Background: L-A totivirus furnishes its transcript by a novel cap-snatching mechanism.
Results: L-BC virus transfers m7Gp from mRNA to the diphosphorylated 5′ end of its transcript to form a cap structure.
Conclusion: L-BC totivirus also can perform cap snatching.
Significance: The novel cap-snatching mechanism is shared among fungal totiviruses.
Keywords: Double-stranded RNA, mRNA, RNA Polymerase, RNA viruses, Transcription, Yeast, Gag, Cap Snatching, Capping
Abstract
Yeast L-A double-stranded RNA virus furnishes its transcript with a 5′ cap structure by a novel cap-snatching mechanism in which m7Gp from a host mRNA cap structure is transferred to the 5′-diphosphate terminus of the viral transcript. His-154 of the coat protein Gag forms an m7Gp adduct, and the H154R mutation abolishes both m7Gp adduct formation and cap snatching. Here we show that L-BC, another totivirus closely related to L-A, also synthesizes 5′-diphosphorylated transcripts and transfers m7Gp from mRNA to the 5′ termini of the transcripts. L-BC Gag also covalently binds to the cap structure and the mutation H156R, which corresponds to H154R of L-A Gag, abolishes cap adduct formation. Cap snatching of the L-BC virus is very similar to that of L-A; N7 methylation of the mRNA cap is essential for cap donor activity, and only 5′-diphosphorylated RNA is used as cap acceptor. L-BC cap snatching is also activated by viral transcription. Furthermore, both viruses require Mg2+ and Mn2+ for cap snatching. These cations are not only required for transcription activation but also directly involved in the cap transfer process. These findings support our previous proposal that the cap-snatching mechanism of the L-A virus is shared by fungal totiviruses closely related to L-A. Interestingly, L-A and L-BC viruses accept either viral transcript as cap acceptor in vitro. Because L-A and L-BC viruses cohabit in many yeast strains, it raises the possibility that their cohabitation in the same host may be beneficial for their mutual cap acquisition.
Introduction
Eukaryotic mRNA is capped at the 5′ end by a 7-methyl GMP moiety via an inverted 5′-5′-triphosphate linkage (m7GpppN-) (1). The cap structure has important roles in promoting the splicing of pre-mRNAs in the nucleus, transport of mRNA to the cytoplasm, and its translation and stability in the cytoplasm (2–6). In eukaryotic cells and for many viruses, the cap structure is installed onto the 5′ end of PolII transcripts through three enzymatic reactions (7, 8). The 5′-γ-phosphate at the 5′ end of the transcript is removed by RNA triphosphatase. Then Gp is transferred from GTP to the 5′ end of the transcript by a guanylyltransferase, thus forming a trisphosphate linkage. During the transfer, Gp is covalently attached to the guanylyltransferase through Lys as an intermediate. Finally, a methyl group from S-adenosyl methionine is transferred to N7 of G by a methyltransferase.
Influenza virus, instead of synthesizing the cap structure de novo, steals the structure from host mRNAs (cap snatching) (9). The trimeric viral polymerase binds host mRNAs, cleaves the RNA endonucleolytically 10–13 nucleotides (nt)2 downstream, and uses the capped fragment as a primer to synthesize its transcript (10). Recently, we found a novel cap-snatching mechanism in the yeast Saccharomyces cerevisiae totivirus L-A (11). L-A virus has a nonsegmented double-stranded RNA (dsRNA) genome of 4.6 kb (12). The genome contains two overlapping genes, gag and pol, that are decoded as Gag (76 kDa) and Gag-Pol (170 kDa) (13, 14). Gag-Pol is made by a −1 ribosomal frameshifting (15), a mechanism similar to that of retroviruses. The genome is packed inside of a 39-nm icosahedral capsid consisting of 60 asymmetric Gag dimers (16, 17), in which one or two Gag molecules are replaced by Gag-Pol. The L-A virion synthesizes the positive strand transcript conservatively (18), and the newly made transcript is released from the virion. The transcript has diphosphate at the 5′ end (19). Totivirus L-BC has a dsRNA genome of 4.6 Kb and is closely related to L-A. Like L-A virus, L-BC has two overlapping genes, gag and pol (20). L-A and L-BC are independent replicons, and they reside stably in the same cells. L-A requires MAK3, MAK10, and MAK31, the N-acetyltransferase C (NatC)-encoding genes (21, 22), but L-BC does not, and the clo1 mutation that results in the loss of L-BC does not affect L-A (23). Two decades ago, Sonenberg and colleagues (24) found that L-A Gag as well as L-BC Gag covalently bound mRNA. Subsequent studies indicated that the m7Gp moiety of the cap structure was attached to His-154 of L-A Gag (25). A mutant L-A Gag (H154R) failed to form the cap adduct (25). We have shown recently that L-A virus, in the presence of polyethylene glycol, which causes crowding of macromolecules in the aqueous solution (26), transfers m7Gp from mRNA to the 5′ end of the L-A transcript, thus forming a cap structure at the 5′ terminus (11) (Fig. 1A). The α- and β-phosphates of the triphosphate linkage are derived from the 5′ terminus of the L-A transcript and the γ-phosphate from mRNA. The H154R mutation abolishes cap snatching, indicating that the ability of Gag to form a cap adduct is essential for the cap transfer reaction (11). L-A virus can form a cap structure not only on an ongoing L-A transcript (cap snatching in cis) but also on an externally added L-A transcript (cap snatching in trans) (27). L-A cap snatching requires the N7 methylation of the mRNA cap structure for cap donor activity and the 5′-diphosphate of the L-A transcript for cap acceptor activity. The reaction is activated by transcription (27). The coupling between transcription and cap snatching may ensure the efficient capping of the L-A transcript.
FIGURE 1.
A, schematic diagram of cap-snatching mechanism in L-A and L-BC viruses. Gag decaps mRNA and forms an intermediate with m7Gp through His (His-154 for L-A and His-156 for L-BC). Then m7Gp is transferred to the diphosphorylated 5′ end of the emerging viral transcript to form a 5′-5′-triphosphate linkage (cap snatching in cis). m7Gp also can be transferred to an externally added viral transcript (cap snatching in trans). B, comparison of selected L-A sequences with those of other fungal totiviruses. His-154 and crucial residues for cap recognition are boxed and indicated by the arrow and vertical bars, respectively. GenBank accession numbers are: L-A (AAA50320.1), TaV1 (ADQ54105.1), XdL1A (NC_020903.1), XdL1B (AFH09413.1), L-BC (NP 042580.1), and SgVL (AGG68772.1)
Fungal totiviruses (including L-BC) closely related to L-A share His-154 and other amino acid residues of L-A Gag important for cap binding, at comparative positions in their coat proteins (Fig. 1B). We proposed that these viruses also acquire a cap structure on their transcripts by a mechanism similar to that of L-A (11). In this work, we demonstrate that the L-BC transcript has diphosphate at its 5′ end and that, like L-A, L-BC virus can install a cap structure on it by the cap-snatching mechanism. Thus these findings support our proposal. Interestingly, L-A and L-BC viruses can utilize either viral transcript as cap acceptor in vitro. Because these viruses cohabit in many yeast strains, it raises the possibility that their cohabitation is beneficial for their mutual cap acquisition.
EXPERIMENTAL PROCEDURES
L-BC Virus Preparation
L-BC virus was prepared from stationary phase cells of strain RE458 (Matα ski2-2 L-A-o, L-BC). The purification procedure was similar to that for L-A virus (28), with modifications. Cells grown in 2 liters of YPAD (2% peptone, 1% yeast extract, 2% glucose, and 0.04% adenine) for 3 days at 28 °C were harvested, washed once with deionized water, suspended in 1 volume/g of cells of buffer B (0.1 m Tris-HCl, pH 7.5, 1.4 m sorbitol, 1% 2-mercaptoethanol, and 2 mg/ml Zymolyase 20T (Seikagaku Corp.)), and incubated at 30 °C for 45 min. All subsequent steps were carried out at 0–4 °C. The spheroplasts were collected, suspended in buffer A (50 mm Tris-HCl, pH 7.5, 0.2 m NaCl, 2.5 mm EDTA, and 1 mm spermidine), and lysed by a single passage through a French pressure cell (14,000 p.s.i.). Cell debris was removed by centrifugation (18,500 × g, 20 min). To the supernatant, 0.25 volume of 25% PEG 4000 and 2.5 m NaCl was added. 30 min later, the suspension was centrifuged again (18,500 × g 10 min). The pellet was washed once with 50 ml of buffer A containing 5% PEG 4000 and an additional 0.5 m NaCl. The pellet was dissolved in buffer A. The density of the solution was adjusted to 1.36 g/ml by the addition of CsCl, and viral particles were banded by centrifugation at 34,000 rpm for 15 h in a Beckman 60Ti rotor. Fractions containing the virions were pooled and dialyzed against buffer A containing 10% (v/v) glycerol for 2 h. Finally, L-BC virions were concentrated by centrifugation at 65,000 rpm for 1 h in a Beckman TLA 100.3 rotor, suspended in 200 μl of buffer A containing 20% (v/v) glycerol, and kept at −70 °C before use.
Cap-snatching Reactions
The standard cap-snatching reaction in trans (25 μl) contains 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 20 mm NaCl, 5 mm KCl, 0.1 mm EDTA, 0.5 mm m7GpppG as cap donor, 4.5 mg/ml bentonite, 20% PEG 4000, and 0.5 mm ATP, 32P-labeled cap acceptor (either 16-nt L-A or 8-nt L-BC transcript, 5,000–10,000 cpm), and L-A or L-BC virions. The reaction was carried out at 30 °C for 20 min for L-BC virions or for 30 min for L-A. After phenol extraction, the products were analyzed on a urea-containing polyacrylamide gel as described previously (27). The cap-snatching reaction in cis is the same as the standard reaction in trans, except that m7GpppG and the 32P-labeled cap acceptor were replaced by GTP and UTP, 0.5 mm each, and a 32P-labeled 6-nt cap donor (3,000–5,000 cpm). We used the 6-nt m7Gp*ppGGGCGp (labeled with 32P, indicated by the asterisk) instead of m7GpppGGGCp (27) because of its ease in handling. The reaction was kept at 30 °C for 1 h, and the products were analyzed as described above. The amount of L-BC virions used was 13 μg of protein per reaction, approximately half of that of L-A (25 μg), except that in Fig. 5, we used fewer L-BC particles (7 μg of protein).
FIGURE 5.

Cap snatching in trans. A cap acceptor, 16-nt L-A transcript (lanes 2 and 3) or 8-nt L-BC transcript (lanes 5 and 6), was incubated with L-A (lanes 2 and 5) or L-BC (lanes 3 and 6) virions along with the cap donor m7GpppG. After incubation, capped products were separated in a denaturing acrylamide gel and visualized by autoradiography. The L-A (lane 1) or L-BC (lane 4) transcript without incubation was run in parallel as control. The acceptors (Acceptor), capped products (Capped), and their nucleotide sizes are indicated. The low capping efficiency by L-BC is due to a lower amount of L-BC virions used, as described under “Experimental Procedures.”
Cap Adduct Formation
The standard reaction mixture (25 μl) contains 50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 20 mm NaCl, 5 mm KCl, 0.1 mm EDTA, 3.6 mg/ml bentonite, 20% PEG 4000, 5,000 cpm of the 6-nt cap donor (0.4 pmol), and L-A or L-BC virions (10 μg of protein). The reaction was carried out at 30 °C for 20 min. After removing bentonite by brief centrifugation, proteins were analyzed by SDS-PAGE.
Expression of L-BC Gag
The open reading frame of L-BC Gag was amplified from L-BC dsRNA by RT-PCR and inserted into the pI2-based vector under the PGK1 promoter (29). The L-BC Gag expressed has an extra 14-amino acid sequence derived from the vector at the C terminus (-GGRHRGGAPILNQS). L-BC Gag with H156R mutation was generated by site-directed mutagenesis. Strain 2927 (Mata ura3 trp1 his3 ski2-2, L-BC-o, 20 S RNA) transformed with the vector containing the wild type or mutant L-BC Gag was grown overnight at 28 °C in H-Trp synthetic medium (30). The cells were harvested from 3 ml of the culture, washed once with lysis buffer (20 mm Tris-HCl, pH 7.5, 0.1 m NaCl), and suspended in 100 μl of lysis buffer. The cells were broken with glass beads by vortex mixing (15 s, 10 times). After removing cell debris and unbroken cells in an Eppendorf centrifuge at maximum speed (13,000 rpm) for 10 min at 4 °C, the lysates (∼20 μg of protein) were incubated in the reaction mixture for cap adduct formation as described above, and proteins were analyzed by SDS-PAGE.
Miscellaneous
TLC was done as described previously (19). L-A virions were purified from yeast strain TF229 (Mata his(3,4) leu2 ski2-2, L-A-HN) as described (28). SP6 transcription was carried out as described in Ref. 27. Site-directed mutagenesis was done as described (31). Radioactive nucleotides were obtained from PerkinElmer Life Sciences. S1 nuclease was from Promega. Tobacco acid pyrophosphatase (TAP) was from Epicenter. Bacterial alkaline phosphatase was from Invitrogen. ATPγS, m7GpppG, and GpppG were from Sigma-Aldrich, Ambion, and Amersham Biosciences, respectively.
RESULTS
Cap Adduct Formation by L-BC Gag
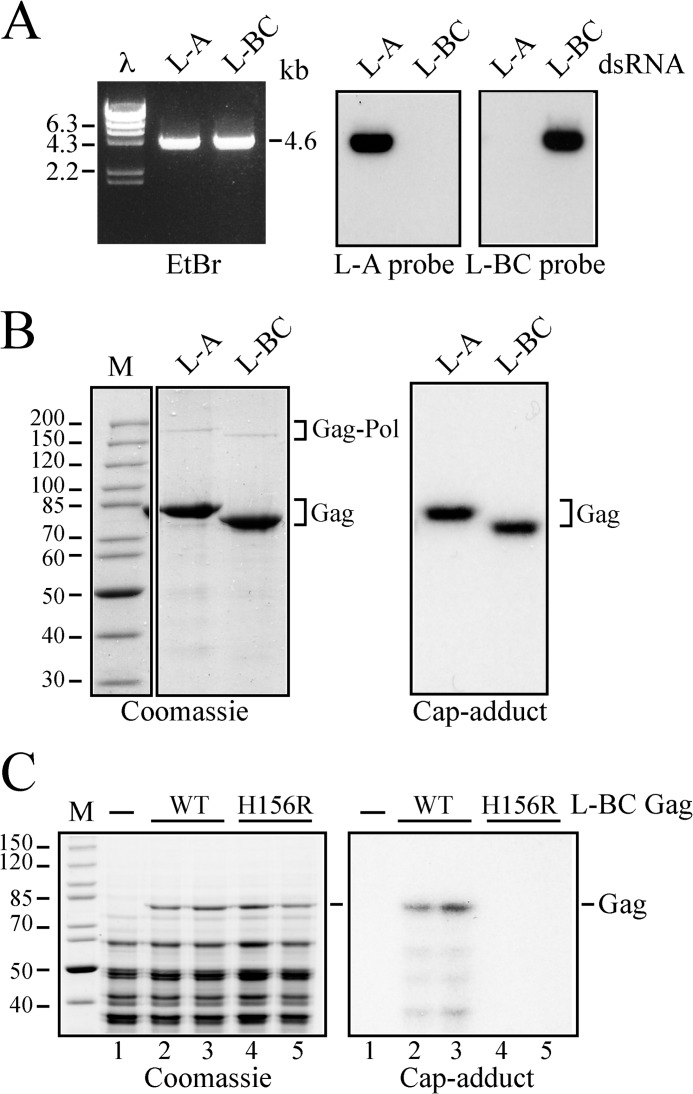
L-BC virions were isolated and purified from an L-A virus-free strain. The purification procedure was essentially the same as that of the L-A virus. Due to the fragility and the low copy number of the L-BC virus, however, we adopted several measures, including milder conditions for CsCl gradient centrifugation and the addition of a final high speed centrifugation step to concentrate purified virions as described under “Experimental Procedures.” The L-BC preparations are free from L-A virus as confirmed by Northern hybridization (Fig. 2A). L-BC Gag ran as a 75-kDa polypeptide and moved slightly faster than L-A Gag in SDS gels (Fig. 2B, left panel) as reported previously (24). When the virions were incubated with a 32P-labeled 6-nt cap donor, L-BC Gag formed a cap adduct (Fig. 2B, right panel) as demonstrated previously (24).
FIGURE 2.
L-BC Gag has cap adduct-forming activity. A, RNAs extracted from L-A and L-BC virus preparations were separated on an agarose gel and visualized by ethidium bromide staining (left panel, EtBr). λ HindIII markers (λ) and some of their sizes in kb are also shown. Autoradiograms of the Northern blots hybridized with L-A- and L-BC-specific probes are shown on the middle panel (L-A probe) and on the right panel (L-BC probe). The L-A probe recognizes the positive strand of L-A from nt 1323 to 1786. The L-BC probe hybridizes the positive strand of L-BC from nt 63 to 502. B, L-A and L-BC virions were incubated with the 6-nt cap donor, and then their proteins were analyzed by SDS-PAGE. The left panel (Coomassie) shows Coomassie Brilliant Blue staining of the gel with protein markers (M) whose sizes in kDa are indicated on the left. An autoradiogram of the same gel is shown on the right panel (Cap adduct). Gag and Gag-Pol of L-A and L-BC viruses are marked on the right of the panels. C, cell lysates from two independent transformants with a plasmid expressing WT (lanes 2 and 3) or H156R mutant (lanes 4 and 5) L-BC Gag were incubated with the 6-nt cap donor and then analyzed by SDS-PAGE. Coomassie Brilliant Blue staining of the gel (Coomassie) and an autoradiogram of the same gel (Cap-adduct) are shown. A lysate from untransformed cells was run in parallel as control (lane 1). M, protein markers. The position of L-BC Gag is indicated.
His-156 of L-BC Gag Is Essential for Cap Adduct Formation
In L-A virus, Gag covalently binds to the m7Gp moiety of the cap structure through His-154. This residue corresponds to His-156 in L-BC Gag (Fig. 1B). We expressed from vectors L-BC Gag with the WT sequence or with the mutation H156R and tested whether His-156 is involved in cap adduct formation. Crude lysates were incubated with the labeled cap donor, and the proteins were separated through an SDS gel. As shown in Fig. 2C, WT Gag formed the cap adduct, whereas H156R Gag failed to do so. Thus these results indicate that His-156 of L-BC Gag is essential for cap adduct formation. Although L-BC virus cannot be generated from L-BC cDNA expression vectors yet, it is now possible to perform reverse genetics to analyze cap adduct formation in L-BC Gag.
L-BC Transcript Has Diphosphate at the 5′ End
In the 5′ terminus, the L-BC-positive strand possesses the first C at position 9 and the second one at position 16 (Fig. 3C). When L-BC virions were incubated in a CTP-omitted transcription mixture containing [α-32P]GTP, the virions made an 8-nt transcript as a major product and a minor 15-nt transcript (Fig. 3A). The 8-nt transcript was isolated from the gel, digested with S1 nuclease, and then analyzed on TLC. The majority of the label was released as GDP, and a minority was released as GTP (Fig. 3B). Because the 8-nt transcript has a single G at the 5′ terminus, the result indicates that the L-BC transcript has diphosphate at its 5′ terminus. It is likely that the minor production of the 15-nt transcript is due to the presence of a trace amount of cytidine nucleotide in the viral preparation. We were, however, unable to eliminate it from the preparation by repeated dialysis or washing with additional centrifugation.
FIGURE 3.
L-BC transcript has diphosphate at the 5′ end. A, transcripts made by L-A or L-BC viruses in CTP-omitted reactions were separated on a 15% acrylamide/8 M urea gel. An autoradiogram of the gel is shown. The sizes of transcripts are indicated. B, a gel-purified 8-nt L-BC transcript labeled with [α-32P]GTP in a CTP-omitted reaction was treated with S1 nuclease and analyzed on PEI-cellulose (S1). None indicates the nontreated 8-nt transcript. The positions of nonlabeled guanine nucleotides are indicated on the right. C, the 5′-terminal sequences from L-BC- and L-A-positive strands. Cs relevant to the transcripts observed in A are marked by the arrows and arrowhead. Identical nucleotides between the first 8-nt L-A and L-BC sequences are underlined. Note that the 8-nt L-BC transcript has a single G at the 5′ terminus.
Cap Snatching in cis
Previously, we demonstrated that L-A virus could transfer m7Gp* (labeled with 32P, indicated by the asterisk) from a cap donor to an ongoing transcript in a CTP-omitted transcription reaction resulting in the production of a 17-nt 5′ capped product (cap snatching in cis) (Fig. 4A) (11). In the same reaction conditions, L-BC transferred the label into the minor 15-nt transcript, thus producing a 16-nt product (Fig. 4A). Although the 8-nt transcript was the major product in transcription, it did not incorporate the label efficiently. To confirm that the product was 5′-capped, we isolated the 16-nt product from the gel and treated it with S1 or TAP. As shown in Fig. 4B, S1 and TAP digestions produced m7Gp*ppG and m7Gp*, respectively. Because S1 does not work on the triphosphate linkage of the cap structure and TAP cleaves anhydrous bonds between α- and β-, and also β- and γ-phosphates of the triphosphate linkage, these results indicate that, like cap snatching in the L-A virus, the L-BC virus also can transfer the m7Gp moiety from the cap donor to the diphosphorylated 5′ terminus of the transcripts, thus forming a cap structure at the 5′ ends (Fig. 1A).
FIGURE 4.
L-BC cap snatching in cis. A, L-A or L-BC virions were incubated with the cap donor m7Gp*ppGGGCGp (* indicates the position of 32p) in a CTP-omitted reaction, and the capped products were separated on a denaturing acrylamide gel. An autoradiogram of the gel is shown. The positions of the donor and the capped products are indicated. Lane − indicates the cap donor alone without incubation as control. B, the 16-nt L-BC capped product was isolated from the gel, treated with S1 nuclease (S1) or tobacco acid pyrophosphatase (TAP), and analyzed on PEI-cellulose. The cap donor was also analyzed in parallel. None indicates nontreated sample. The deduced transfer reaction is shown on the bottom with the S1 and TAP cleavage sites.
Cap Snatching in trans
L-A virus can transfer m7Gp from the cap analog m7GpppG to an externally added L-A transcript (cap acceptor) to form a cap structure at the 5′ terminus (cap snatching in trans) (27). We tested whether L-BC virus also can perform cap snatching in trans. The 8-nt L-BC transcript was isolated and then incubated with L-BC virions in the presence of m7GpppG. As shown in Fig, 5, lane 6, L-BC virions converted a part of the transcript to a slower moving band. The same band was also produced in a reaction with L-A virus (Fig. 5, lane 5). Furthermore, when the 16-nt L-A transcript was incubated with L-BC virus, L-BC converted a part of it to a slower moving band (Fig. 5, lane 3), which comigrated with the 17-nt capped species produced by the L-A virus (Fig. 5, lane 2). These results suggest firstly that L-BC virus can perform cap snatching in trans, and secondly that both L-A and L-BC viruses can use either viral transcript as cap acceptor. Because the 16-nt L-A transcript can be produced with ease (Fig. 3A), hereafter we used this L-A transcript as acceptor for cap snatching in trans with L-BC virus. We confirmed cap snatching in trans by the L-BC virus as shown in Fig. 6. The label located at the 5′-α-phosphate of the 16-nt L-A transcript (acceptor) can be removed by bacterial alkaline phosphatase (Fig. 6A, lane 2), whereas the label in the 17-nt product generated by the L-BC virus became resistant to the treatment due to its internalization (Fig. 6A, lane 4). Bacterial alkaline phosphatase is a phosphomonoesterase that hydrolyzes 5′- and 3′-phosphates from RNA and DNA. We isolated the bacterial alkaline phosphatase-resistant 17-nt transcript from the gel. S1 treatment of the 17-nt product generated m7GpppG, thus confirming the formation of cap structure at the 5′ end (Fig. 6B, lane 4). The 17-nt product treated with TAP remained at the origin (Fig. 6B, lane 5) because the remaining 16-nt RNA body was intact. Further S1 treatment generated GMP (Fig. 6B, lane 6). These results thus indicate that the label was located at the α-phosphate of the triphosphate linkage in the cap structure.
FIGURE 6.
Proof of L-BC cap snatching in trans. A, a 16-nt L-A transcript (Acceptor) 5′-labeled with [α-32P]GTP was incubated with L-BC virions in the presence of the cap donor m7GpppG. The product (lanes 3 and 4) or the acceptor (lanes 1 and 2) was nontreated (lanes 1 and 3) or treated with bacterial alkaline phosphatase (BAP, lanes 2 and 4) and separated on a denaturing acrylamide gel. An autoradiogram of the gel is shown. B, the bacterial alkaline phosphatase-resistant product shown in A, lane 4, was isolated from the gel, treated with S1 nuclease (S1) or TAP alone, or sequentially treated with TAP and S1, and analyzed on PEI-cellulose. The acceptor was also treated with S1 and analyzed on the same plate. None: nontreated sample. The deduced cap transfer reaction is shown on the bottom with S1 and TAP cleavage sites.
Characterization of L-BC Cap Snatching in trans
Cap-snatching activity of the L-BC virus has characteristics very similar to those of the L-A virus. Like in the case of L-A, the L-BC virus could utilize m7GpppG but not GpppG as the cap donor (Fig. 7B), indicating that N7 methylation is essential for cap donor activity. The reaction was completed within 20 min of incubation (Fig. 7A). L-A cap snatching requires ATP, and none of the other three nucleotides can replace ATP for the reaction (27). In addition to ATP, L-A cap snatching in trans required a primer for transcription, either a guanine nucleotide or the cap analog m7GpppG, resulting in the synthesis of 5′-XAAAAA-OH (X indicates primer). This led to the conclusion that the cap transfer process is activated by viral transcription. In accordance, a nonhydrolyzable ATP analog ATPγS can replace ATP for cap snatching because the viral polymerase efficiently utilizes this analog for transcription (27). As shown in Fig. 7C, L-BC cap snatching also requires ATP, and none of the other three nucleotides could support the L-BC cap-snatching reaction (Fig. 7D). Furthermore, ATPγS could replace ATP for the reaction (Fig. 7C). These results suggest that even the synthesis of the small fragment m7GpppGAA-OH by transcription is sufficient to activate L-BC cap snatching. The addition of EDTA inhibited the cap snatching of the L-BC virus (Fig. 7D), indicating that the reaction requires divalent cations. Mg2+ and Mn2+ could support both cap snatching (Fig. 8, A and B) and transcription (Fig. 8C) in L-A and L-BC viruses, although Mn2+ is less active than Mg2+ in transcription. These results raise the possibility that the divalent cations support cap snatching indirectly through the activation of transcription. We further analyzed the effects of divalent cations on cap adduct formation by L-BC Gag. As shown in Fig. 8D, cap adduct formation also required Mg2+ and Mn2+. Because this reaction is closely related with the cap transfer process and proceeds in the absence of ATP (without transcription activation), we conclude that these cations are directly involved in the cap transfer reaction itself.
FIGURE 7.
Requirements for L-BC cap snatching in trans. A, time course of L-BC cap snatching. The incubation times are indicated below the panel. m7GpppG and the 16-nt L-A transcript were used as cap donor and acceptor, respectively. B, the cap-snatching reaction was carried out in the presence of 0.5 mm m7GpppG or GpppG or in their absence (−) as indicated. The 16-nt L-A transcript was used as cap acceptor. The presence (+) or absence (−) of L-BC virions (L-BC) in the reaction is also indicated. C, the cap-snatching reaction was carried out in the presence of 0.5 mm ATP (+) or ATPγS or in their absence (−) as indicated. 0.5 mm m7GpppG was used as cap donor. The presence (+) or absence (−) of 5 mm MgCl2 in the reaction is also shown. D, ATP requirements. The cap-snatching reaction was carried out in the presence of one of nucleotide triphosphates (0.5 mm) or in their absence (−). In the rightmost lane, 10 mm EDTA was also added to the reaction.
FIGURE 8.
Effects of divalent cations on cap snatching and cap adduct formation. A and B, effects of divalent cations (5 mm) on L-BC (A) or L-A (B) cap snatching. C, effects of divalent cations (5 mm) on L-A or L-BC transcription. The transcripts were analyzed on an agarose gel and visualized by ethidium bromide staining (EtBr, upper panel). An autoradiogram of the gel is also shown in the lower panel. D, cap adduct formation of L-BC Gag in the presence of 5 mm divalent cations or in their absence (−). L-BC virions were incubated with the labeled 6-nt cap donor in the presence or absence of divalent cations. Then the protein was separated by SDS-PAGE and visualized by Coomassie Brilliant Blue staining (Coomassie). An autoradiogram of the gel (Cap-adduct) is shown on the right. M: protein mobility markers (kDa).
Specificity for Cap Acceptor
Previously, we demonstrated that L-A virus accepted 5′-diphosphorylated RNA but not tri- or monophosphorylated molecules as cap acceptors in the cap-snatching reaction (27). Here we synthesized 16-nt L-A 5′-terminal RNAs using SP6 RNA polymerase from an L-A cDNA fused to the SP6 promoter. Using GTP, GDP, and GMP as a primer, we generated RNAs with tri-, di-, and monophosphate, respectively, at their 5′ termini. Like L-A virus, L-BC admitted only 5′-diphosphorylated RNA as cap acceptor (Fig. 9A). As demonstrated in Fig. 5, L-A virus accepted not only its 16-nt transcript but also the 8-nt L-BC transcript as cap acceptor. This indicates that the L-A sequence from 9 to 16 is not essential for cap acceptor activity for L-A virus. L-BC virus also used not only its 8-nt transcript but also the 16-nt L-A transcript as cap acceptor. In the first 8 nucleotides, L-A and L-BC transcripts share 1GAA3 and 7UU8 sequences (Fig. 3C). Therefore, 4AAA6 of the L-A transcript is not needed for L-A cap snatching. Likewise 4UUU6 of L-BC transcript is not necessary for L-BC cap snatching. We modified the second or third A of the L-A sequence to U by site-directed mutagenesis. SP6 RNA polymerase, however, made very few transcripts with the A2U mutation because the polymerase requires GA at the 5′ end of the transcript as part of the promoter. When RNA with the A3U mutation was tested, L-A and L-BC viruses used this RNA as cap acceptor (Fig. 9B). Therefore, L-A and L-BC viruses require diphosphate at the 5′ termini of the viral transcripts but apparently no specific viral nucleotide sequences for cap acceptor activity. The role of the 5′-terminal GA sequence in the activity, however, remains to be clarified.
FIGURE 9.
A diphosphate status at the 5′ end is required for cap acceptor activity in L-BC cap snatching. A, 16-nt L-A 5′-terminal fragments bearing tri (ppp-), di (pp-), and mono (p-) phosphate at their 5′ ends were synthesized by SP6 RNA polymerase using GTP, GDP, and GMP, respectively, as primers, and their cap acceptor activity in the L-BC cap-snatching reaction was tested. B, effects of nucleotide sequence on cap acceptor activity. A 16-nt L-A 5′-terminal fragment having the WT sequence or the A3U mutation was synthesized by SP6 RNA polymerase using GDP as a primer, and its cap acceptor activity in the L-A or L-BC cap-snatching reaction was tested. The A3U mutation is indicated by the dot.
In many strains, L-A and L-BC viruses cohabit in the same cells. Because these viruses can use either viral transcript as cap acceptor in the in vitro cap-snatching reactions, this raises the possibility that their cohabitation may be beneficial for their mutual cap acquisition in vivo. M1, a satellite RNA of the L-A virus, requires L-A-encoded proteins for encapsidation and replication. M1 encodes the K1 killer toxin and its immunity (32). Efficient expression of the toxin requires capping of M1 transcripts (11). Cells carrying M1 maintained by capping-deficient L-A Gag (H154R) showed a negative phenotype in killer assay (11), and the presence or absence of L-BC viruses apparently did not affect this phenotype (Fig. 10). These results suggest that M1 (or L-A) and L-BC preferentially install the cap structure on their ongoing transcript by cap snatching in cis in the cell. Alternatively, because the copy number of the L-BC virus is 1 or 2 orders of magnitude lower than that of L-A, the effects of the L-BC virus on M1 capping may be subtle, and thus we may need to develop a more sensitive capping assay
FIGURE 10.

Effects of L-BC virus on M1 killer phenotype. A, diagram of L-A expression plasmid. Gag (closed dot) and Gag-Pol (closed dot-closed square) are expressed from the constitutive PGK1 promoter (PGK1). The thick line represents the L-A cDNA sequence, and the thin lines are vector sequences. The position of His-154 in Gag is indicated by the vertical bar. B, wild type (WT) or mutated (H154R) virions were expressed from the vectors in L-BC or L-BC-o strains. Satellite M1 RNA can be encapsidated into virions produced by L-A proteins expressed from the plasmids (29). Mutated (H154R) virions can also maintain M1 RNA, although M1 transcripts produced by the mutant virions lacked the cap structure at the 5′ ends and thus expressed the secreted killer toxin poorly (11). C, killer toxin assay. Colonies separated by the vertical line harbor M1 satellite RNA maintained by a vector expressing either WT (left) or H154R (right) L-A Gag. Colonies harboring L-BC virus (L-BC) or no virus (L-BC-o) are shown below or above the horizontal line. The expression of the killer toxin is visualized by clear halos resulting from killing the toxin-sensitive lawn cells.
DISCUSSION
In this work, we have demonstrated that yeast L-BC virus can perform cap snatching to furnish its transcript with a cap structure and that the capping reaction is very similar to that found in the L-A virus. L-BC virus synthesizes transcripts with diphosphate at their 5′ termini and transfers m7Gp from mRNA to the 5′ terminus of the transcript. L-BC virus can form a cap structure not only on an ongoing transcript (cap snatching in cis) but also on an externally added transcript (cap snatching in trans), indicating that the cap-snatching site is externally accessible. The cap-snatching site of the L-A virus is located in a trench on the outer surface of Gag (33). L-A transcripts are made inside the virion and presumably released to the cytoplasm through one of pores located at its icosahedral five-fold symmetric axes. The trenches are positioned in the Gag asymmetric dimer close to the two-fold and three-fold axes, far from the pores. There are no structural studies on the L-BC virus. Considering the similarities between L-A and L-BC viruses, however, it is likely that the cap-snatching site of L-BC is also located on the surface of Gag far from its five-fold axes. This could explain why L-BC virus preferentially capped a 15-nt transcript over a shorter 8-nt transcript in cap snatching in cis. The 8-nt transcript might be too short to interact at the 5′ end with the cap-snatching site while anchored at the 3′ end in the pore. L-BC cap snatching requires ATP, and its analog ATPγS can replace ATP for the activity. This suggests that as in the case of L-A, L-BC cap snatching is also activated by transcription. L-BC cap snatching requires a divalent cation Mg2+ or Mn2+. Both transcription and cap adduct formation require the same cations. Therefore, these cations have dual roles in cap snatching: one through transcription activation and the other a direct involvement in the cap transfer reaction itself.
The capping reaction of the L-BC virus uses N7 methylated cap molecules but not nonmethylated ones as cap donors. Like L-A virus, L-BC requires 5′-terminal diphosphate for cap acceptor activity. Interestingly, L-A and L-BC viruses can admit either viral transcript as cap acceptor in vitro. It raises the possibility that their cohabitation in the same cell is beneficial for their cap acquisition. Furthermore, because host RNA with 5′-terminal diphosphate is rare in the cytoplasm, it also suggests that this requirement alone is sufficient to discriminate host RNA as cap acceptor in cap snatching. mRNA degradation in eukaryotes usually begins with the shortening of the poly(A) tail at the 3′ end followed by decapping at the 5′ end by the Dcp1-Dcp2 decapping enzyme to generate an RNA body bearing 5′-monophosphate (34, 35). Then decapped RNA is digested by the SKI1/XRN1 5′-exonuclease. Alternatively, the deadenylated RNA is digested by the 3′-exonuclease activity of the exosome. It is not clear how RNAs with 5′-diphosphate are produced in the cytoplasm and how stable they are. The scavenger decapping enzyme removes m7Gp from dinucleotide- or oligonucleotide-capped molecules generated by the exosome, leaving the 3′ fragment with 5′-diphosphate (36, 37). The CTL1 gene product has RNA triphosphatase activity (38). It localizes in the cytoplasm, but its physiological role is unknown. We have also observed that ski1Δ cells accumulate L-A transcripts, most of which are 5′-monophosphorylated (19), thus indicating the presence of RNA diphosphatase activity in the cytoplasm. If this activity operates efficiently enough to convert the 5′-diphosphorylated viral transcripts, once released from the virions, to 5′-monophosphorylated ones, then it might suppress cross capping in vivo.
In L-A virus, His-154 of Gag forms a phosphoamide bond with m7Gp. His-154 is located at the tip of a loop (residues 144–163) that is part of the upper rim of the trench. Upon m7GDP binding, the rim moves inwardly and forms a closed conformation (39). In the trench, Tyr-150, Asp-152, Tyr-452, and Tyr-538 are located close to the bound m7GDP, suggesting their involvement in cap recognition. Mutagenesis studies indicate that these residues are crucial for cap adduct formation (39). Guanylyltransferase also contains a trench that can adopt either an open or a closed conformation during the canonical mRNA capping reaction (40, 41). This enzyme forms a Gp-enzyme intermediate with GTP and transfers Gp to the diphosphorylated 5′ termini of PolII transcripts. Therefore, the cap-snatching reaction of L-A resembles that of guanylyltransferase. Because His-156 of L-BC Gag is essential for cap adduct formation, it is likely that L-BC Gag also forms an intermediate with m7Gp through His-156 and transfers m7Gp to the 5′ terminus of its transcript. Guanylyltransferase relies on Lys but not on His to form a Gp-enzyme intermediate. DNA and T4 RNA ligases also use Lys to form an AMP (NAD)-enzyme intermediate and transfer the Ap moiety to a DNA/RNA 5′ end, resulting in the formation of an activated A(5′)pp(5′)DNA/RNA intermediate (42, 43). Then a terminal DNA deoxyribose (RNA ribose) 3′-OH attacks the 5′ α-phosphate of A(5′)pp(5′)DNA/RNA to produce a 3′,5′-phosphodiester at the splice junction. In the nonconventional capping reaction of vesicular stomatitis virus, however, L protein covalently binds the 5′-monophosphorylated pre-mRNA through His and transfers the bound pre-mRNA to GDP (44). Members of the alphavirus-like superfamily methylate GTP first and then appear to form an m7Gp-enzyme intermediate through His and transfer m7Gp to the 5′-diphosphate end of the viral transcript during the capping reaction (45). Recently, it has been demonstrated that the RNA ligase RtcB from Pyrococcus horikoshii or Escherichia coli forms a Gp-enzyme intermediate through His with GTP and transfers Gp to a polynucleotide 3′-phosphate to form polynucleotide-(3′)pp(5′)G (46, 47). Then attack of an RNA 5′-OH on the -N(3′)pp(5′)G end completes the ligation reaction. RNA cyclase RtcA from E. coli reacts with ATP to form an Ap-enzyme intermediate through His and transfers Ap to an RNA 3′-phosphate terminus to form RNA(3′)pp(5′)A (48). Then the terminal RNA ribose 2′-OH attacks the 3′-phosphate of RNA(3′)pp(5′)A to generate RNA 2′,3′-cyclic phosphate. Interestingly, this enzyme can also transfer Ap to an RNA 5′-phosphate terminus, resulting in the formation of A(5′)pp(5′)RNA, reminiscent of the intermediates produced by DNA and T4 RNA ligases, although RtcA cannot promote the ligation reaction (49). Therefore, many enzymes use not only Lys but also His to form (poly)nucleotide-enzyme intermediates for chemical transformations at polynucleotide termini.
Transcription of influenza virus occurs in the nucleus in the form of a ribonucleoprotein complex consisting of viral RNA, RNA-dependent RNA polymerase, and NP proteins. The polymerase is a hetero-trimer composed of PA, PB1, and PB2 subunits. The polymerase is accessible to pre-mRNA in the nucleus; PB2 binds to the cap structure of pre-mRNA, PA cleaves the bound RNA endonucleolytically 10–13 nt downstream of the cap, and finally, PB1 uses the capped fragment as a primer to transcribe the viral RNA. By contrast, transcription and replication of L-A and L-BC totiviruses occur inside the virion. Once assembled, the Pol domain of Gag-Pol is confined inside the virion and is not accessible to mRNA in the cytoplasm. In this context, it is quite reasonable that the cap-snatching site of the totivirus is located at the cytoplasmic side of the capsid protein Gag. This raises another interesting question of how L-A and L-BC viruses can activate cap snatching at the surface of the virion by transcription that occurs inside the virion. The answer to this will not only provide insights on virus structure and function but also contribute to the understanding of how these simplest dsRNA viruses with only two encoded proteins can live and propagate in their eukaryotic host.
Acknowledgment
We thank “Fundación Ramón Areces” for financial support of our Institute.
This work was supported by Grant BFU2010-15768 from the Spanish Ministry of Education and Science.
- nt
- nucleotide(s)
- TAP
- tobacco acid pyrophosphatase
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Shatkin A. J. (1976) Capping of eucaryotic mRNAs. Cell 9, 645–653 [DOI] [PubMed] [Google Scholar]
- 2. Konarska M. M., Padgett R. A., Sharp P. A. (1984) Recognition of cap structure in splicing in vitro of mRNA precursors. Cell 38, 731–736 [DOI] [PubMed] [Google Scholar]
- 3. Wickens M. P., Gurdon J. B. (1983) Post-transcriptional processing of simian virus 40 late transcripts in injected frog oocytes. J. Mol. Biol. 163, 1–26 [DOI] [PubMed] [Google Scholar]
- 4. Izaurralde E., Lewis J., Gamberi C., Jarmolowski A., McGuigan C., Mattaj I. W. (1995) A cap-binding protein complex mediating U snRNA export. Nature 376, 709–712 [DOI] [PubMed] [Google Scholar]
- 5. Gingras A. C., Raught B., Sonenberg N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 6. Furuichi Y., Shatkin A. J. (2000) Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55, 135–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Venkatesan S., Moss B. (1980) Donor and acceptor specificities of HeLa cell mRNA guanylyltransferase. J. Biol. Chem. 255, 2835–2842 [PubMed] [Google Scholar]
- 8. Shuman S. (1995) Capping enzyme in eukaryotic mRNA synthesis. Prog. Nucleic Acid Res. Mol. Biol. 50, 101–129 [DOI] [PubMed] [Google Scholar]
- 9. Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. (1981) A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 [DOI] [PubMed] [Google Scholar]
- 10. Boivin S., Cusack S., Ruigrok R. W., Hart D. J. (2010) Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J. Biol. Chem. 285, 28411–28417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fujimura T., Esteban R. (2011) Cap-snatching mechanism in yeast L-A double-stranded RNA virus. Proc. Natl. Acad. Sci. U.S.A. 108, 17667–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wickner R. B. (2007) in Fields Virology (Knipe D. M., Howley P. M., eds) 5th Ed., pp. 737–768, Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 13. Icho T., Wickner R. B. (1989) The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 264, 6716–6723 [PubMed] [Google Scholar]
- 14. Fujimura T., Wickner R. B. (1988) Gene overlap results in a viral protein having an RNA binding domain and a major coat protein domain. Cell 55, 663–671 [DOI] [PubMed] [Google Scholar]
- 15. Dinman J. D., Icho T., Wickner R. B. (1991) A-1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. U.S.A. 88, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng R. H., Caston J. R., Wang G. J., Gu F., Smith T. J., Baker T. S., Bozarth R. F., Trus B. L., Cheng N., Wickner R. B., Steven A. C. (1994) Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J. Mol. Biol. 244, 255–258 [DOI] [PubMed] [Google Scholar]
- 17. Castón J. R., Trus B. L., Booy F. P., Wickner R. B., Wall J. S., Steven A. C. (1997) Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J. Cell Biol. 138, 975–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujimura T., Esteban R., Wickner R. B. (1986) In vitro L-A double-stranded RNA synthesis in virus-like particles from Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 83, 4433–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujimura T., Esteban R. (2010) Yeast double-stranded RNA virus L-A deliberately synthesizes RNA transcripts with 5′-diphosphate. J. Biol. Chem. 285, 22911–22918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park C. M., Lopinski J. D., Masuda J., Tzeng T. H., Bruenn J. A. (1996) A second double-stranded RNA virus from yeast. Virology 216, 451–454 [DOI] [PubMed] [Google Scholar]
- 21. Tercero J. C., Wickner R. B. (1992) MAK3 encodes an N-acetyltransferase whose modification of the L-A gag NH2 terminus is necessary for virus particle assembly. J. Biol. Chem. 267, 20277–20281 [PubMed] [Google Scholar]
- 22. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 23. Wesolowski M., Wickner R. B. (1984) Two new double-stranded RNA molecules showing non-Mendelian inheritance and heat inducibility in Saccharomyces cerevisiae. Mol. Cell Biol. 4, 181–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Blanc A., Goyer C., Sonenberg N. (1992) The coat protein of the yeast double-stranded RNAvirus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol. Cell Biol. 12, 3390–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blanc A., Ribas J. C., Wickner R. B., Sonenberg N. (1994) His-154 is involved in the linkage of the Saccharomyces cerevisiae L-A double-stranded RNA virus Gag protein to the cap structure of mRNAs and is essential for M1 satellite virus expression. Mol. Cell Biol. 14, 2664–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Minton A. P. (1983) The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol. Cell Biochem. 55, 119–140 [DOI] [PubMed] [Google Scholar]
- 27. Fujimura T., Esteban R. (2012) Cap snatching of yeast L-A double-stranded RNA virus can operate in trans and requires viral polymerase actively engaging in transcription. J. Biol. Chem. 287, 12797–12804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujimura T., Wickner R. B. (1992) Interaction of two cis sites with the RNA replicase of the yeast L-A virus. J. Biol. Chem. 267, 2708–2713 [PubMed] [Google Scholar]
- 29. Wickner R. B., Icho T., Fujimura T., Widner W. R. (1991) Expression of yeast L-A double-stranded RNA virus proteins produces derepressed replication: a ski− phenocopy. J. Virol. 65, 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wickner R. B. (1980) Plasmids controlled exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell 21, 217–226 [DOI] [PubMed] [Google Scholar]
- 31. Esteban R., Fujimura T., Wickner R. B. (1989) Internal and terminal cis-acting sites are necessary for in vitro replication of the L-A double-stranded RNA virus of yeast. EMBO J. 8, 947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tipper D. J., Bostian K. A. (1984) Double-stranded ribonucleic acid killer systems in yeasts. Microbiol. Rev. 48, 125–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Naitow H., Tang J., Canady M., Wickner R. B., Johnson J. E. (2002) L-A virus at 3.4-Å resolution reveals particle architecture and mRNA decapping mechanism. Nat. Struct. Biol. 9, 725–728 [DOI] [PubMed] [Google Scholar]
- 34. Parker R., Song H. (2004) The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127 [DOI] [PubMed] [Google Scholar]
- 35. Wilusz C. J., Wormington M., Peltz S. W. (2001) The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237–246 [DOI] [PubMed] [Google Scholar]
- 36. Wang Z., Kiledjian M. (2001) Functional link between the mammalian exosome and mRNA decapping. Cell 107, 751–762 [DOI] [PubMed] [Google Scholar]
- 37. Liu H., Rodgers N. D., Jiao X., Kiledjian M. (2002) The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21, 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodriguez C. R., Takagi T., Cho E. J., Buratowski S. (1999) A Saccharomyces cerevisiae RNA 5-triphosphatase related to mRNA capping enzyme. Nucleic Acids Res. 27, 2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang J., Naitow H., Gardner N. A., Kolesar A., Tang L., Wickner R. B., Johnson J. E. (2005) The structural basis of recognition and removal of cellular mRNA 7-methyl G 'caps' by a viral capsid protein. A unique viral response to host defense. J. Mol. Recognit. 18, 158–168 [DOI] [PubMed] [Google Scholar]
- 40. Håkansson K., Doherty A. J., Shuman S., Wigley D. B. (1997) X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell 89, 545–553 [DOI] [PubMed] [Google Scholar]
- 41. Chu C., Das K., Tyminski J. R., Bauman J. D., Guan R., Qiu W., Montelione G. T., Arnold E., Shatkin A. J. (2011) Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc. Natl. Acad. Sci. U.S.A. 108, 10104–10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shuman S., Lima C. D. (2004) The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 14, 757–764 [DOI] [PubMed] [Google Scholar]
- 43. Pascal J. M. (2008) DNA and RNA ligases: structural variations and shared mechanisms. Curr. Opin. Struct. Biol. 18, 96–105 [DOI] [PubMed] [Google Scholar]
- 44. Ogino T., Yadav S. P., Banerjee A. K. (2010) Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 3463–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang Y. L., Hsu Y. H., Han Y. T., Meng M. (2005) mRNA guanylation catalyzed by the S-adenosylmethionine-dependent guanylyltransferase of bamboo mosaic virus. J. Biol. Chem. 280, 13153–13162 [DOI] [PubMed] [Google Scholar]
- 46. Chakravarty A. K., Subbotin R., Chait B. T., Shuman S. (2012) RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 109, 6072–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., Eom S. H., Konigsberg W. H., Söll D., Wang J. (2012) Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 109, 15235–15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chakravarty A. K., Smith P., Shuman S. (2011) Structures of RNA 3′-phosphate cyclase bound to ATP reveal the mechanism of nucleotidyl transfer and metal-assisted catalysis. Proc. Natl. Acad. Sci. U.S.A. 108, 21034–21039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chakravarty A. K., Shuman S. (2011) RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J. Biol. Chem. 286, 4117–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]