Background: Whether a change in protein biosynthesis modulates protein degradation is unknown.
Results: Inhibition of protein biosynthesis induces activation of AKT (protein kinase B), leading to activation of E3 ubiquitin ligase and thus degradation of its substrate proteins.
Conclusion: Protein degradation is regulated by protein biosynthesis.
Significance: This study reveals a novel regulation of protein degradation.
Keywords: AKT, Protein Degradation, Protein Synthesis, Protein Turnover, Signal Transduction
Abstract
The homeostasis of protein metabolism is maintained and regulated by the rates of protein biosynthesis and degradation in living systems. Alterations of protein degradation may regulate protein biosynthesis through a feedback mechanism. Whether a change in protein biosynthesis modulates protein degradation has not been reported. In this study, we found that inhibition of protein biosynthesis induced phosphorylation/activation of AKT and led to phosphorylation of AKT target substrates, including FoxO1, GSK3α/β, p70S6K, AS160, and the E3 ubiquitin ligase MDM2. Phosphorylation of ribosomal protein S6 was also modulated by inhibition of protein biosynthesis. The AKT phosphorylation/activation was mediated mainly through the PI3K pathway because it was blocked by the PI3K inhibitor LY294002. The activated AKT phosphorylated MDM2 at Ser166 and promoted degradation of the tumor suppressor p53. These findings suggest that inhibition of protein biosynthesis can alter degradation of some proteins through activation of AKT. This study reveals a novel regulation of protein degradation and calls for caution in blocking protein biosynthesis to study the half-life of proteins.
Introduction
The most common approach to determine the turnover or half-life of a protein in cultured cells is to measure the degradation of the protein after blocking its biosynthesis. Cycloheximide, a protein synthesis inhibitor that acts specifically on the 60 S subunit of eukaryotic ribosomes (1), is widely used for this purpose. However, inhibition of protein synthesis also confers cellular stress. Cells respond to stress in a variety of ways, ranging from activation of survival pathways to initiation of programmed cell death, eventually eliminating damaged cells. Whether cells provide a protective or destructive stress response depends on the nature and duration of the stress as well as the cell type (2). Treatment of rat hepatocytes or astrocytes, human leukocytes, and Burkitt lymphoma cells with cycloheximide can induce apoptosis (3–5). Whether the responses of cells to protein synthesis inhibitors alter protein turnover is not known.
The serine/threonine kinase AKT (transforming murine retrovirus AKT8-related oncogene), also known as PKB, is a downstream effector of PI3K/PDK1 (3-phosphoinositide-dependent protein kinase 1) and plays key roles in the regulation of cell survival, the cell cycle, cell growth, and cell metabolism (6). AKT is activated through its phosphorylation at Thr308 and Ser473 (7, 8). There are three isoforms of AKT in mammals, termed AKT1/PKBα, AKT2/PKBβ, and AKT3/PKBγ. These isoforms are products of distinct genes but are highly related, exhibiting >80% protein sequence homology, and share the same structural organization (9). However, the AKT isoforms distribute differently and have different functions despite the high level of homology. AKT1 is expressed in most tissues; AKT2 is highly expressed in insulin-responsive tissues, including adipose tissue, skeletal muscle, and liver; and AKT3 expression is prominent in the brain and testes (10). Animal models deficient in AKT1, AKT2, or AKT3 have indicated that the three AKT isoforms differ in physiological functions. Mice lacking AKT1 demonstrate increased perinatal mortality and reductions in body weight (11, 12). In contrast, AKT2-deficient mice exhibit a diabetes-like syndrome with an elevated fasting plasma glucose level, elevated hepatic glucose output, and peripheral insulin resistance (10, 13). Although the functions of AKT3 are not well known, AKT3-deficient mice exhibit a reduction in brain weight resulting from decreases in both cell size and cell number but maintain normal glucose homeostasis and body weight (14, 15).
During investigation of the regulation and turnover of AKT, we observed that inhibition of protein biosynthesis by cycloheximide induced phosphorylation/activation of AKT and altered its degradation. Furthermore, we found that the degradation of several other proteins was also altered when protein biosynthesis was inhibited and that this phenomenon involved AKT activation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
LY294002, rapamycin, cycloheximide, and anti-p53 antibody were products of Sigma-Aldrich. NSC119889 was purchased from EMD Chemicals. Antibodies against GAPDH, MDM2 (SMP14), and phospho-MDM2 (Ser166) were products of Santa Cruz Biotechnology. Antibodies against AKT, phospho-AKT (Ser473 or Thr308), FoxO1, phospho-FoxO1 (Ser256), GSK3α/β, phospho-GSK3α/β (Ser21/Ser9), AS160, phospho-AS160 (Thr642), S6, phospho-S6 (Ser235/Ser236), p70S6K, and phospho-p70S6K (Thr389) were purchased from Cell Signaling Technology.
Cell Culture
The human embryonic kidney cell line HEK-293FT and the mouse neuroblastoma cell line (N2a) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 units/ml streptomycin and incubated in a humidified atmosphere of 5% CO2 at 37 °C.
Plasmid Construction and DNA Mutagenesis
The expression construct for human AKT1 was generated by RT-PCR from RNA isolated from normal human neuronal progenitor cells and confirmed by DNA sequence analysis and was obtained from SignalChem (Richmond, British Columbia, Canada). AKT tagged with HA was cloned into the pCI-Neo vector via XhoI and SalI sites. Mutation of Thr308 or Ser473 to Ala in AKT was achieved using the QuikChange II site-directed mutagenesis kit (Stratagene) with primers 5′-ggtgccaccatgaagGccttttgcggcacacctg-3′ (forward) and 5′-caggtgtgccgcaaaaggCcttcatggtggcacc-3′ (reverse) for T308A and 5′-ccacttcccccagttcGcctactcggccagcggc-3′ (forward) and 5′-gccgctggccgagtaggCgaactgggggaagtgg-3′ (reverse) for S473A. The mutations at Thr308 and Ser473 to Ala (T308A/S473A) were produced by PCR from the vector pCI-Neo-HA-AKT-T308A with the same forward primers used for S473A. The mutations were confirmed by DNA sequence analysis.
Transfection
All transfections of HEK-293FT cells were performed with FuGENE 6 (Roche Applied Science) in 12-well plates. HEK-293FT cells were transfected with 0.5 μg of plasmid DNA and 1.5 μl of FuGENE 6 in 50 μl of Opti-MEM I (Invitrogen) for 16 h and then treated with the protein synthesis inhibitor cycloheximide or NSC119889. LipofectamineTM 2000 reagent (Invitrogen) was used for transfection in N2a cells according to the manufacturer's instructions.
Cycloheximide Chase Analysis
For cycloheximide chase analysis, after transfection with HA-tagged wild-type or mutant AKT for 16 h, HEK-293FT cells were incubated with 100 μm cycloheximide (diluted from a 355.4 mm stock solution in Me2SO) for various times. Cell lysates were prepared, and the expression of HA-AKT was analyzed by Western blotting.
Cell Lysis and Western Blotting
Whole cell extracts were prepared using buffer consisting of 50 mm Tris-HCl (pH 7.4), 150 mm sodium chloride, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1.0 mm PMSF, and Roche cOmplete mini protease inhibitor mixture. Protein concentrations of cell lysates were determined using the Pierce 660nm protein assay reagent (Thermo Fisher Scientific). Western blotting was carried out after 10% SDS-PAGE. After incubation with HRP-conjugated secondary antibodies, the protein-antibody complexes were visualized using the Pierce ECL Western blotting substrate (Thermo Scientific Inc.) and exposed to Kodak medical x-ray film. Specific immunostaining was quantified using the MultiGauge version 3.0 software (Fujifilm).
Statistical Analysis
All experiments were repeated at least three times. To determine differences between groups, analysis of variance or Student's t test was performed, and a p value of <0.05 was defined to be statistically significant.
RESULTS
Inhibition of Protein Synthesis Promotes Phosphorylation and Alters Degradation of AKT
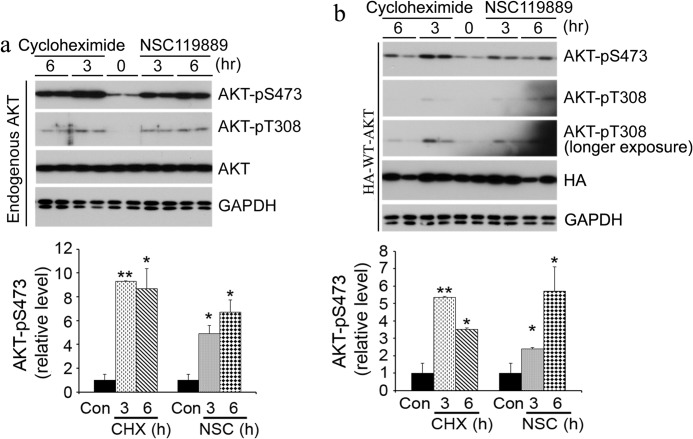
When we tried to determine the half-lives of HA-tagged WT and mutant (T308A/S473A, T308A, and S473A) AKT by measuring the level of HA-AKT with anti-HA antibody at various time points after treatment with the protein synthesis inhibitor cycloheximide at the concentration (100 μm) that is used in many studies (16, 17), we found that AKT with a single mutation (T308A or S473A) was more stable than WT AKT, whereas the turnover of the T308A/S473A mutant was more rapid compared with WT AKT (Fig. 1, a–e). To investigate what made such apparent differences in their turnover, we determined AKT phosphorylation after cycloheximide treatment because the turnover of some protein is regulated by phosphorylation. We found that cycloheximide dramatically induced AKT phosphorylation at Ser473 in HEK-293FT cells transfected with HA-WT AKT (Fig. 1a). Phosphorylation of AKT at Thr308 was also detected 36 h after the cycloheximide treatment. The same phenomenon was seen for endogenous AKT when HA-AKT-T308A/S473A, in which no phosphorylation could occur on residue 308 or 473 of exogenous AKT, was expressed in HEK-293FT cells (Fig. 1b). Phosphorylation of AKT at Ser473 and Thr308 was also seen in cells expressing the AKT single mutant T308A or S473A (Fig. 1, c and d). Treatment of the cells with various concentrations of cycloheximide ranging from 0.1 to 300 μm, which covered almost all of the concentrations used in published studies, indicated a dose-dependent induction of AKT phosphorylation (Fig. 1f). Dose-dependent inhibition of protein biosynthesis was also evident, as the total amounts of cellular proteins were decreased along with increasing concentrations of cycloheximide (Fig. 1f, lower left panel). Correlation analysis indicated a strong negative correlation (r = −0.99) between AKT phosphorylation at Ser473 and the total amounts of cellular proteins (Fig. 1f, lower right panel), suggesting that AKT phosphorylation correlates positively with inhibition of protein biosynthesis.
FIGURE 1.
Inhibition of protein synthesis induces AKT phosphorylation in HEK-293FT cells. a–d, HEK-293FT cells were transfected with plasmids expressing HA-WT AKT (a), HA-AKT-T308A/S473A (HA-AKT-2A) (b), HA-AKT-T308A (c), or HA-AKT-S473A (d) for 16 h, followed by treatment with 100 μm cycloheximide for the indicated periods of time. The levels and site-specific phosphorylation of AKT in the cell lysates were determined by Western blotting. e, quantification of AKT levels (HA blots) after normalization with GAPDH. f, upper left panel, after transfection with plasmids expressing HA-WT AKT for 16 h, equal amounts of HEK-293FT cells were treated with the indicated concentrations of cycloheximide for 3 h. The levels of AKT and its phosphorylation in the cell lysates were determined by Western blotting. Upper right panel, quantification of AKT phospho-Ser473 blots after normalization with the HA blots. Lower left panel, the total amounts of cellular proteins were also determined and are presented relative to those of untreated cells. Lower right panel, the linear correlation between AKT phospho-Ser473 and the total protein amounts is shown. g, HEK-293FT cells with (right panel) or without (left panel) FuGENE 6 pretreatment for 16 h were treated with 100 μm cycloheximide and harvested at the indicated time points. h, HEK-293FT cells were treated with 50 μm NSC119889 (NSC) for 3 h, and the cell lysates were analyzed by Western blotting. The experiments were repeated three to four times, and the graphs in e–h are quantifications of the blots and are presented as means ± S.E. *, p < 0.05; **, p < 0.01 versus 0 h or 0 μm controls.
To investigate whether the AKT phosphorylation we observed above was the cells' response to the stress induced by the transfection reagent FuGENE 6, we studied cycloheximide-induced AKT phosphorylation in HEK-293FT cells both with and without FuGENE 6 pretreatment for 16 h. We found that cycloheximide induced an increase in AKT phosphorylation at Ser473 under both conditions (Fig. 1g). No phosphorylation of AKT at Thr308 was detected within 12 h after cycloheximide treatment. These results suggest that the AKT phosphorylation we observed in HEK-293FT cells was induced by cycloheximide, rather than by FuGENE 6.
To determine whether the cycloheximide-induced AKT phosphorylation results from inhibition of protein synthesis, we treated HEK-293FT cells with NSC119889, another protein synthesis inhibitor that acts by preventing mRNA-ribosome interaction and inhibiting 5′-mediated/cap-dependent initiation of protein synthesis (18). A marked increase in AKT phosphorylation at Ser473 was also observed (Fig. 1h). Because cycloheximide and NSC119889 are two structurally different compounds and inhibit protein synthesis through different mechanisms, our results suggest that inhibition of protein synthesis induces AKT phosphorylation.
We also investigated the effect of protein synthesis inhibition on AKT phosphorylation in mouse neuroblastoma N2a cells. We found that, as in HEK-293FT cells, inhibition of protein synthesis in N2a cells by either cycloheximide or NSC119889 promoted Ser473 phosphorylation of both endogenous AKT (Fig. 2a) and transfected AKT (Fig. 2b). AKT phosphorylation at Thr308 was also seen as early as 3 h after the treatments with the inhibitors in N2a cells. Taken together, these data suggest that inhibition of protein synthesis induces AKT phosphorylation, which does not appear to be a cell type-specific phenomenon.
FIGURE 2.
Inhibition of protein synthesis induces AKT phosphorylation in N2a cells. Untransfected N2a cells (a, upper panel) or N2a cells transfected with HA-tagged WT AKT for 16 h (b, upper panel) were treated with 100 μm cycloheximide (CHX) or 50 μm NSC119889 (NSC), and the cells were then harvested at the indicated time points. The levels and site-specific phosphorylation of AKT in the cell lysates were determined by Western blotting. The lower panels show quantifications of the blots. Con, control. *, p < 0.05; **, p < 0.01 versus 0 h controls.
AKT Phosphorylation Mediated by Inhibition of Protein Synthesis Leads to Phosphorylation of Multiple AKT Substrates
AKT is activated through its phosphorylation at Thr308 and Ser473 in response to various stimuli or stresses (19). To study whether protein synthesis inhibition-induced phosphorylation of AKT indeed leads to activation of its kinase activity, we measured phosphorylation of several well known AKT substrates, including AKT1 downstream substrates FoxO1, GSK3α/β, p70S6K, and ribosomal protein S6 and the AKT2 downstream substrate AS160. We found that treatment of the WT AKT-expressing HEK-293FT cells with cycloheximide resulted in a marked increase in phosphorylation of both the AKT1 and AKT2 substrates (Fig. 3a). To determine whether these substrates are also phosphorylated by endogenous AKT under these conditions, we treated the untransfected HEK-293FT cells with cycloheximide and found a similar increase in phosphorylation of all of the AKT substrates studied (Fig. 3b). When the cells were treated with NSC119889, a marked increase in phosphorylation of FoxO1, p70S6K, and AS160, but not of GSK3α/β, was observed (Fig. 1c). In contrast, phosphorylation of ribosomal protein S6 was significantly decreased with NSC119889 treatment. Taken together, these results indicate that inhibition of protein synthesis induces both phosphorylation and activation of AKT and that this phenomenon is not AKT isoform-dependent.
FIGURE 3.
Inhibition of protein synthesis induces phosphorylation of AKT substrates. a, upper panel, HEK-293FT cells were transfected with HA-WT AKT for 16 h, followed by treatment with 100 μm cycloheximide (CHX) for 3 h. The levels and phosphorylation of AKT and its substrates were analyzed by Western blotting. b and c, upper panels, HEK-293FT cells were treated with 100 μm cycloheximide (b) or 50 μm NSC119889 (NSC; c) for the indicated periods of time, and the cell lysates were analyzed. The lower panels show the densitometric quantification (mean ± S.D.) of the phosphorylation of individual AKT substrates calculated after being normalized to the levels of the corresponding proteins. Con, control. *, p < 0.05; **, p < 0.01 versus 0-h controls.
Inhibition of Protein Synthesis Induces AKT Phosphorylation/Activation Mainly through the PI3K/PDK1 Pathway
AKT can be activated through phosphorylation at Thr308 and/or Ser473 by its upstream kinases, i.e. PI3K/PDK1 or mTORC2 (mammalian target of rapamycin complex 2) (7, 20). To investigate through which upstream pathway the protein synthesis inhibitors induce AKT phosphorylation/activation, we treated cultured cells with cycloheximide in the presence of LY294002, a selective inhibitor of PI3K, or rapamycin, an inhibitor of mTOR. We observed that 10 μm LY294002, which is commonly used for selective inhibition of PI3K (21, 22), not only blocked the cycloheximide-induced AKT Ser473 phosphorylation but also blocked basal phosphorylation (Fig. 4, a and b), suggesting that cycloheximide-induced AKT phosphorylation/activation in HEK-293FT cells is mediated mainly through the PI3K/PDK1 pathway.
FIGURE 4.

LY294002 and rapamycin inhibit AKT phosphorylation induced by cycloheximide. a, HEK-293FT cells were treated with 100 μm cycloheximide (CHX), 10 μm LY294002 (LY), or 100 nm rapamycin (Rap) alone or in combination for 3 or 24 h, followed by analysis of the levels and phosphorylation of AKT by Western blotting. The GAPDH blot was included as a loading control. b, quantification of the blots shown in a. The relative AKT phosphorylation (mean ± S.E.) after being normalized to the total AKT levels is shown. c, HEK-293FT cells transfected with WT AKT for 16 h were treated with cycloheximide, LY294002, or rapamycin for 3 or 24 h, followed by analysis of the levels and phosphorylation of AKT by Western blotting. exp, exposure; Con, control. *, p < 0.05; **, p < 0.01.
Treatment of HEK-293FT cells with 100 nm rapamycin for 24 h inhibited AKT Ser473 phosphorylation, but treatment for 3 h instead increased AKT Ser473 phosphorylation (Fig. 4, a and b). The latter might have resulted from the known rapamycin-induced feedback activation of PI3K/PDK1 (8, 23, 24). Cycloheximide-induced AKT Ser473 phosphorylation was also inhibited by rapamycin at 24 h post-treatment. Additive effects were seen when both LY294002 and rapamycin were used. Similar results were seen in HEK-293FT cells transfected with WT AKT (Fig. 4c). As cycloheximide did not induce detectable phosphorylation of AKT at Thr308 during 24 h of treatment (Fig. 1a), phosphorylation at this site could not be studied here. Taken together, these results suggest that inhibition of protein synthesis induces AKT phosphorylation/activation mainly through the PI3K/PDK1 pathway, but the mTOR pathway could also play a minor role.
Inhibition of Protein Synthesis Induces Activation of the E3 Ubiquitin Ligase MDM2 and Degradation of Its Substrate Proteins
Most cellular proteins are degraded through the ubiquitin-proteasome system, in which the E3 ubiquitin ligases are critical. MDM2 is an E3 ligase and is known to be regulated by AKT (25, 26). Phosphorylation of MDM2 by activated AKT at Ser166 and Ser186 activates MDM2 and promotes its nuclear entry (25, 26). To determine whether cycloheximide treatment increases MDM2 phosphorylation via AKT activation, we treated HEK-293FT cells transfected with HA-WT AKT with cycloheximide and observed time-dependent increases in phosphorylation of both AKT at Ser473 and MDM2 at Ser166 (Fig. 5, a–c). The cycloheximide-induced phosphorylation of both proteins could be blocked by the PI3K inhibitor LY294002 (20 μm).
FIGURE 5.

Cycloheximide induces p53 turnover through activation of AKT. a, HEK-293FT cells were pretreated with or without 20 μm LY294002 for 1 h, followed by treatment with 100 μm cycloheximide for the indicated periods of time. The cell lysates were then analyzed by Western blotting. b and c, quantification of the blots shown in a (left panel) in the absence of LY294002. d, quantification of p53 levels. The data are representative of three independent experiments with similar results.
The tumor suppressor p53 is a known substrate of MDM2, and MDM2 plays a central role in p53 ubiquitination and rapid degradation by the 26 S proteasome (27, 28). To study whether cycloheximide-induced phosphorylation/activation of MDM2 leads to p53 degradation, we determined the p53 level and found a rapid decrease in the p53 level (Fig. 5d) along with phosphorylation/activation of AKT and MDM2 (Fig. 5, a–c). This decrease was not the consequence of merely protein synthesis inhibition by cycloheximide because inhibition of AKT and MDM2 phosphorylation with LY294002 in the presence of cycloheximide completely blocked the p53 reduction (Fig. 5d). Because the level of AKT and MDM2 was also decreased after cycloheximide treatment for 60–120 min, which is consistent with self-ubiquitination and degradation upon their activation (29), the total phosphorylated MDM2 and AKT decreased slightly after those time points (Fig. 5, a and b). However, after normalization with the corresponding protein level, clear time-dependent increases in phosphorylation of these two proteins were seen with the cycloheximide treatments (Fig. 5, a and c). These data indicate that inhibition of protein synthesis can alter the degradation of some proteins via activation of AKT and its downstream E3 ligase.
DISCUSSION
The homeostasis of protein metabolism is maintained and regulated by the rates of protein biosynthesis and degradation in living system. In some cases, alterations of protein degradation regulate protein biosynthesis through a feedback mechanism. Whether a change in protein biosynthesis modulates protein degradation has not been reported. In this study, we found that inhibition of protein synthesis by cycloheximide or NSC119889 induced AKT phosphorylation/activation mainly through the PI3K pathway. AKT signaling regulates many aspects of biological functions, including cell survival, proliferation, metabolism, migration, and metastasis. Activated AKT can phosphorylate the E3 ubiquitin ligase MDM2 and promote MDM2 nuclear entry and p53 degradation via the ubiquitin-proteasome pathway (25, 26). We thus postulate that cycloheximide alters protein turnover via activation of AKT and its downstream substrates, including MDM2. Our results show the increased degradation of two proteins (MDM2 and p53) when protein biosynthesis was inhibited with cycloheximide or NSC119889.
Accumulating evidence has shown that AKT can be activated by various cellular stresses, such as heat, oxidative stress, hyperosmotic stress, and sodium arsenite (30). AKT contains a PH (pleckstrin homology) domain that binds to phosphatidylinositol 3,4,5-triphosphate with high affinity. Once at the correct position in the plasma membrane, AKT can be phosphorylated by PDK1 mainly at Thr308 (7) and by mTORC2 mainly at Ser473 (8). In this study, we found that the cellular stress of protein biosynthesis inhibition activated AKT. As the phosphorylation/activation of AKT was completely blocked by the PI3K inhibitor LY294002 and was also partially affected by the mTORC inhibitor rapamycin, we conclude that the protein biosynthesis inhibition-induced AKT phosphorylation/activation is mediated mainly through the PI3K pathway. Consistent with our observations, Hemi et al. (19) reported that translational inhibitors activate the transactivation of ErbB2/ErbB2 receptors, leading to activation of the PI3K-regulated pathway. AKT activation is also induced by oxidative stress via the EGF receptor/PI3K pathway (30).
Inhibition of protein synthesis has been shown to be protective in various apoptosis models (31, 32). We speculate that AKT phosphorylation/activation under this condition might underlie this protective role because AKT signaling is anti-apoptotic (33). Activated AKT can phosphorylate and inactivate the pro-apoptotic proteins Bad and caspase-9 (34–36), leading to inhibition of apoptosis and promotion of cell survival.
The AKT kinase family includes three highly homologous isoforms: AKT1, AKT2, and AKT3. The development of AKT isoform-specific null mice has proven a functional diversity of AKT isoforms in physiology and in disease, although they have overlapping functions. In this study, we observed increased phosphorylation of both AKT1 and AKT2 substrates when protein synthesis was inhibited. These results suggest that protein synthesis inhibition-induced phosphorylation/activation of AKT may not be AKT isoform-dependent.
Rapamycin is a potent inhibitor of mTORC1 (37). We observed that treatment of HEK-293FT cells with rapamycin for 24 h partially inhibited protein synthesis inhibition-induced phosphorylation/activation of AKT, which is consistent with previous studies showing that prolonged rapamycin treatment also inhibits mTORC2 and AKT/PKB (38). Interestingly, we observed AKT activation after treatment of cells with rapamycin for 3 h. This short-term phenomenon could be mediated by feedback activation of AKT signaling through an insulin-like growth factor-1 receptor mechanism (8, 23, 24).
Ubiquitination of proteins occurs through three steps that require ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin ligases (E3) (39, 40). Ubiquitin itself contains seven lysine residues, and each of these can be further conjugated to the C terminus of another ubiquitin to form a polyubiquitin chain (41). As the three-step process advances, specificity increases: E1 interacts with all E2 enzymes, which then interact with a more limited subset of E3 enzymes. Each E3 in turn targets only a limited array of protein substrates based on a shared recognition motif within the proteins to be labeled. This enables the ubiquitination-proteasome pathway to be specific in the selection of the proteins to be labeled (42). The human proteome contains two ubiquitin E1 enzymes, ∼50 E2 enzymes, and 600 E3 enzymes (43). It is the specificity of the E3 enzyme that determines which proteins in the cell are to be marked for destruction in the proteasome (43).
Although MDM2 originally functions as an E3 ligase to degrade p53 (44–46), MDM2 also targets other proteins, such as the androgen receptor (47) and Numb (a protein that plays an important role in specifying cell fate during development) (48, 49), for ubiquitination and degradation. Because we observed an increase in the phosphorylation/activation of MDM2 despite its elevated degradation after treatment of cells with cycloheximide, we speculate the altered degradation or half-lives of the proteins that are substrates of MDM2, when protein biosynthesis is inhibited.
Activation of AKT in response to inhibition of protein synthesis could alter degradation of other cellular proteins. It has been reported that activated AKT phosphorylates a deubiquitinating enzyme called USP8 at Thr907, resulting in its activation and stabilization, which in turn stabilizes the ubiquitin ligase Nrdp1 and promotes degradation of the growth factor receptor ErbB3 (50). Activated AKT1, but not AKT2, phosphorylates the ubiquitin ligase Skp2 at Ser72 and protects it from Cdh1-mediated degradation through disruption of the interaction between Skp2 and its E3 ligase Cdh1 (51). Importantly, the E3 ubiquitin ligase Skp2 targets a growing number of proteins, including the transcription factors E2F-1 (52) and c-Myc (53, 54), the Cdk inhibitors p21Cip1 (55) and p27Kip1 (56), and the Forkhead transcription factor FoxO1 (57). Collectively, inhibition of protein synthesis may alter turnover of many proteins via activation of AKT and its downstream ubiquitin ligases.
Our observations of increased p53 degradation with cycloheximide treatment are consistent with a previous study showing that the chemopreventive agent apigenin inhibits ubiquitination of p53 and increases its stability (58). Apigenin can actually lead to a dose- and time-dependent decrease in AKT phosphorylation at Ser473 (59). In light of our observations, apigenin-induced AKT inhibition may underlie the apigenin-mediated p53 stabilization. AKT activation also promotes proteasome-dependent degradation of the AKT substrates tuberin, FoxO1, and FoxO3a (60).
It is worth noting that inhibition of protein biosynthesis by either cycloheximide or NSC119889 increased phosphorylation of AKT and its substrates FoxO1, p70S6K, and AS160. However, cycloheximide, but not NSC119889, led to increased phosphorylation of GSK3α/β and S6. The exact mechanism leading to these differential consequences with these two inhibitors remains to be investigated. These observations suggest that besides inhibition of protein biosynthesis, NSC119889 might have an additional activity that would stimulate other signaling pathways and in turn counteract the AKT-induced phosphorylation of GSK3α/β and S6 when protein biosynthesis is inhibited. This additional activity might also affect turnover of some proteins.
A common approach to determine a protein's half-life is to measure the remaining amount of the protein throughout a period of time after new protein biosynthesis is completely blocked. Cycloheximide is most commonly used for this purpose in biological research. It blocks protein synthesis through interfering with the translocation step (movement of two tRNA molecules and mRNA in relation to the ribosome) and thus blocking translation elongation (61). Our results indicate that inhibition of protein synthesis, such as with cycloheximide, alters turnover of some proteins via activation of AKT. Therefore, for determination of the half-lives of these affected proteins, the approach using protein synthesis inhibitors should be avoided. Pulse-chase radiolabeling analysis, which does not require inhibition of protein biosynthesis, should be used instead.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AG027429, R03 TW008123, R01 AG031969, and R01 AG019158. This work was also supported by the New York State Office for People with Developmental Disabilities, Alzheimer's Association Grants IIRG-10-170405 and IIRG-10-173154, National Natural Science Foundation of China Grant 81030059, and Jiangsu Natural Science Foundation Grant BK2011387.
REFERENCES
- 1. Baliga B. S., Pronczuk A. W., Munro H. N. (1969) Mechanism of cycloheximide inhibition of protein synthesis in a cell-free system prepared from rat liver. J. Biol. Chem. 244, 4480–4489 [PubMed] [Google Scholar]
- 2. Fulda S., Gorman A. M., Hori O., Samali A. (2010) Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010, 214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baskić D., Popović S., Ristić P., Arsenijević N. N. (2006) Analysis of cycloheximide-induced apoptosis in human leukocytes: fluorescence microscopy using annexin V/propidium iodide versus acridine orange/ethidium bromide. Cell Biol. Int. 30, 924–932 [DOI] [PubMed] [Google Scholar]
- 4. Ishii H. H., Etheridge M. R., Gobé G. C. (1995) Cycloheximide-induced apoptosis in Burkitt lymphoma (BJA-B) cells with and without Epstein-Barr virus infection. Immunol. Cell Biol. 73, 463–468 [DOI] [PubMed] [Google Scholar]
- 5. Tsuchida T., Kato T., Yamada A., Kawamoto K. (2002) Cycloheximide induces apoptosis of astrocytes. Pathol. Int. 52, 181–185 [DOI] [PubMed] [Google Scholar]
- 6. Buzzi F., Xu L., Zuellig R. A., Boller S. B., Spinas G. A., Hynx D., Chang Z., Yang Z., Hemmings B. A., Tschopp O., Niessen M. (2010) Differential effects of protein kinase B/Akt isoforms on glucose homeostasis and islet mass. Mol. Cell. Biol. 30, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alessi D. R., Andjelkovic M., Caudwell B., Cron P., Morrice N., Cohen P., Hemmings B. A. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–6551 [PMC free article] [PubMed] [Google Scholar]
- 8. Sun S. Y., Rosenberg L. M., Wang X., Zhou Z., Yue P., Fu H., Khuri F. R. (2005) Activation of Akt and eIF4E survival pathways by rapamycin-mediated mammalian target of rapamycin inhibition. Cancer Res. 65, 7052–7058 [DOI] [PubMed] [Google Scholar]
- 9. Woodgett J. R. (2005) Recent advances in the protein kinase B signaling pathway. Curr. Opin. Cell Biol. 17, 150–157 [DOI] [PubMed] [Google Scholar]
- 10. Garofalo R. S., Orena S. J., Rafidi K., Torchia A. J., Stock J. L., Hildebrandt A. L., Coskran T., Black S. C., Brees D. J., Wicks J. R., McNeish J. D., Coleman K. G. (2003) Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKBβ. J. Clin. Invest. 112, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen W. S., Xu P. Z., Gottlob K., Chen M. L., Sokol K., Shiyanova T., Roninson I., Weng W., Suzuki R., Tobe K., Kadowaki T., Hay N. (2001) Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15, 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cho H., Thorvaldsen J. L., Chu Q., Feng F., Birnbaum M. J. (2001) Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J. Biol. Chem. 276, 38349–38352 [DOI] [PubMed] [Google Scholar]
- 13. Cho H., Mu J., Kim J. K., Thorvaldsen J. L., Chu Q., Crenshaw E. B., 3rd, Kaestner K. H., Bartolomei M. S., Shulman G. I., Birnbaum M. J. (2001) Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292, 1728–1731 [DOI] [PubMed] [Google Scholar]
- 14. Easton R. M., Cho H., Roovers K., Shineman D. W., Mizrahi M., Forman M. S., Lee V. M., Szabolcs M., de Jong R., Oltersdorf T., Ludwig T., Efstratiadis A., Birnbaum M. J. (2005) Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol. Cell. Biol. 25, 1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tschopp O., Yang Z. Z., Brodbeck D., Dummler B. A., Hemmings-Mieszczak M., Watanabe T., Michaelis T., Frahm J., Hemmings B. A. (2005) Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132, 2943–2954 [DOI] [PubMed] [Google Scholar]
- 16. Yoshiura S., Ohtsuka T., Takenaka Y., Nagahara H., Yoshikawa K., Kageyama R. (2007) Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc. Natl. Acad. Sci. U.S.A. 104, 11292–11297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xue Z., He Y., Ye K., Gu Z., Mao Y., Qi L. (2011) A conserved structural determinant located at the interdomain region of mammalian inositol-requiring enzyme 1α. J. Biol. Chem. 286, 30859–30866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Novac O., Guenier A. S., Pelletier J. (2004) Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 32, 902–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hemi R., Paz K., Wertheim N., Karasik A., Zick Y., Kanety H. (2002) Transactivation of ErbB2 and ErbB3 by tumor necrosis factor-α and anisomycin leads to impaired insulin signaling through serine/threonine phosphorylation of IRS proteins. J. Biol. Chem. 277, 8961–8969 [DOI] [PubMed] [Google Scholar]
- 20. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 21. Bae I. H., Park M. J., Yoon S. H., Kang S. W., Lee S. S., Choi K. M., Um H. D. (2006) Bcl-w promotes gastric cancer cell invasion by inducing matrix metalloproteinase-2 expression via phosphoinositide 3-kinase, Akt, and Sp1. Cancer Res. 66, 4991–4995 [DOI] [PubMed] [Google Scholar]
- 22. Chandrasekharan U. M., Yang L., Walters A., Howe P., DiCorleto P. E. (2004) Role of CL-100, a dual specificity phosphatase, in thrombin-induced endothelial cell activation. J. Biol. Chem. 279, 46678–46685 [DOI] [PubMed] [Google Scholar]
- 23. Wan X., Harkavy B., Shen N., Grohar P., Helman L. J. (2007) Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 26, 1932–1940 [DOI] [PubMed] [Google Scholar]
- 24. O'Reilly K. E., Rojo F., She Q. B., Solit D., Mills G. B., Smith D., Lane H., Hofmann F., Hicklin D. J., Ludwig D. L., Baselga J., Rosen N. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mayo L. D., Donner D. B. (2001) A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou B. P., Liao Y., Xia W., Zou Y., Spohn B., Hung M. C. (2001) HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 3, 973–982 [DOI] [PubMed] [Google Scholar]
- 27. Honda R., Yasuda H. (1999) Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18, 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 29. Feng J., Tamaskovic R., Yang Z., Brazil D. P., Merlo A., Hess D., Hemmings B. A. (2004) Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J. Biol. Chem. 279, 35510–35517 [DOI] [PubMed] [Google Scholar]
- 30. Wang X., McCullough K. D., Franke T. F., Holbrook N. J. (2000) Epidermal growth factor receptor-dependent Akt activation by oxidative stress enhances cell survival. J. Biol. Chem. 275, 14624–14631 [DOI] [PubMed] [Google Scholar]
- 31. Coxon F. P., Benford H. L., Russell R. G., Rogers M. J. (1998) Protein synthesis is required for caspase activation and induction of apoptosis by bisphosphonate drugs. Mol. Pharmacol. 54, 631–638 [PubMed] [Google Scholar]
- 32. Cohen J. J., Duke R. C. (1984) Glucocorticoid activation of a calcium-dependent endonuclease in thymocyte nuclei leads to cell death. J. Immunol. 132, 38–42 [PubMed] [Google Scholar]
- 33. Cantley L. C. (2002) The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 [DOI] [PubMed] [Google Scholar]
- 34. Porter A. G., Jänicke R. U. (1999) Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 6, 99–104 [DOI] [PubMed] [Google Scholar]
- 35. del Peso L., González-García M., Page C., Herrera R., Nuñez G. (1997) Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science 278, 687–689 [DOI] [PubMed] [Google Scholar]
- 36. Cardone M. H., Roy N., Stennicke H. R., Salvesen G. S., Franke T. F., Stanbridge E., Frisch S., Reed J. C. (1998) Regulation of cell death protease caspase-9 by phosphorylation. Science 282, 1318–1321 [DOI] [PubMed] [Google Scholar]
- 37. Benjamin D., Colombi M., Moroni C., Hall M. N. (2011) Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat. Rev. Drug Discov. 10, 868–880 [DOI] [PubMed] [Google Scholar]
- 38. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 39. Lee J. C., Peter M. E. (2003) Regulation of apoptosis by ubiquitination. Immunol. Rev. 193, 39–47 [DOI] [PubMed] [Google Scholar]
- 40. Yang Y., Yu X. (2003) Regulation of apoptosis: the ubiquitous way. FASEB J. 17, 790–799 [DOI] [PubMed] [Google Scholar]
- 41. Chen Z. J. (2005) Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7, 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hershko A., Heller H., Elias S., Ciechanover A. (1983) Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 43. Bhoj V. G., Chen Z. J. (2009) Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 [DOI] [PubMed] [Google Scholar]
- 44. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 45. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 46. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 47. Lin H. K., Wang L., Hu Y. C., Altuwaijri S., Chang C. (2002) Phosphorylation-dependent ubiquitylation and degradation of androgen receptor by Akt require Mdm2 E3 ligase. EMBO J. 21, 4037–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yogosawa S., Miyauchi Y., Honda R., Tanaka H., Yasuda H. (2003) Mammalian Numb is a target protein of Mdm2, ubiquitin ligase. Biochem. Biophys. Res. Commun. 302, 869–872 [DOI] [PubMed] [Google Scholar]
- 49. Juven-Gershon T., Shifman O., Unger T., Elkeles A., Haupt Y., Oren M. (1998) The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol. Cell. Biol. 18, 3974–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cao Z., Wu X., Yen L., Sweeney C., Carraway K. L., 3rd (2007) Neuregulin-induced ErbB3 down-regulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol. Cell. Biol. 27, 2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gao D., Inuzuka H., Tseng A., Chin R. Y., Toker A., Wei W. (2009) Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat. Cell Biol. 11, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marti A., Wirbelauer C., Scheffner M., Krek W. (1999) Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1, 14–19 [DOI] [PubMed] [Google Scholar]
- 53. Kim S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., Tansey W. P. (2003) Skp2 regulates Myc protein stability and activity. Mol. Cell 11, 1177–1188 [DOI] [PubMed] [Google Scholar]
- 54. von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I., Söderberg O., Kerppola T. K., Larsson L. G. (2003) The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 11, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 55. Bornstein G., Bloom J., Sitry-Shevah D., Nakayama K., Pagano M., Hershko A. (2003) Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J. Biol. Chem. 278, 25752–25757 [DOI] [PubMed] [Google Scholar]
- 56. Nakayama K., Nagahama H., Minamishima Y. A., Matsumoto M., Nakamichi I., Kitagawa K., Shirane M., Tsunematsu R., Tsukiyama T., Ishida N., Kitagawa M., Nakayama K., Hatakeyama S. (2000) Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J. 19, 2069–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang H., Regan K. M., Wang F., Wang D., Smith D. I., van Deursen J. M., Tindall D. J. (2005) Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. U.S.A. 102, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McVean M., Xiao H., Isobe K., Pelling J. C. (2000) Increase in wild-type p53 stability and transactivational activity by the chemopreventive agent apigenin in keratinocytes. Carcinogenesis 21, 633–639 [DOI] [PubMed] [Google Scholar]
- 59. Kaur P., Shukla S., Gupta S. (2008) Plant flavonoid apigenin inactivates Akt to trigger apoptosis in human prostate cancer: an in vitro and in vivo study. Carcinogenesis 29, 2210–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Plas D. R., Thompson C. B. (2003) Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J. Biol. Chem. 278, 12361–12366 [DOI] [PubMed] [Google Scholar]
- 61. Schneider-Poetsch T., Ju J., Eyler D. E., Dang Y., Bhat S., Merrick W. C., Green R., Shen B., Liu J. O. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]