Background: The mechanism by which MIC-1 expression is regulated post-transcriptionally is not clear.
Results: MIC-1 transcript can be stabilized and mediate cell growth suppression via RNPC1.
Conclusion: RNPC1 can increase MIC-1 expression and inhibit cell growth by binding its 3′-UTR.
Significance: A novel post-transcriptional mechanism is found to regulate MIC-1 expression by RNPC1.
Keywords: Cell Proliferation, Gene Regulation, mRNA Decay, p53, RNA-binding Protein, GDF-15, MIC-1, RNPC1, p21
Abstract
Macrophage inhibitory cytokine-1 (MIC-1), a secreted cytokine, is a direct target of p53 and known to play a role in cell proliferation, apoptosis, cell metastasis, and angiogenesis through autocrine and paracrine signaling. Previous studies have shown that serum levels of MIC-1 closely parallel cancer progression and are being explored as a diagnostic tool. MIC-1 has also shown potential as a therapeutic agent as it has exhibited several anti-carcinogenic activities. Thus, MIC-1 displays two opposing effects: tumor suppression versus promotion. However, it remains unclear whether MIC-1 is regulated by a mechanism other than transcription and how MIC-1 exerts its tumor suppression. In this study, we show that overexpression of RNA-binding protein RNPC1 can increase, whereas knockdown or knock-out of RNPC1 decreases, MIC-1 transcript and protein levels. Additionally, we demonstrate that RNPC1 can bind to MIC-1 mRNA via an AU-rich element within MIC-1 3′-UTR and then enhances MIC-1 mRNA stability. Finally, to explore the functional significance of MIC-1, we showed that knockdown of MIC-1 can decrease RNPC1-induced cell growth suppression. Altogether, we uncover a novel mechanism by which MIC-1 can be regulated through RNPC1 via mRNA stability.
Introduction
Macrophage inhibitory cytokine-1 (MIC-1),3 also known as GDF-15, NAG-1, and PTGF-β, is a divergent member of the TGF-β family (1, 2) and a direct target of wild-type p53 (3). As a secreted protein, MIC-1 has been shown to play a role in cell proliferation, apoptosis, metastasis, cell migration, and angiogenesis through autocrine and paracrine signaling. Although normally undetectable under physiologic conditions aside from the placenta and some nervous tissues (4–6), MIC-1 becomes highly up-regulated under conditions of stress and acute injury including cancer (7), myocardial infarction, and ischemia (8). MIC-1 has been determined to be a marker of mortality (9) with high serum levels being a predictor of death particularly due to cancer or cardiovascular disease. Levels of MIC-1 have also been shown to parallel tumor progression closely with increased serum levels correlating to the stage and severity of tumors especially for colon and prostate cancer (7, 10, 11). Due to this close correlation, MIC-1 has shown the potential to act as a biomarker of tumor progression and metastasis. Although MIC-1 exhibits mostly anti-carcinogenic activity including promotion of apoptosis, cell cycle arrest, and inhibition of cell motility, reports have indicated that this is not always the case (12–15). The actual functional role of MIC-1 seems to vary with tumor origin, stage, and bioavailability of MIC-1. For instance, it has been recently reported within mouse models that overexpression of MIC-1 results in smaller tumors and longer survival times but also higher levels of distant organ metastases (16). Generally, within early stages MIC-1 tends to display anti-cancer properties, but this effect is altered in later stage cancers (7). MIC-1 expression has also been found to be subject to epigenetic regulation in some tumor cell lines (17, 18).
p53, a crucial tumor suppressor, plays an important role in the prevention of cancer development by mediating cell cycle arrest and apoptosis through its downstream targets upon DNA damage or cellular stress (19, 20). The importance of p53 is seen because more than half of all human cancers lack a functional p53 (21). As a direct target of p53, MIC-1 has been shown to become up-regulated upon treatment with anti-cancer drugs (22). Interestingly, in some cases an increase in MIC-1 can be induced in cells either with or without p53 (23). Additionally, MIC-1 has also been shown to be able seemingly to induce apoptosis in the absence of p53, making MIC-1 a potential therapeutic as well as a diagnostic agent against cancer. Inhibition of cell death can also occur if MIC-1 expression is diminished by disrupting p53 binding to the MIC-1 promoter (24).
RNPC1, also known as RBM38, is an RNA-binding protein belonging to the RNA recognition motif-containing family of RNA-binding proteins. It is expressed as two isoforms, RNPC1a, and RNPC1b (25). RNPC1 is a direct target of p53 and has been shown to interact with other members of the p53 family including stabilization of p21 and p73 transcripts and destabilization of p63 transcripts (25–27). RNPC1 also functions in a negative feedback loop against p53 by repressing p53 mRNA translation (28). As an RNA-binding protein, RNPC1 regulates expression of various genes through binding of AU-rich elements (AREs) usually located within the 3′-UTR. The MIC-1 transcript carries an AU-rich region in its 3′-UTR. Therefore, it is likely that MIC-1 is a target of RNPC1. As it has been shown previously that MIC-1 can have anti-tumorigenic properties in the presence or absence of p53, we hypothesize that RNPC1 can regulate MIC-1 post-transcriptionally by binding to and modulating the MIC-1 transcript, leading to increased MIC-1 expression and cell growth suppression.
EXPERIMENTAL PROCEDURES
Cell Culture
RKO, MCF7, and H1299 cells were cultured using DMEM with 8% FBS supplied by Hyclone and Benchmark. Mouse embryonic fibroblast (MEF) cells were cultured with DMEM containing 10% FBS (Hyclone), 55 μm β-mercaptoethanol, and 1× minimum Eagle's medium nonessential amino acids.
Cell Line Generation
Inducible RNPC1a-expressing cell lines were generated previously with RKO, MCF7, HCT116, and H1299 cells using the Tet-on inducible system (25). Two plasmids were used including pcDNA4/TO containing two Tet operators within a human CMV promoter as well as our gene of interest RNPC1, and pcDNA6/TR which contains a Tet repressor under the control of the CMV promoter. Without tetracycline, the Tet repressor binds to the promoter within the plasmid containing RNPC1a, preventing gene expression. Upon treatment, the tetracycline binds the repressor, leading to RNPC1a expression.
Lentiviral Knockdown
Lentiviral infection was done by first obtaining a lentiviral vector expressing shRNA against RNPC1 and another against luciferase to be used as a negative control. 10 μg of either shRNPC1 or shLuciferase was added to three other plasmids including pCMV-VSVG (5 μg), pMDL-g/pRRE (5 μg), and pRSV-REV (5 μg) and cotransfected into 293T cells using Expressfect transfection reagent. Lentiviral particles were collected from the medium and concentrated through ultracentrifugation (25,000 rpm, 4 °C, 1.5 h). Cells were then infected using our concentrated lentiviral particles and selected using puromycin for 3 days.
Western Blot Analysis
Whole cell lysates were obtained using 1× SDS sample buffer and run on 8–11% acrylamide gel. The blots were transferred to a nitrocellulose membrane and probed with the appropriate antibodies. Antibodies used include anti-p53 (mouse monoclonal), anti-NAG-1/PTGFβ (rabbit polyclonal; Millipore), anti-p21 SX-118 (mouse monoclonal; Santa Cruz Biotechnology), anti-RNPC1(rabbit polyclonal), anti-actin (Sigma), and anti-GAPDH FL-335 (rabbit polyclonal; Santa Cruz Biotechnology). For detection of secreted MIC-1, conditioned media were collected and immunoprecipitated using 1 μg of anti-MIC-1 antibody (FL-308, Santa Cruz Biotechnology). Immunoprecipitated proteins were brought down with Sepharose beads and eluted with SDS-PAGE and analyzed through Western blotting as above.
RT-PCR
RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer's protocol. RT-PCR was performed using MMLV-reverse transcriptase (Promega). PCR was run using GoTaq DNA Polymerase (Promega). The primers used to detect human and mouse mRNA and premature mRNA are summarized in Table 1. Primers used to amplify p21, GAPDH, and RNPC1 were used as described previously (29).
TABLE 1.
Primers used for RT-PCR
F, forward; R, reverse.
| Primer name | Sequence |
|---|---|
| MIC-F | 5′-CCAGAGCTGGGAAGATTCGAACAAC-3′ |
| MIC-R | 5′-AGATTCTGCCAGCAGTTGGTCCGAC-3′ |
| Pre-MIC-F | 5′-ACGCTACGAGGACCTGCTAA-3′ |
| Pre-MIC-R | 5′-CCAAGGGGATCCAGGATATT-3′ |
| mMIC-F | 5′-GCTGTCCGGATACTCAGTCC-3′ |
| mMIC-R | 5′-CCTGCCACAGTCTCCAAGTGA-3′ |
| Pre-mMIC-F | 5′-CTGGCACACACCCTAAGGACAT-3′ |
| Pre-mMIF-R | 5′-CCTGCACAGTCTCCAAGTGA-3′ |
RNA Immunoprecipitation
RNA immunoprecipitation was carried out as described previously (25). The cytosolic extracts from RKO cells were prepared with polysome lysis buffer (10 mm HEPES, pH 7.0, 100 mm KCl, 5 mm MgCl2, 0.5% Nonidet P-40, 1 mm DTT) and then incubated with 2 μg of rabbit polyclonal anti-RNPC1 or nonimmunized rabbit IgG at 4 °C for 4 h. The RNA-protein immunocomplexes were brought down by protein G beads followed by RT-PCR analysis. The primers to detect human RNPC1 mRNA are the same as those described previously (29).
RNA Electrophoretic Mobility Shift Assay (RNA EMSA)
32P-Labeled RNA probes were generated by in vitro transcription using cDNA containing T7 promoter and various regions of MIC-1 3′-UTR. For the RNA EMSA assay, 250 nm recombinant GST or GST-RNPC1 fusion protein, 100 μg/ml yeast tRNA, and 100,000 cpm of 32P-labeled RNA probe were mixed in binding buffer (10 mm Tris-Cl, pH 8.0, 25 mm KCl, 10 mm MgCl2, 2 mm DTT) for 20 min at 25 °C. RNA-protein complexes were digested by adding 100 units of RNase-T1 for 15 min at 37 °C and then separated in 6% of native polyacrylamide gel. RNA-protein complexes were visualized by autoradiography.
Colony Formation Assay
RKO cells were plated in triplicate at 300 cells/well in a 6-well plate. Cells were cultured either with or without tetracycline (0.25 μg/ml) to induce RNPC1 expression for 10 days. Cells were then fixed using a 7:1 mixture of ethanol to acetic acid and then stained using 0.2 g/liter crystal violet overnight. The results were quantified through densitometry, and statistical relevance was determined by the t test. p < 0.05 was considered significant.
RESULTS
MIC-1 Expression Is Increased by Ectopic Expression of RNPC1
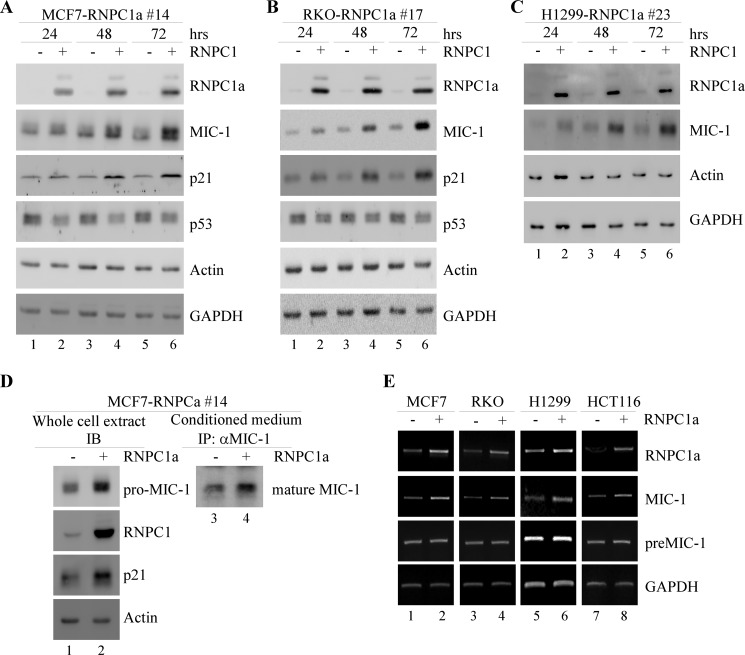
First, to examine whether there is any correlation between MIC-1 and RNPC1, protein and RNA samples were gathered from inducible RNPC1a-overexpressing cell lines under the control of a tetracycline-regulated promoter. RKO, MCF7, and H1299 cells that can be induced to express RNPC1a were treated with 0.5 μg/ml tetracycline for 24, 48, or 72 h, and the results were analyzed by Western blotting. Without RNPC1a induction, MIC-1 expression remained low (Fig. 1, A–C). Upon induction, increased levels of MIC-1 were detected at 24 h and were further increased at 48 and 72 h in a time-dependent manner (Fig. 1, A–C). Increases in MIC-1 were also seen in H1299 cells, but the overall basal expression levels were lower than in RKO and MCF7 cells perhaps due to the absence of p53 in H1299 (Fig. 1, A–C). Consistent with previous reports, MIC-1 levels closely paralleled p21 levels (13, 22). Additionally, secreted mature MIC-1 was increased upon induction of RNPC1a within MCF7 cells (Fig. 1D).
FIGURE 1.
MIC-1 expression is increased by RNPC1a. A–C, MCF7, RKO, and H1299 cells were induced or uninduced with 0.5 μg/ml tetracycline to express RNPC1a for 24, 48, and 72 h. The levels of RNPC1a, MIC-1, p21, p53, actin, and GAPDH were measured by Western blotting. D, MCF7 cells were induced or uninduced with tetracycline for 48 h. Levels of RNPC1a, p21, pro-MIC-1, and actin were analyzed through Western blotting. Secreted MIC-1 was detected by Western blotting after immunoprecipitation (IP) with conditioned medium gathered from induced or uninduced MCF7 cells. IB, immunoblotting. E, levels of RNPC1a, MIC-1, and GAPDH mRNA were analyzed by RT-PCR. RNA was isolated from cell extracts obtained from MCF7, RKO, H1299, and HCT116 cells that were uninduced or induced to express RNPC1a for 36 h. preMIC-1, premature MIC-1 mRNA.
Second, to determine how RNPC1 regulates MIC-1 expression, the MIC-1 transcript was measured by RT-PCR. RKO, MCF7, H1299, and HCT116 cells were induced or uninduced to express RNPC1a for 36 h. Upon induction of RNPC1a, an increase in MIC-1 mRNA was detected (Fig. 1E). We also examined pre-mRNA levels of MIC-1 to check whether the increased expression was due to post- or pre-transcriptional changes. Whereas mature mRNA levels were affected by changing RNPC1 levels, pre-mRNA levels remained relatively even (Fig. 1E), suggesting that MIC-1 is regulated mainly at a post-transcriptional level.
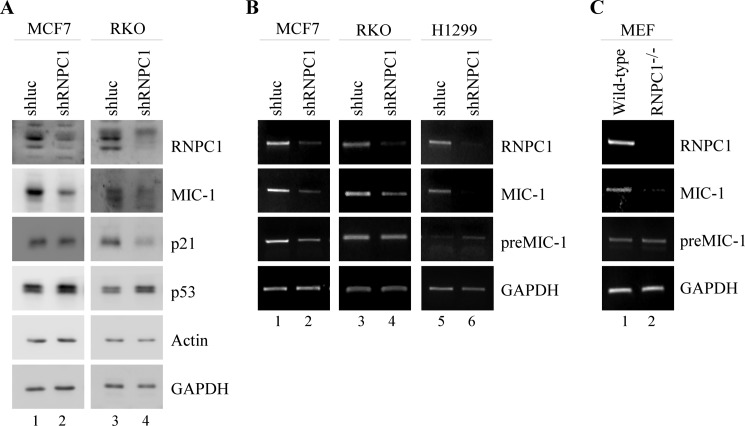
Third, we wanted to verify whether knockdown of RNPC1 can negatively impact MIC-1 expression in RKO, MCF7, and H1299 cells. RNPC1 expression was knocked down with shRNA targeting RNPC1 through lentiviral transduction. shRNA targeting luciferase was used as a negative control. We found that knockdown of RNPC1 through shRNA was able to decrease levels of MIC-1 in RKO and MCF7 (Fig. 2, A and B). This effect was seen at both mRNA and protein levels. In addition, p53 expression was increased, whereas p21 expression was decreased (Fig. 2, A and B), consistent with previous reports (25, 28). However, MIC-1 protein levels were difficult to detect in H1299 cells likely due to the lack of p53. Nevertheless, the decrease in MIC-1 mRNA could be detected by RT-PCR (Fig. 2B). Once again, we found that MIC-1 pre-mRNA remained relatively unchanged, suggesting that MIC-1 is regulated by RNPC1 primarily at a post-transcriptional level (Fig. 2B).
FIGURE 2.
MIC-1 expression is decreased by knockdown of RNPC1. A, RKO and MCF7 cells were transduced with a lentivirus containing either control luciferase shRNA (shluc) or RNPC1 shRNA (shRNPC1) and then selected by puromycin (1 μg/ml) for 3 days prior to Western blot analysis. B, MCF7, RKO, and H1299 cells were transduced as in A and underwent puromycin selection for 3 days. Transcript levels were measured through RT-PCR. C, levels of RNPC1, MIC-1, and GAPDH in wild-type and RNPC1−/− MEFs were analyzed through RT-PCR.
Fourth, we examined the effect of RNPC1 on MIC-1 expression in wild-type and RNPC1−/− MEFs using RT-PCR. Whereas MIC-1 mRNA expression was seen clearly within wild-type MEF cells, the levels of MIC-1 were markedly reduced in MEFs without RNPC1 (Fig. 2C). Additionally, MIC-1 pre-mRNA remained unchanged (Fig. 2C). Together, our data indicate that RNPC1 can positively affect MIC-1 expression.
RNPC1 Can Enhance MIC-1 mRNA Stability
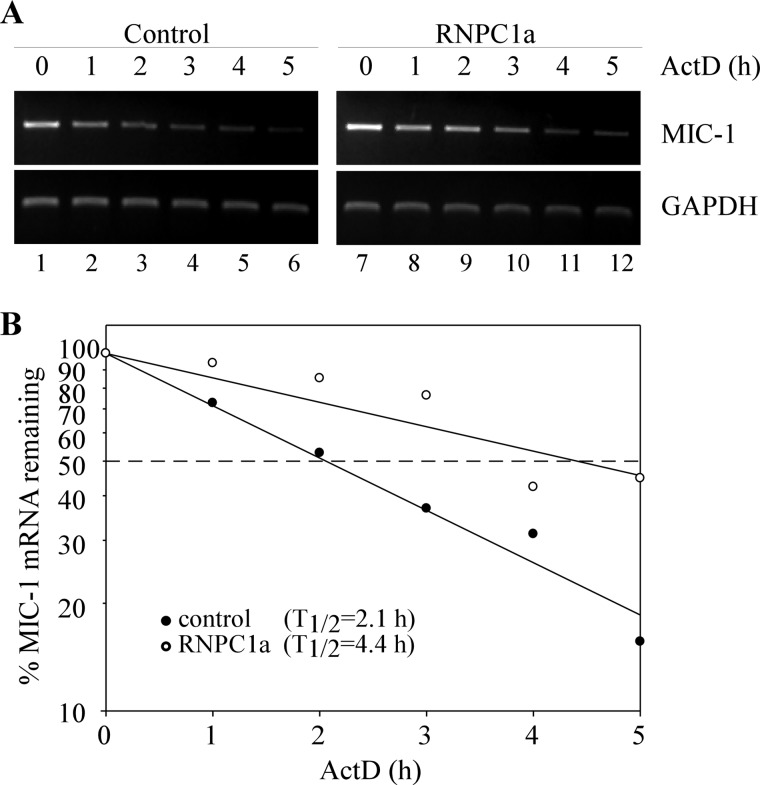
As an RNA-binding protein, RNPC1 is known to regulate gene expression through mRNA stability. Thus, we examined whether RNPC1 regulates MIC-1 expression by altering the stability of its mRNA. To test this, MCF7 cells were treated with tetracycline (0.5 μg/ml) to induce RNPC1a expression for 36 h prior to treatment with 5 μg/ml actinomycin D, a transcription inhibitor. Total RNAs were harvested from the cells over a 5-h period at 1-h intervals and then analyzed by RT-PCR (Fig. 3A). With induced expression of RNPC1, the half-life of MIC-1 transcript was increased from ∼2.1 to ∼ 4.4 h (Fig. 3), suggesting that MIC-1 mRNA stability is regulated by RNPC1.
FIGURE 3.
MIC-1 mRNA stability is regulated by RNPC1. A, MCF7 cells were treated with actinomycin D (ActD, 5 μg/ml) over a 5-h period at 1-h intervals. Total RNAs were isolated, and MIC-1 and GAPDH transcript levels were analyzed using RT-PCR. B, MIC-1 levels were normalized with GAPDH transcript levels and plotted along a time course to calculate the relative half-life of MIC-1 in the presence or absence of RNPC1.
MIC-1 3′-UTR Contains an AU-rich Element, Which Can Be Recognized by RNPC1
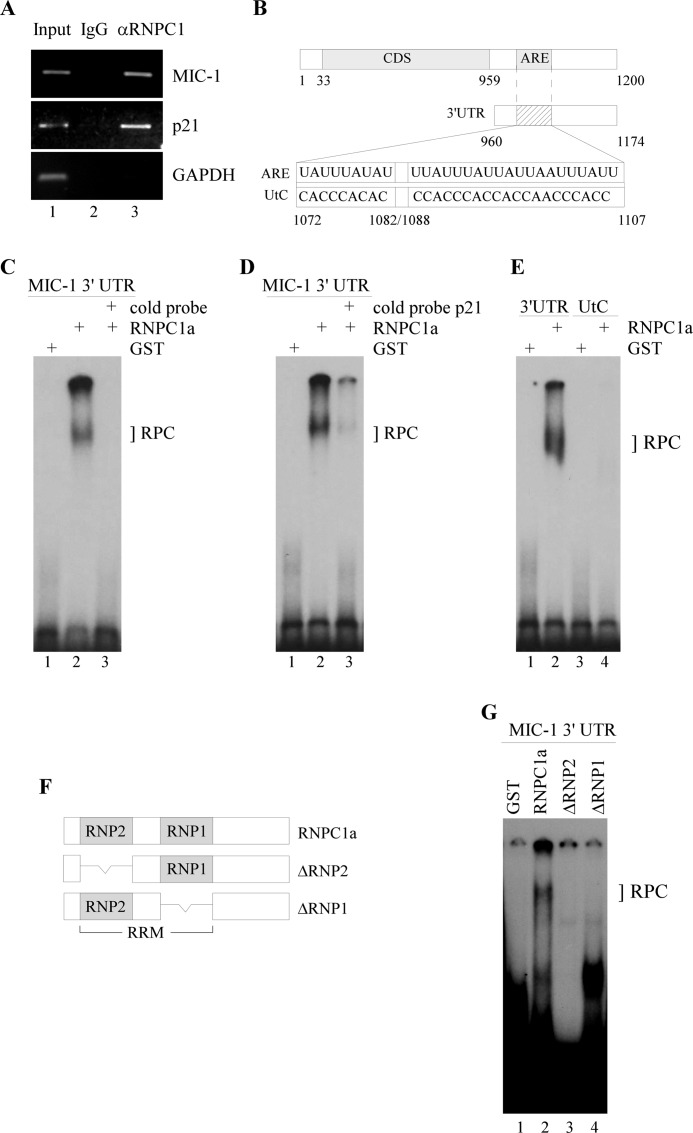
Next, we wanted to determine whether RNPC1 physically associates with MIC-1 transcripts in RKO cells. To test this, we performed an RNA immunoprecipitation assay followed by RT-PCR and found that MIC-1 transcript was detectable in RNPC1, but not control IgG immunocomplexes (Fig. 4A). p21 transcript was measured as a positive control as it has previously been shown to form immunocomplexes with RNPC1 (25). No noticeable GAPDH band was seen to associate with either RNPC1 or IgG as expected. These results indicate that RNPC1 can physically bind to MIC-1 transcripts.
FIGURE 4.
RNPC1 can directly bind to MIC-1 transcript at its 3′-UTR. A, RKO cell lysates were immunoprecipitated with RNPC1 antibody or control IgG followed by RT-PCR to measure transcript levels of MIC-1, p21, and GAPDH within RNPC1 or IgG immunocomplexes. B, schematic diagram of MIC-1 transcript includes the MIC-1 coding region, 5′-UTR, 3′-UTR, and the ARE. MIC-1 3′-UTR containing an ARE was used as an RNA probe and designated as ARE. The mutant probe was designated as UtC. C, RNA-EMSA was performed by mixing 32P-labeled MIC-1 3′-UTR with either GST alone or GST-fused RNPC1a. Unlabeled wild-type MIC-1 3′-UTR was used as cold probe for competition. The bracket labeled RPC represents an RNA-protein complex. D, the experiment was performed as in C except p21 cold probe was used for competition. E, the experiment was performed as in C except that 32P-labeled wild-type and mutant MIC-1 probes were used for RNA-EMSA. F, schematic diagram details RNPC1a and ΔRNP2 and ΔRNP1 mutants. RRM, RNA recognition motif region in RNPC1. G, the experiment was performed as in C except that RNA-EMSA was performed with GST alone, GST-RNPC1a, GST-ΔRNP2, or GST-ΔRNP1.
To map out the RNPC1 binding region in MIC-1, we performed RNA-EMSA using radiolabeled RNA probes. RNA fragments containing wild-type MIC-1 3′-UTRs and an altered ARE were generated (Fig. 4B). We found that whereas GST alone did not bind to the MIC-1 3′-UTR, GST-fused RNPC1a formed a complex with wild-type MIC-1 3′-UTR (Fig. 4C, lane 2). Formation of this complex was abrogated with the addition of a cold probe, indicating that the binding was specific (Fig. 4C, lane 3). Addition of cold p21, which is known to contain an ARE recognized by RNPC1 (29), also mitigated the binding between MIC-1 and GST-RNPC1a (Fig. 4D, compare lanes 2 and 3). Mutant MIC-1 3′-UTR, designated as UtC, was also probed with GST-RNPC1a (Fig. 4B). The UtC fragment was unable to form a complex with RNPC1, indicating that the ARE is indeed necessary for binding of RNPC1 (Fig. 4E, lane 4). Finally, we examined whether RNPC1 mutants (ΔRNP2 and ΔRNP1), which lack a subdomain in the RNPC1 RNA recognition motif, are capable of binding to MIC-1 3′-UTR (Fig. 4F). Whereas a binding complex was detected between MIC-1 and GST-RNPC1a (Fig. 4G, lane 2), little if any binding was detected if either RNP site was absent (Fig. 4G, lanes 3 and 4). Altogether, this suggests that MIC-1 requires the ARE within its 3′-UTR to physically associate with RNPC1. The RNA binding domains in RNPC1 are also required for binding to the MIC-1 transcript.
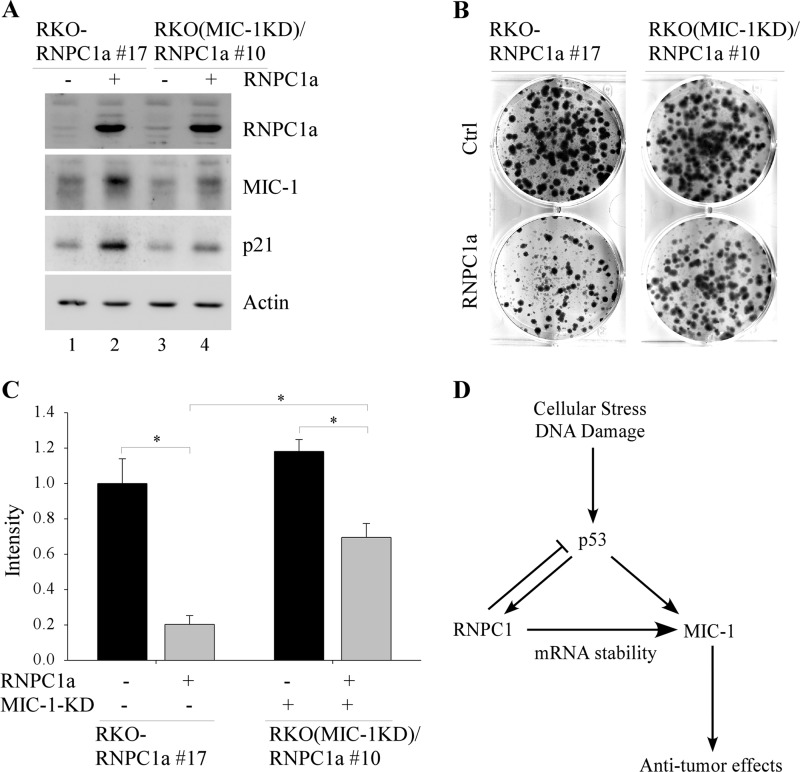
MIC-1 Knockdown Attenuates RNPC1-induced Inhibition of Cell Growth
Previous reports have indicated that MIC-1 exhibits both anti-tumorigenic and oncogenic activity. To examine the role of MIC-1 in cancer cell growth, we generated MIC-1 knockdown RKO cell lines in which RNPC1 can be inducibly expressed. The resulting cell line was designated RKO(MIC-1KD)/RNPC1a. Clone 10 was selected for further study. RKO-RNPC1 17, which is capable of inducibly expressing RNPC1a (25), was used for control. Whereas MIC-1 was induced by RNPC1 (Fig. 5A, compare lanes 1 and 2), increased MIC-1 expression was not as prominent in MIC-1 knockdown cells despite similar levels of RNPC1 (Fig. 5A, compare lanes 3 and 4). Next, colony formation assay was performed and showed that upon induction of RNPC1, cell growth was markedly inhibited. However, by knocking down MIC-1, cell growth inhibition was greatly mitigated (Fig. 5B). Quantitative analysis showed that RNPC1-mediated growth inhibition was diminished from 80% in the presence of MIC-1 to 38% upon knockdown of MIC-1 (Fig. 5C). Previous studies showed that overexpression of MIC-1 suppresses cell growth by signaling downstream through the TGF-β pathway, which can increase p21 expression (3, 30). Furthermore, targeting MIC-1 expression using siRNA has been observed to down-regulate p21 (31). Consistently, we found that RNPC1 induction of p21 was decreased when MIC-1 was knocked down (Fig. 5A, compare lanes 2 and 4). Altogether, this suggests that MIC-1 mediates RNPC1-induced cell growth suppression at least in part via p21.
FIGURE 5.
MIC-1 knockdown reduces RNPC1-induced cell growth suppression. A, RKO cells were uninduced or induced to express RNPC1a for 48 h. Cell lysates were collected and analyzed by Western blotting. B, colony formation assay was performed using the same cells and treatment as in A. The assay was done in triplicate, and a representative image is shown. C, quantified data were obtained from colony formation plates. Cell density for uninduced cell line RKO-RNPC1a clone 17 was set at 1 as a reference point. *, p = 0.005. Error bars, S.D. D, a model shows RNPC1 regulation of MIC-1 in response to a stress signal.
DISCUSSION
Previous studies have shown that the RNA-binding protein RNPC1 can regulate several p53 family members through binding of their AREs. In this study, we have shown that induction of RNPC1 can increase MIC-1 transcript and protein expression. Conversely, lack of RNPC1 negatively affects MIC-1 levels. Pre-mRNA levels in both cases, however, are unaffected by RNPC1, indicating that regulation of MIC-1 occurs at a post-transcriptional level. Consistent with this, we have identified an ARE within MIC-1 3′-UTR which acts as a potential binding site for RNPC1. Binding of MIC-1 ARE within its 3′-UTR by RNPC1 leads to enhanced stability of the MIC-1 transcript and cell growth suppression. This binding can only occur if MIC-1 ARE is present. Altogether, our data indicate that RNPC1 is a positive post-transcriptional regulator of MIC-1 expression.
Expression of MIC-1 can be induced in response to a variety of factors including cellular stress, inflammation, and chemicals such as nonsteroidal anti-inflammatory drugs, DNA-damaging agents, and anti-cancer drugs through either p53-dependent or -independent pathways (18, 32). Whereas reports thus far have investigated mainly regulation of MIC-1 at a transcriptional level, here we have uncovered a novel post-transcriptional mechanism by which MIC-1 expression can be regulated. Due to its high levels within certain cancers such as gastric, colorectal, prostate, ovarian, and breast cancer and its close association with the presence particularly of premalignant colonic diseases, MIC-1 is being explored as a marker of tumor progression and a prognostic serum biomarker (10, 33, 34). However, our understanding of the signaling pathways involving MIC-1 within the context of tumor development is still incomplete. Similar to other members of the TGF-β family, MIC-1 displays conflicting roles during early and late stage cancers. Through this study, we propose that under nonstressed conditions, p53 expression remains low, resulting in negligible levels of MIC-1. Wild-type p53 activation in response to a stress signal leads to increased levels of RNPC1 and transcriptional activation of MIC-1. RNPC1 can then function simultaneously in a negative feedback loop to inhibit further p53 expression while binding and stabilizing MIC-1 transcripts, which results in the overall increased expression of MIC-1 seen in early stage and premalignant cancers (Fig. 5D). This post-transcriptional regulation of MIC-1 serves to amplify tumor cell death and growth inhibition signals that are mediated in part by p21. As the cancer progresses into later stages, wild-type p53 may no longer be present. Given that p53 acts as an up-regulator of MIC-1, loss of wild-type p53 should result in a corresponding decrease in MIC-1. However, MIC-1 levels tend to increase rather than decrease as cancers progress. We may speculate that this is due to another transcriptional activator. Regardless, the high levels of MIC-1 observed in late stage cancers suggest that whatever anti-tumorigenic properties MIC-1 displayed early on are no longer functional. Furthermore, additional mutations that may occur within downstream targets of MIC-1 in cancer cells can render them immune to regulation by MIC-1. More studies are required to clarify the dual pro- and anti-tumorigenic roles of MIC-1 and its signaling pathway, which may provide an insight into possible therapeutic as well as diagnostic uses for MIC-1.
Footnotes
- MIC-1
- macrophage inhibitory cytokine-1
- ARE
- AU-rich element
- MEF
- mouse embryonic fibroblast
- Tet
- tetracycline.
REFERENCES
- 1. Bootcov M. R., Bauskin A. R., Valenzuela S. M., Moore A. G., Bansal M., He X. Y., Zhang H. P., Donnellan M., Mahler S., Pryor K., Walsh B. J., Nicholson R. C., Fairlie W. D., Por S. B., Robbins J. M., Breit S. N. (1997) MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-β superfamily. Proc. Natl. Acad. Sci. U.S.A. 94, 11514–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lawton L. N., Bonaldo M. F., Jelenc P. C., Qiu L., Baumes S. A., Marcelino R. A., de Jesus G. M., Wellington S., Knowles J. A., Warburton D., Brown S., Soares M. B. (1997) Identification of a novel member of the TGF-β superfamily highly expressed in human placenta. Gene 203, 17–26 [DOI] [PubMed] [Google Scholar]
- 3. Tan M., Wang Y., Guan K., Sun Y. (2000) PTGF-β, a type β transforming growth factor (TGF-β) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-β signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 97, 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fairlie W. D., Moore A. G., Bauskin A. R., Russell P. K., Zhang H. P., Breit S. N. (1999) MIC-1 is a novel TGF-β superfamily cytokine associated with macrophage activation. J. Leukoc. Biol. 65, 2–5 [DOI] [PubMed] [Google Scholar]
- 5. Böttner M., Suter-Crazzolara C., Schober A., Unsicker K. (1999) Expression of a novel member of the TGF-β superfamily, growth/differentiation factor-15/macrophage-inhibiting cytokine-1 (GDF-15/MIC-1) in adult rat tissues. Cell Tissue Res. 297, 103–110 [DOI] [PubMed] [Google Scholar]
- 6. Schober A., Böttner M., Strelau J., Kinscherf R., Bonaterra G. A., Barth M., Schilling L., Fairlie W. D., Breit S. N., Unsicker K. (2001) Expression of growth differentiation factor-15/macrophage inhibitory cytokine-1 (GDF-15/MIC-1) in the perinatal, adult, and injured rat brain. J. Comp. Neurol. 439, 32–45 [DOI] [PubMed] [Google Scholar]
- 7. Welsh J. B., Sapinoso L. M., Kern S. G., Brown D. A., Liu T., Bauskin A. R., Ward R. L., Hawkins N. J., Quinn D. I., Russell P. J., Sutherland R. L., Breit S. N., Moskaluk C. A., Frierson H. F., Jr., Hampton G. M. (2003) Large-scale delineation of secreted protein biomarkers overexpressed in cancer tissue and serum. Proc. Natl. Acad. Sci. U.S.A. 100, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kempf T., Eden M., Strelau J., Naguib M., Willenbockel C., Tongers J., Heineke J., Kotlarz D., Xu J., Molkentin J. D., Niessen H. W., Drexler H., Wollert K. C. (2006) The transforming growth factor-β superfamily member growth-differentiation factor-15 protects the heart from ischemia/reperfusion injury. Circ. Res. 98, 351–360 [DOI] [PubMed] [Google Scholar]
- 9. Wiklund F. E., Bennet A. M., Magnusson P. K., Eriksson U. K., Lindmark F., Wu L., Yaghoutyfam N., Marquis C. P., Stattin P., Pedersen N. L., Adami H. O., Grönberg H., Breit S. N., Brown D. A. (2010) Macrophage inhibitory cytokine-1 (MIC-1/GDF15): a new marker of all-cause mortality. Aging Cell 9, 1057–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown D. A., Ward R. L., Buckhaults P., Liu T., Romans K. E., Hawkins N. J., Bauskin A. R., Kinzler K. W., Vogelstein B., Breit S. N. (2003) MIC-1 serum level and genotype: associations with progress and prognosis of colorectal carcinoma. Clin. Cancer Res. 9, 2642–2650 [PubMed] [Google Scholar]
- 11. Brown D. A., Lindmark F., Stattin P., Bälter K., Adami H. O., Zheng S. L., Xu J., Isaacs W. B., Grönberg H., Breit S. N., Wiklund F. E. (2009) Macrophage inhibitory cytokine 1: a new prognostic marker in prostate cancer. Clin. Cancer Res. 15, 6658–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng J. C., Chang H. M., Leung P. C. (2011) Wild-type p53 attenuates cancer cell motility by inducing growth differentiation factor-15 expression. Endocrinology 152, 2987–2995 [DOI] [PubMed] [Google Scholar]
- 13. Li P. X., Wong J., Ayed A., Ngo D., Brade A. M., Arrowsmith C., Austin R. C., Klamut H. J. (2000) Placental transforming growth factor-β is a downstream mediator of the growth arrest and apoptotic response of tumor cells to DNA damage and p53 overexpression. J. Biol. Chem. 275, 20127–20135 [DOI] [PubMed] [Google Scholar]
- 14. Bauskin A. R., Brown D. A., Kuffner T., Johnen H., Luo X. W., Hunter M., Breit S. N. (2006) Role of macrophage inhibitory cytokine-1 in tumorigenesis and diagnosis of cancer. Cancer Res. 66, 4983–4986 [DOI] [PubMed] [Google Scholar]
- 15. Bauskin A. R., Brown D. A., Junankar S., Rasiah K. K., Eggleton S., Hunter M., Liu T., Smith D., Kuffner T., Pankhurst G. J., Johnen H., Russell P. J., Barret W., Stricker P. D., Grygiel J. J., Kench J. G., Henshall S. M., Sutherland R. L., Breit S. N. (2005) The propeptide mediates formation of stromal stores of PROMIC-1: role in determining prostate cancer outcome. Cancer Res. 65, 2330–2336 [DOI] [PubMed] [Google Scholar]
- 16. Husaini Y., Qiu M. R., Lockwood G. P., Luo X. W., Shang P., Kuffner T., Tsai V. W., Jiang L., Russell P. J., Brown D. A., Breit S. N. (2012) Macrophage inhibitory cytokine-1 (MIC-1/GDF15) slows cancer development but increases metastases in TRAMP prostate cancer prone mice. PLoS One 7, e43833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kadowaki M., Yoshioka H., Kamitani H., Watanabe T., Wade P. A., Eling T. E. (2012) DNA methylation-mediated silencing of nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) in glioma cell lines. Int. J. Cancer 130, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoshioka H., Kamitani H., Watanabe T., Eling T. E. (2008) Nonsteroidal anti-inflammatory drug-activated gene (NAG-1/GDF15) expression is increased by the histone deacetylase inhibitor trichostatin A. J. Biol. Chem. 283, 33129–33137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harms K., Nozell S., Chen X. (2004) The common and distinct target genes of the p53 family transcription factors. Cell Mol. Life Sci. 61, 822–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 21. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 22. Yang H., Filipovic Z., Brown D., Breit S. N., Vassilev L. T. (2003) Macrophage inhibitory cytokine-1: a novel biomarker for p53 pathway activation. Mol. Cancer Ther. 2, 1023–1029 [PubMed] [Google Scholar]
- 23. Kim I. Y., Park S. Y., Kang Y., Thapa D., Choi H. G., Kim J. A. (2011) Role of nonsteroidal anti-inflammatory drug-activated gene-1 in docetaxel-induced cell death of human colorectal cancer cells with different p53 status. Arch. Pharm. Res. 34, 323–330 [DOI] [PubMed] [Google Scholar]
- 24. Qian Y., Jung Y. S., Chen X. (2012) Differentiated embryo-chondrocyte expressed gene 1 regulates p53-dependent cell survival versus cell death through macrophage inhibitory cytokine-1. Proc. Natl. Acad. Sci. U.S.A. 109, 11300–11305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shu L., Yan W., Chen X. (2006) RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 20, 2961–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J., Jun Cho S., Chen X. (2010) RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc. Natl. Acad. Sci. U.S.A. 107, 9614–9619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan W., Zhang J., Zhang Y., Jung Y. S., Chen X. (2012) p73 expression is regulated by RNPC1, a target of the p53 family, via mRNA stability. Mol. Cell. Biol. 32, 2336–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J., Cho S. J., Shu L., Yan W., Guerrero T., Kent M., Skorupski K., Chen H., Chen X. (2011) Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 25, 1528–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cho S. J., Zhang J., Chen X. (2010) RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 38, 2256–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soto-Cerrato V., Viñals F., Lambert J. R., Pérez-Tomás R. (2007) The anticancer agent prodigiosin induces p21WAF1/CIP1 expression via transforming growth factor-β receptor pathway. Biochem. Pharmacol. 74, 1340–1349 [DOI] [PubMed] [Google Scholar]
- 31. Kim J. S., Baek S. J., Sali T., Eling T. E. (2005) The conventional nonsteroidal anti-inflammatory drug sulindac sulfide arrests ovarian cancer cell growth via the expression of NAG-1/MIC-1/GDF-15. Mol. Cancer Ther. 4, 487–493 [DOI] [PubMed] [Google Scholar]
- 32. Lee S. H., Krisanapun C., Baek S. J. (2010) NSAID-activated gene-1 as a molecular target for capsaicin-induced apoptosis through a novel molecular mechanism involving GSK3β, C/EBPβ and ATF3. Carcinogenesis 31, 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown D. A., Hance K. W., Rogers C. J., Sansbury L. B., Albert P. S., Murphy G., Laiyemo A. O., Wang Z., Cross A. J., Schatzkin A., Danta M., Srasuebkul P., Amin J., Law M., Breit S. N., Lanza E. (2012) Serum macrophage inhibitory cytokine-1 (MIC-1/GDF15): a potential screening tool for the prevention of colon cancer? Cancer Epidemiol. Biomarkers Prev. 21, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vaňhara P., Hampl A., Kozubík A., Souček K. (2012) Growth/differentiation factor-15: prostate cancer suppressor or promoter? Prostate Cancer Prostatic Dis. 15, 320–328 [DOI] [PubMed] [Google Scholar]