the timing of two time courses are consistent with the sequential activation of first visual and then somatomotor cortices in this visuomotor task.
Fig. 9.
Detection of relative timing of the group average K-InI reconstructions at visual (blue trace) and somatomotor (red trace) cortices. The time-to-peak and the timing of reaching 50% of the peak values are reported. The figure inlet includes the zoomed time courses between -1 and 4 s after the onset of the visual stimulus. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Discussion
In this study, we introduce the k-space InI (K-InI) reconstruction method to restore the volumetric spatial distribution of dynamic changes of brain activity and the associated statistical inference in a visuomotor fMRI experiment. K-InI is a generalization of GRAPPA reconstruction. K-InI estimates the missing k-space data in the accelerated scan by interpolating the acquired k-space data in the accelerated scan with coefficients estimated from the k-space ACS collected in the reference scan. The results of the detecting functional brain activity (Figs. 4–7) show that K-InI generally outperforms the MNE. LCMV reconstruction can offer a more sensitive detection of brain activity in the visual cortex, but it cannot generate an instantaneous image, since there is no data covariance matrix for each instantaneous measurement and such a data covariance is a necessary parameter in LCMV reconstruction. The better reconstructions from K-InI can be attributed to the ACS, which enforces the data consistency within the data across different channels in a coil array and different k-space locations. On the contrary, the MNE and LCMV reconstructions do not have such an auto-calibrating procedure. For the analysis of fMRI time series, we reconstructed the HRF basis coefficients. The chosen FIR basis actually corresponded to one instantaneous measurement. We were aware that the deconvolution in the GLM can actually correlate (or un-correlate) the measurements across time points implicitly. We were also aware that it is possible to use HRF basis functions showing stronger correlations (for example, the canonical hemodynamic response function and its temporal derivative). However, we want to indicate that the selection of the basis function in the analysis of fMRI time series is independent of the K-InI reconstruction in the spatial domain. And K-InI can actually provide instantaneous reconstruction. This was clearly demonstrated in Fig. 3, where one time point reconstructed volume was shown. From this demonstration we can see that K-InI reconstruction is not restricted to reconstructing the coefficients of the HRFs at each time point relative to a stimulus.
The assumption of K-InI
The derivation of K-InI assumes that the power of the reconstructing coefficients (Eq. (5)) is minimal, which is a pure mathematical constraint. This assumption enables us an analytical and computationally efficient algorithm to estimate these coefficients (Eq. (6)).In Eq. (7) the regularization coefficient was theoretically derived from the tradeoff between the “signal” quantified by the auto-calibrating data and the “noise” C. And we have empirically validated that the regularization parameter suggested in Eq. (7) marginally modulated the reconstructed images (Lin et al., 2008b). K-InI reconstruction, like other InI reconstructions, further assumes that there is no motion between the measurements of the reference auto-calibrating data and the accelerated data. This sensitivity to motion is further discussed in the following “Spatiotemporal coupling of the K-InI reconstruction” section.
Applying K-InI to fMRI time series, we used the same FIR basis functions at all spatial locations in the projection images. As expected, it is possible that two functionally active areas were spatially overlapped in the projection images. However, given the simultaneous measurements from all channels of the coil array, we separated the spatially collapsed functional activity into a 3D space. This was practically achieved by estimating different coefficients for these FIR basis functions for different channels of the coil array individually using the reference auto-calibrating data (across all channels) and the accelerated InI data (for each individual channel separately). Repeating the reconstruction for different channels, K-InI provided 3D whole-brain HRF estimates at each individual coil (coil-by-coil reconstruction). We then combined different channels by calculating the sum-of-squares. Here we assume that the spatial information afforded from the coil array can spatially resolved the projection image along the partition encoding direction. And this spatially information was time-invariant during the experiment. From our data reported here using a 32-channel coil array at 3 T, this assumption was empirically valid.
Relationship between K-InI, MNE and LCMV
Mathematically, the estimation of interpolation coefficients in the K-InI reconstruction (Eq. (4)) is identical to the InI MNE image reconstruction. We attempt to estimate the unknown coefficients (in K-InI) or image (in InI MNE) in an under-determined linear system. Since there are infinite sets of solutions satisfying the forward problem, additional constraint must be imposed to obtain a unique solution. Here the constraint in K-InI is minimizing the power of the coefficients interpolating the missing k-space data. And the reason for choosing this constraint is that it gives a robust solution in an analytic form. Alternatively, we may apply LCMV spatial filtering to estimating the coefficient βj,m in Eqs. (3) and (4) The performance of such k-space LCMV spatial filtering needs further investigation to quantify its spatial resolution and localization accuracy.
While closely related to each other, the MNE, LCMV, and K-InI are still different in a few aspects. First, the K-InI can reconstruct individual coil images, which include phase and magnitude. Both MNE and LCMV reconstructions are image space methods and they offer only one composite image reconstruction. This distinction is not prominent in this study because fMRI is mostly interested in the BOLD-contrast, which differentiates between the baseline and the active states of the human brain based on the difference in image magnitude (Kwong et al., 1992). The phase information, however, can be crucial for other experiments. For example, in our preliminary report on a spectroscopic imaging experiment (Lin et al., 2008a), where repetitive measurements are required to collect off-resonance free induction decays from different metabolites or macromolecules, only K-InI can achieve reasonable reconstruction. The MNE fails to reconstruct spectroscopic imaging data because the phases derived from the ratio between the reconstructed real and imaginary parts of the image are distorted significantly. This is consistent with our spatial resolution analysis (Figs. 1 and 2) and reconstructed images (Fig. 3). The other difference between K-InI and MNE/LCMV is that K-InI offers the flexibility to obtain reconstructed image for individual coil in the array. However, the coil-by-coil reconstruction can be computationally demanding since the amount of calculation is in proportion to the number of channels in an coil array. As the number of channels in an RF coil array increases from conventional 8, 12, 32, and even 96 channels (Wiggins et al., 2005), this computation can be particularly demanding. This technical challenge can be partially mitigated by the array compression approach (Buehrer et al., 2007): Via a few linear combinations of channels in the array, the spatial information can be effectively preserved, while the total number of “channels” can be reduced dramatically to improve the computational efficiency. K-InI is also closely related to the consistency GRAPPA reconstruction (Lustig et al., 2009), which requires the k-space data to be locally consistent across the whole k-space using multiple array measurements. Differently, K-InI estimates and applies this consistency property globally along all data points in the kz (partition encoding) direction.
The dependence of K-InI on the regularization parameter
Estimating the reconstruction coefficient in Eq. (6) requires a regularization parameter during the derivation of the inverse operator. This is because the matrixAACCH AACC can be rank deficient as the result of few channels and/or less independent spatial information offered by the coil array. A rank-deficientAACCH AACC prohibits the calculation of coefficients in Eq. (6) since the necessary matrix inversion does not exist. To address this issue, a fully ranked noise covariance matrix is introduced as a remedy for the rank deficiency. Eq. (7) uses an empirical formula to estimate the regularization parameter. Alternative regularization parameter estimates have been studied extensively. For example, we can use L-curve (Hansen, 1998), or generalized cross validation (Golub et al., 1979) to estimate different λ. However, from empirical data (Fig. 8), we found that the results are marginally depending on the regularization parameter. This is because we estimated 64 unknowns from 32 channels, which was not as ill-posed as electroencelphalograph (EEG) and magnetoencephalography (MEG) source localization, where around 100 sensors/channels are available to estimate current sources of approximately 10,000 locations. We hypothesize that under the electromagnetic approximation of infinite wave, the K-InI reconstructions are less dependent on the regularization parameter at higher fields and using an RF coil array with more channels, since a higher field has a corresponding shorter wavelength and more channels can offer more disparate spatial information to resolve the projection images in the accelerated scan. Nevertheless, further theoretical and empirical studies are required to test this hypothesis and to quantify the upper bound of the maximal channel number in a given field strength for the optimal reconstruction.
Spatiotemporal coupling of the K-InI reconstruction
The K-InI reconstruction is a pure spatial domain reconstruction. It is un-coupled from the temporal domain processing, which was implemented by the General Linear Model (GLM) with the FIR base functions in the analysis of the fMRI time series. However, such separation between spatial and temporal domain is only an ideal case, Practically, the acquisition and reconstruction can be correlated spatiotemporally. For example, consider that there is some motion during the InI data acquisition. Motion artifacts can cause the inconsistency between the fully gradient encoded auto-calibrating data and the accelerated InI data. Such an inconsistency can eventually lead to the increased residuals energy. And consequently the reconstructions can be deteriorated by showing either strong aliasing artifacts or noisy images. In fact, motion artifacts are critical to all InI and parallel MRI time series reconstructions. If there is no auto-calibrating procedure at each instantaneous measurement, the reconstruction kernel (fitting coefficients in K-InI) cannot be accurately used to reconstruct the instantaneous data after the motion. The sensitivity to motion can be empirically controlled by avoiding lengthy measurements (we collected one run for 4 min in this study) and using cushion filling inside the coil array to reduce the explicit motions.
Relative onset latency in K-InI reconstruction
This study first demonstrates the volumetric dynamic brain activity at different cortical areas with a high temporal resolution. Our previous studies only showed the InI time courses at visual cortex in 2D (Lin et al., 2006) and 3D (Lin et al., 2008b; Lin et al., 2008c). And time courses at both visual and somatomotor cortices with clear activation are shown here. Given such a high temporal resolution, we are interested in the relative timing onset across cortical areas. This timing difference is particularly interesting to neurophysiologists to reveal the complex orchestration between brain areas to accomplish tasks and cognition. The result in Fig. 9 shows that for a visuomotor task, the hemodynamic activity at the visual cortex actually precedes that in the motor cortex in a group. While this result is logical since it follows the intuitive causal relationship in a visuomotor task, the ability to resolve sequential brain activity in visual and motor cortices using the “sluggish” BOLD-contrast hemodynamic response is still surprising, since the BOLD-contrast fMRI is a secondary measure of the neuronal activity (Logothetis et al., 2001) and the local vasculature variety may overwhelm the timing information elicited from the neuronal activity. Our preliminary result only finds this sequential activation in the group average data, but not individual data. This is consistent with the previous report (Miezin et al., 2000). Earlier studies show that the hemodynamic responses can actually carry timing information (Huettel and McCarthy, 2001; Menon et al., 1998). Nowadays we are equipped with improved RF coil array, a higher field magnet, and improved data acquisitions and reconstructions. Thus it is possible to resolve this minor latency difference from the BOLD-contrast fMRI. Still, further detail studies are required to ascertain the reliability and the sensitivity to monitor neuronal activity via fMRI vascular responses.
Future development
In our implementation, the InI data always acquired at the same central partition in the Kz direction. This sampling pattern in k-space is time invariant. Other GRAPPA reconstructions have been coupled with the time-varying sampling patterns in order to achieve the optimal spatiotemporal resolutions. For example, T-GRAPPA (Breuer et al., 2005) uses an interleave lattice structure over time to collect the ACS data. k-t GRAPPA further modifies the ACS acquisition to achieve a higher efficiency (Huang et al., 2005). The advantage of these two approaches is that the ACS data can be obtained along the accelerated scans. And such interleave sampling strategies have the benefit of reduced reconstruction noises as the result of trading-off the distribution of the point spread function in both time and spatial domains. The same principle can be applied to K-InI acquisitions and reconstructions: By repetitively collecting data using the multi-shot EVI sequence in the reference scans, each shot consists of an accelerated InI acquisition, and the composite of different shots over time can generate the fully gradient encoded reference data for K-InI reconstruction coefficients estimation.
Recently, sparse MRI (Lustig et al., 2007) has been proposed as an alternative approach to accelerate data acquisitions using either single receiver or multiple receivers. Such compressed sensing (CS) approach attempts to restore randomly sampled data, which generate intrinsically incoherent aliasing artifacts. Subsequently a nonlinear algorithm is used to solve the underlying image. Due to insufficient data samples to satisfy the Nyquist theorem, similar to InI, the CS pMRI is also dealing with an under-determined linear system. Using a cost function formulation, CS image reconstruction solves the image by minimizing the residual errors arising from the random sampling and a prior cost measuring the sparsity of the underlying image via the wavelet transform. In fact, this formulation can be incorporated to either MNE InI or K-InI reconstruction in the future. Specifically, we can replace the constraint of minimizing the ℓ2 norm power of the image (MNE) or the fitting coefficients (K-InI) with a constraint of minimizing the ℓ1 norm in order to achieve more sparse estimates in image space and k-space respectively. However, the detection power, sensitivity to noises, and the localization accuracy needs to be further studied quantitatively in order to select the optimal reconstruction method for different experiments.
Acknowledgments
This work was supported by National Institutes of Health Grants by R01DA14178, R01HD040712, R01NS037462, P41 RR14075, R01EB006847, RO1EB000790, R21EB007298, and the Mental Illness and Neuroscience Discovery Institute (MIND), NSC97-2320-B-002-058-MY3, NSC 98-2320-B-002-004-MY3 (National Science Council, Taiwan), and NHRI-EX99-9715EC (National Health Research Institute, Taiwan).
References
- Brainard DH. The Psychophysics Toolbox. Spat Vis. 1997;10:433–436. [PubMed] [Google Scholar]
- Breuer FA, Kellman P, Griswold MA, Jakob PM. Dynamic autocalibrated parallel imaging using temporal GRAPPA (TGRAPPA) Magn Reson Med. 2005;53:981–985. doi: 10.1002/mrm.20430. [DOI] [PubMed] [Google Scholar]
- Buehrer M, Pruessmann KP, Boesiger P, Kozerke S. Array compression for MRI with large coil arrays. Magn Reson Med. 2007;57:1131–1139. doi: 10.1002/mrm.21237. [DOI] [PubMed] [Google Scholar]
- Dale AM. Optimal experimental design for event-related fMRI. Hum Brain Mapp. 1999;8:109–114. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<109::AID-HBM7>3.0.CO;2-W. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dale AM, Liu AK, Fischl BR, Buckner RL, Belliveau JW, Lewine JD, Halgren E. Dynamic statistical parametric mapping: combining fMRI and MEG for high-resolution imaging of cortical activity. Neuron. 2000;26:55–67. doi: 10.1016/s0896-6273(00)81138-1. [DOI] [PubMed] [Google Scholar]
- de Zwart JA, van Gelderen P, Kellman P, Duyn JH. Reduction of gradient acoustic noise in MRI using SENSE-EPI. NeuroImage. 2002;16:1151–1155. doi: 10.1006/nimg.2002.1119. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Glover GH. Deconvolution of impulse response in event-related BOLD fMRI. NeuroImage. 1999;9:416–429. doi: 10.1006/nimg.1998.0419. [DOI] [PubMed] [Google Scholar]
- Golub GH, Heath MT, Wahba G. Generalized cross-validation as a method of choosing a good ridge parameter. Technometrics. 1979;21:215–223. [Google Scholar]
- Griswold MA, Jakob PM, Nittka M, Goldfarb JW, Haase A. Partially parallel imaging with localized sensitivities (PILS) Magn Reson Med. 2000;44:602–609. doi: 10.1002/1522-2594(200010)44:4<602::aid-mrm14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- Hansen PC. Rank-Deficient and Discrete Ill-Posed Problems: Numerical Aspects of Linear Inversion. SIAM; Philadelphia: 1998. [Google Scholar]
- Hennig J, Zhong K, Speck O. MR-Encephalography: fast multi-channel monitoring of brain physiology with magnetic resonance. NeuroImage. 2007;34:212–219. doi: 10.1016/j.neuroimage.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Huang F, Akao J, Vijayakumar S, Duensing GR, Limkeman M. k-t GRAPPA: k-space implementation for dynamic MRI with high reduction factor. Magn Reson Med. 2005;54:1172–1184. doi: 10.1002/mrm.20641. [DOI] [PubMed] [Google Scholar]
- Huettel SA, McCarthy G. Regional differences in the refractory period of the hemodynamic response: an event-related fMRI study. NeuroImage. 2001;14:967–976. doi: 10.1006/nimg.2001.0900. [DOI] [PubMed] [Google Scholar]
- Hyde JS, Jesmanowicz A, Froncisz W, Kneeland B, Grist TM, Campagna NF. Parallel image acquisition from noninteracting local coils. J Magn Reson. 1986;70:512–517. [Google Scholar]
- Jakob PM, Griswold MA, Edelman RR, Sodickson DK. AUTO-SMASH: a self-calibrating technique for SMASH imaging. SiMultaneous Acquisition of Spatial Harmonics Magma. 1998;7:42–54. doi: 10.1007/BF02592256. [DOI] [PubMed] [Google Scholar]
- Kwong KK, Belliveau JW, Chesler DA, Goldberg IE, Weisskoff RM, Poncelet BP, Kennedy DN, Hoppel BE, Cohen MS, Turner R, Cheng H, Brady TJ, Rosen BR. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakos WE, Panych LP, Kacher DF, Westin CF, Bao SM, Mulkern RV, Jolesz FA. Sensitivity profiles from an array of coils for encoding and reconstruction in parallel (SPACE RIP) Magn Reson Med. 2000;44:301–308. doi: 10.1002/1522-2594(200008)44:2<301::aid-mrm18>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Lin FH, Wald LL, Ahlfors SP, Hamalainen MS, Kwong KK, Belliveau JW. Dynamic magnetic resonance inverse imaging of human brain function. Magn Reson Med. 2006;56:787–802. doi: 10.1002/mrm.20997. [DOI] [PubMed] [Google Scholar]
- Lin FH, Tsai SY, Lin YR, Otazo R, Wiggins G, Wald LL, Belliveau JW, Posse S. Proc Intl Soc Mag Reson Med. Vol. 595. Toronto, Canada: 2008a. Single-shot proton MR spectroscopic inverse imaging. [Google Scholar]
- Lin FH, Witzel T, Mandeville JB, Polimeni JR, Zeffiro TA, Greve DN, Wiggins G, Wald LL, Belliveau JW. Event-related single-shot volumetric functional magnetic resonance inverse imaging of visual processing. NeuroImage. 2008b;42:230–247. doi: 10.1016/j.neuroimage.2008.04.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FH, Witzel T, Zeffiro TA, Belliveau JW. Linear constraint minimum variance beamformer functional magnetic resonance inverse imaging. NeuroImage. 2008c;43:297–311. doi: 10.1016/j.neuroimage.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AK, Belliveau JW, Dale AM. Spatiotemporal imaging of human brain activity using functional MRI constrained magnetoencephalography data: Monte Carlo simulations. Proc Natl Acad Sci U S A. 1998;95:8945–8950. doi: 10.1073/pnas.95.15.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AK, Dale AM, Belliveau JW. Monte Carlo simulation studies of EEG and MEG localization accuracy. Hum Brain Mapp. 2002;16:47–62. doi: 10.1002/hbm.10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412:150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Lustig M, Donoho D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- Lustig M, Alley M, Vasanawala S, Donoho DL, Pauly JM. L1SPIR-iT: Autocalibrating Parallel Imaging Compressed Sensing. ISMRM Workshop on Data Sampling and Image Reconstruction; Sedona AZ, USA. 2009. p. 334. [Google Scholar]
- Mansfield P. Multi-planar image formation using NMR spin echos. J Physics. 1977;C10:L55–L58. [Google Scholar]
- Menon RS, Luknowsky DC, Gati JS. Mental chronometry using latency-resolved functional MRI. Proc Natl Acad Sci U S A. 1998;95:10902–10907. doi: 10.1073/pnas.95.18.10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miezin FM, Maccotta L, Ollinger JM, Petersen SE, Buckner RL. Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. NeuroImage. 2000;11:735–759. doi: 10.1006/nimg.2000.0568. [DOI] [PubMed] [Google Scholar]
- Ohliger MA, Grant AK, Sodickson DK. Ultimate intrinsic signal-to-noise ratio for parallel MRI: electromagnetic field considerations. Magn Reson Med. 2003;50:1018–1030. doi: 10.1002/mrm.10597. [DOI] [PubMed] [Google Scholar]
- Pelli DG. The VideoToolbox software for visual psychophysics: transforming numbers into movies. Spat Vis. 1997;10:437–442. [PubMed] [Google Scholar]
- Preibisch C, Pilatus U, Bunke J, Hoogenraad F, Zanella F, Lanfermann H. Functional MRI using sensitivity-encoded echo planar imaging (SENSE-EPI) NeuroImage. 2003;19:412–421. doi: 10.1016/s1053-8119(03)00080-6. [DOI] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16:192–225. doi: 10.1002/mrm.1910160203. [DOI] [PubMed] [Google Scholar]
- Schmidt CF, Degonda N, Luechinger R, Henke K, Boesiger P. Sensitivity-encoded (SENSE) echo planar fMRI at 3T in the medial temporal lobe. NeuroImage. 2005:25, 625–641. doi: 10.1016/j.neuroimage.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Sodickson DK. Tailored SMASH image reconstructions for robust in vivo parallel MR imaging. Magn Reson Med. 2000;44:243–251. doi: 10.1002/1522-2594(200008)44:2<243::aid-mrm11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn Reson Med. 1997;38:591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- Wiesinger F, Boesiger P, Pruessmann KP. Electrodynamics and ultimate SNR in parallel MR imaging. Magn Reson Med. 2004;52:376–390. doi: 10.1002/mrm.20183. [DOI] [PubMed] [Google Scholar]
- Wiggins GC, Potthast A, Triantafyllou C, Lin FH, Benner T, Wiggins CJ, Wald LL. International Society for Magnetic Resonance in Medicine Thirteenth Scientific Meeting and Exhibition. International Society for Magnetic Resonance in Medicine; Miami, Florida, USA: 2005. A 96-channel MRI system with 23- and 90-channel phase array head coils at 1.5 Tesla; p. 671. [Google Scholar]