Background: PEDF has neurotrophic activity and interacts with PEDF-R, a membrane-linked lipase.
Results: A PEDF-binding region of PEDF-R is required for PEDF-R enzymatic stimulation, and peptides derived from this region block PEDF·PEDF-R-mediated retinal survival activities.
Conclusion: A ligand binding domain is identified in PEDF-R, a critical receptor for the survival activity of PEDF.
Significance: The findings provide mechanistic insight into the survival activity of PEDF.
Keywords: Cell Surface Receptor, Ligand-binding Protein, Receptors, Retina, Retinal Degeneration
Abstract
The extracellular pigment epithelium-derived factor (PEDF) displays retina survival activity by interacting with receptor proteins on cell surfaces. We have previously reported that PEDF binds and stimulates PEDF receptor (PEDF-R), a transmembrane phospholipase. However, the PEDF binding site of PEDF-R and its involvement in survival activity have not been identified. The purpose of this work is to identify a biologically relevant ligand-binding site on PEDF-R. PEDF bound the PEDF-R ectodomain L4 (Leu159–Met325) with affinity similar to the full-length PEDF-R (Met1–Leu504). Binding assays using synthetic peptides spanning L4 showed that PEDF selectively bound E5b (Ile193–Leu232) and P1 (Thr210–Leu249) peptides. Recombinant C-terminal truncated PEDF-R4 (Met1–Leu232) and internally truncated PEDF-R and PEDF-R4 (ΔHis203–Leu232) retained phospholipase activity of the full-length PEDF-R. However, PEDF-R polypeptides without the His203–Leu232 region lost the PEDF affinity that stimulated their enzymatic activity. Cell surface labeling showed that PEDF-R is present in the plasma membranes of retina cells. Using siRNA to selectively knock down PEDF-R in retina cells, we demonstrated that PEDF-R is essential for PEDF-mediated cell survival and antiapoptotic activities. Furthermore, preincubation of PEDF with P1 and E5b peptides blocked the PEDF·PEDF-R-mediated retina cell survival activity, implying that peptide binding to PEDF excluded ligand-receptor interactions on the cell surface. Our findings establish that PEDF-R is required for the survival and antiapoptotic effects of PEDF on retina cells and has determinants for PEDF binding within its L4 ectodomain that are critical for enzymatic stimulation.
Introduction
Pigment epithelium-derived factor (PEDF)3 is an important protein for the survival and function of the retina (1–3). This factor is secreted apicolaterally from the retinal pigment epithelium to act on photoreceptor morphogenesis, retinal neuroprotection, and neurite outgrowth (4–8). Its efficacy in delaying photoreceptor cell degeneration and apoptosis is demonstrated in genetic and light-induced damage animal models (6, 9). PEDF also protects cells of the inner retina and retinal ganglion cell layer from death induced by ischemia and cytotoxic agents (5, 10–13). It also has protective activities on CNS neurons, such as motorneurons, cerebellar granule cells, hippocampal neurons, and cortical neurons (14–18). In addition to its well established neurotrophic activities, PEDF has potent antiangiogenic properties that prevent neovessel invasion in the eye and in tumors (19–22). PEDF holds promise in clinical neuroprotection and vascular disease therapy, and understanding the mechanisms of its actions is of key importance for the development of PEDF-based therapies.
PEDF is a 50-kDa glycoprotein and a member of the serpin superfamily without a demonstrable inhibitory activity against serine proteases (23). It has been shown that the biological activities of PEDF depend on binding to cell surfaces of target cells rather than inhibiting serine protease. We and others have reported that PEDF binds to high affinity receptors (KD = 2–8 nm) on retina, neurons, endothelium, and tumor cell surfaces (21, 24, 25). The molecular mechanism of PEDF multifunctionality could be explained by responses to interactions with distinct cell surface receptors. We have identified the novel gene PNPLA2 in the retina that encodes a lipase-linked cell membrane protein with high affinity for PEDF and termed it PEDF-R (26). Later, other PEDF-binding proteins were reported in endothelial and tumor cells (37/67-kDa non-integrin laminin receptor (27) and cell surface F1F0-ATP synthase (28, 29)) and on ARPE-19 cells (LRP6, a Wnt co-receptor (30)). However, it is not yet known if PEDF-R is a functional receptor for PEDF activity on the retina.
The PEDF-R protein specifically binds PEDF with high affinity (KD = 3 nm) and does not have affinity for other serpins like maspin and ovalbumin (26). It is detected in the inner segments of the photoreceptors, at lower levels in the inner retina and retinal ganglion cell layer of the native retina, and also in the retinal pigment epithelium. Interestingly, the distribution of PEDF-R in the retina matches that of PEDF binding sites (25), implying that these cells contain PEDF-R molecules available to interact with PEDF. Most tissues express PNPLA2, including retina and brain (26, 31–33). PEDF-R is a member of the PNPLA2 (patatin-like phospholipase domain-containing 2) family and is also known as adipocyte triglyceride lipase, transport secretion protein-2.2, Ca2+ independent phospholipase A2ζ, and desnutrin (32–34). The structure of the exons of PNPLA2 reflects the proposed domain structure of the protein with three endodomains (L1, L3, and L5) and two ectodomains (L2 and L4) (35). Immunoreactivity of non-permeabilized cells, FACS, antibody capture experiments using antibodies to peptides from intracellular L3 and extracellular L4 domains, and cell surface biotinylation experiments agree with the predicted PEDF-R topology and show that PEDF-R is one of the proteins labeled at the surface of ARPE-19 cells (26). The amino acid sequence reveals a phospholipase A2 (PLA2) domain toward its amino end. Indeed, PEDF-R exhibits PLA2, triglyceride lipase, and acylglycerol transacylase activities (26, 34). Moreover, we have shown that PEDF stimulates the PLA2 activity of the PEDF-R enzyme, resulting in the release of fatty acids from phospholipids (26, 35).
The purpose of this study was to identify regions of PEDF-R necessary for PEDF function. We used human PEDF-R recombinant polypeptide fragments, synthetic peptides designed to span L4, and highly purified human PEDF in binding assays. Enzymatic assays were performed to determine the ability of PEDF to stimulate the PLA2 activity of PEDF-R polypeptide fragments. In experiments with live cells, Pnpla2 siRNA and PEDF-binding peptides were used to explore their potential to block PEDF biological activities using a retinal progenitor cell line derived from the neonatal rat retina. We discuss the discovery of PEDF-R as a neurotrophic receptor for PEDF and a region in PEDF-R that is critical for PEDF binding, enzymatic enhancement, and survival and antiapoptotic activities.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins
Recombinant human PEDF was expressed in baby hamster kidney cells (BHK(pMA-PEDF) cells) and purified by ammonium sulfate precipitation and cation exchange column chromatography (36) followed by anion exchange chromatography. Fluorescein-conjugated PEDF (Fl-PEDF) was prepared from recombinant human PEDF as described before (25). The PNPLA2 gene product will be referred as PEDF-R. Recombinant PEDF-R proteins were expressed by cell-free in vitro protein synthesis using the pEXP-based vectors and Escherichia coli extracts from ExpresswayTM or MembraneMaxTM (Invitrogen) cell-free protein expression kits. PNPLA2-containing expression vectors for PEDF-R and derived polypeptides (PEDF-R(D166A), L4, PEDF-R4, PEDF-R6, PEDF-R7, PEDF-RΔ203–232, and PEDF-R4Δ203–232) were constructed into pEXP1-DEST vectors with N-terminal epitope tags (N-end-His6/Xpress) under the T7 promoter as described previously (26). The primers used to prepare the various expression constructs are given in supplemental Table 1. Fig. 1B shows the schematic representation of the open reading frames in the PEDF-R expression constructs. Transcription/translation reactions were with 1 μg of cDNA for a final volume of 100 μl and incubated at 25 °C for 2 h. Epitope-tagged PEDF-R polypeptides were purified using His tag affinity column chromatography with Ni2+-NTA resin (Invitrogen). The proteins were bound to the resin in binding buffer containing 25 mm sodium phosphate, pH 8.0, 500 mm NaCl, and 6 mm imidazole. This was followed by washing with 10 column volumes of binding buffer containing 20 mm imidazole and elution in binding buffer with 250 mm imidazole. Approximately 10 μg of protein were obtained per 800-μl reaction. The purity of the fractions was about 50%. The samples were immediately used for activity assays or stored at 4 °C.
FIGURE 1.
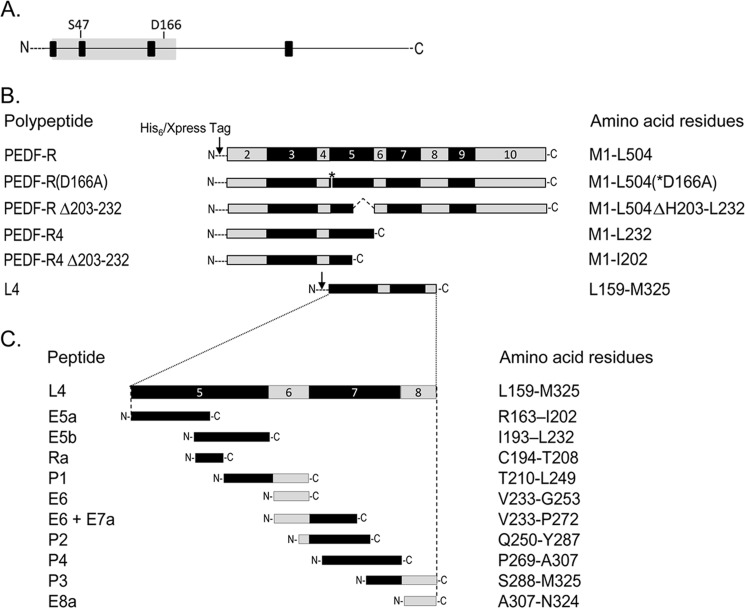
Schematic representation of the PEDF-R constructs and peptides. A, a linear form of the PEDF-R amino acid sequence. Black boxes indicate the locations of the four predicted transmembrane domains, and the gray area indicates the patatin-like phospholipase domain (PNPLA) spanning amino acids 10–179. Locations of the residues that comprise the catalytic site dyad (Ser47 and Asp166) are shown. B, exons (exons 2–10) encoding the PEDF-R polypeptide are illustrated in alternating gray and black code in linear form. The exon numbers are given inside each box. A series of expression constructs for truncated PEDF-R polypeptides is shown. The arrow indicates the position of the His6/Xpress tag fused to the N terminus of PEDF-R fragments. The asterisk denotes the position of the point mutation for D166A. The internally deleted region is indicated by dotted lines. C, design of synthetic peptides spanning L4, the longest extracellular loop of PEDF-R. The names and the amino acid residues comprising each fragment are given to the left and right, respectively, of each fragment. N- and C- indicate the locations of the N and C terminus, respectively.
Peptides
Peptides were designed and chemically synthesized (Aves Labs) based on exons 5, 6, 7, and 8 and the protein sequence of human PEDF-R (see Fig. 1C).
Surface Plasmon Resonance (SPR) Assays
Interactions between PEDF-R-derived polypeptides or P1 peptide and immobilized PEDF were analyzed in real time by SPR using a BIAcore 3000 instrument (BIAcore), as described previously (26). PEDF (5000 response units) was immobilized on a CM5 sensor chip by amine coupling. A reference surface without protein was prepared by the same procedure. Running buffer was HBS-Nonidet P-40 (0.01 m HEPES, pH 7.4, 0.15 m NaCl, 0.1% Nonidet P-40 in HBS). For kinetic analysis, injections with different concentrations of the PEDF-R-derived polypeptide were performed over the surfaces with a flow rate of 10 μl/min for 2 min. This was followed by injections with running buffer at the same flow rate. Sensograms (plot of response units versus time) were recorded. The data were fitted using BIAevaluation software (BIAcore). Data were double referenced such that nonspecific binding as well as buffer contributions to the signal were subtracted. The best fit was with a 1:1 Langmuir model for the PEDF surface-binding assay.
Ligand Blotting
Highly pure solutions of synthetic peptides (1 μg) or the indicated amounts of proteins were applied to wells in a manifold (Invitrogen) containing a nitrocellulose membrane (Bio-Rad, catalog no. 162-0116, 0.45 μm) presoaked in transfer buffer (25 mm Tris, 0.192 m glycine, 20% methanol) using vacuum as a driving force. The membrane was subjected to ligand immunoblot. Briefly, the membranes were blocked with blocking solution (1% BSA in TBS-T (0.05 m Tris, pH 7.5, 0.05 m NaCl, 0.05% Tween 20)) for 1 h at room temperature and then washed three times with TBS-T for 5 min each. This was followed by incubation with the indicated concentrations of Fl-PEDF for 1.5 h at room temperature or overnight at 4 °C. For competition experiments, Fl-PEDF was preincubated with E5b or P1 before addition to the membrane. Membranes were washed three times for 5 min each with TBS-T. Bound Fl-PEDF was detected by immunoblot versus polyclonal anti-fluorescein (Molecular Probes) diluted 1:200,000 in 1% BSA and HRP-conjugated goat anti-rabbit IgG (KPL) diluted 1:200,000 in 1% BSA. When the fluorescein-conjugated or non-conjugated PEDF and BSA (0.5 μg each) were immobilized on the membrane, bound E5b was detected by immunoblot versus anti-RA (Anaspec) (26) diluted 1:75,000 in 1% BSA and HRP-conjugated goat anti-rabbit IgG diluted 1:70,000 in 1% BSA. Alternately, the bound Fl-PEDF was quantified directly using the fluorescence imager Typhoon 9410 (GE Healthcare).
Peptide Affinity Chromatography
Peptide affinity resin was prepared commercially by Aves Labs. A total of 50 μl of peptide affinity beads (2 nmol of peptide) and 300 ng (6 pmol) of Fl-PEDF were mixed in TBS-T binding buffer to a final volume of 60 μl and incubated with gentle rotation at 4 °C for 1.5 h. Bound and unbound material were separated by centrifugation at 800 × g for 1 min. The beads were washed three times with 50 μl of binding buffer. SDS-PAGE sample buffer (100 μl) was added to the beads, mixed, heated at 100 °C, and centrifuged; 25 μl of the supernatant was resolved by SDS-PAGE. Bound and unbound material was analyzed by Western blot.
Phospholipase Activity (PLA2) Assay
The PLA2 activity was spectrophotometrically determined as described previously (26, 37) with some modifications. Briefly, this assay uses 1,2-dilinoleoyl-phosphatidylcholine as the phospholipase substrate and lipoxygenase as the coupling enzyme. The phospholipase activity releases linoleic acid from the substrate, and the lipoxygenase oxidizes the released linoleic acid to form a derivative hydroperoxide. The activity was followed spectrophotometrically by measuring the increase in absorbance at 234 nm as a result of the formation of linoleic acid hydroperoxide. Freshly prepared PEDF-R was incubated in 50 mm Tris-Cl buffer, pH 7.5, containing 3 mm deoxycholate in the presence of 0.26 mm phospholipase substrate (Sigma) and 6000 units/ml lipoxygenase (EC 1.13.11.12; Sigma) in an assay volume of 1 ml. A spectrophotometric scan of the products formed was obtained every minute for 10 min, and the rate of absorbance change at 234 nm/min was determined using a Beckman DU 640 spectrophotometer (Beckman Coulter). The PLA2 activity, expressed as the rate of product formed (A234 nm/min), was obtained using software from the spectrophotometer.
His Tag Pull-down Assays
Binding of PEDF to His6-tagged PEDF-R polypeptides was assayed by His tag pull-down of bound complexes with Ni-NTA resin. For PEDF-R·PEDF interactions, soluble aliquots of in vitro protein synthesis reaction (37.5 μl) containing His6-tagged PEDF-R and PEDF-RΔ203–232 were each mixed with Fl-PEDF protein (0.25 μg) in PLA2 reaction buffer (50 mm Tris-Cl buffer, pH 7.5, containing 3 mm deoxycholate, 50 μl final volume) and soluble aliquots of the in vitro protein synthesis reaction (25 μl) containing His6-tagged PEDF-R4 and PEDF-R4Δ203–232 were each mixed with Fl-PEDF protein (0.25 μg) in reaction buffer (50 mm sodium phosphate buffer, pH 7.5, containing 0.5 m NaCl and 1% Nonidet P-40, 50 μl final volume). The reaction mixtures were incubated at 4 °C with gentle rotation for 2 h. The Ni-NTA resin beads (25 μl), pre-equilibrated in the above buffer, were added, and the suspension was incubated for 1 h at 4 °C with gentle rotation. The resin beads were sedimented by low speed centrifugation (800 × g) and washed three times with the same buffer used for binding, and the proteins were extracted from the beads with SDS-PAGE sample buffer (25 μl). Bound samples (10 μl/lane) were resolved by SDS-PAGE and further analyzed by Western blotting against anti-PEDF antibodies (BioproductsMD) diluted 1:10,000 in 1% BSA and HRP-conjugated goat anti-rabbit IgG (1:100,000 in 1% BSA).
Cell Culture
R28 cells, an immortalized retinal progenitor cell line derived from the neonatal rat retina (kind gift of Dr. Gail Seigel, University of Buffalo), were cultured in DMEM with 10% fetal bovine serum (FBS) at 37 °C with 5% CO2 and 95% humidity. Cells in passage numbers between 47 and 60 were used.
siRNA Transfection
For PEDF-R expression silencing experiments, two silencer® select predesigned siRNAs (Ambion), siPEDF-R1 (ID s167781) and siPEDF-R2 (ID s167782), were used, and silencer® Select Negative Control No. 1 siRNA (ID 4390843) was used as scrambled. These siRNAs are designed using a novel algorithm and have the locked nucleic acid modification (LNA®) that has been shown to reduce off-target effects by up to 90% with improved specificity. Variation was observed in silencing with siPEDF-R2, and only experiments with PEDF-R knockdown were considered for quantitation. Transfections of R28 cells in 24-well plates were carried out using 30 pmol of siRNA and 0.5 μl of LipofectamineTM 2000 reagent (Invitrogen) in antibiotic-free medium containing 5% FBS for 48 h.
RNA Extraction, cDNA Synthesis, and Quantitative RT-PCR
Total RNA was purified using the RNeasyTM minikit (Qiagen) according to the manufacturer's instructions. Total RNA (0.1–1 μg) was used for reverse transcription using the SuperScript III first strand synthesis system (Invitrogen). Pnpla2 or Bcl-2 mRNA levels were normalized to 18 S levels by quantitative RT-PCR using SYBR Green mix (Applied Biosystems) in the Bio-Rad Chromo4 real-time system. The primers used were as follows: rat PEDF-R, 5′-TGT GGC CTC ATT CCT CCT AC-3′ (forward) and 5′-TGA GAA TGG GGA CAC TGT GA-3′ (reverse); rat Bcl-2, 5′-TGG ACA ACA TCG CTC TGT GGA TGA-3′ (forward) and 5′-GGG CCA TAT AGT TCC ACA AAG GCA-3′ (reverse); and 18 S, 5′-GGT TGA TCC TGC CAG TAG-3′ (forward) and 5′-GCG ACC AAA GGA ACC ATA AC-3′ (reverse).
Cell Surface Biotinylation Assay
R28 cells were incubated with 0.25 mg/ml membrane-impermeable, thiol-cleavable, amine-reactive sulfo-N-hydroxysuccinimide-SS-biotin reagent (Pierce) for 1 h. After incubation, the reaction was quenched, and the cells were harvested and subjected to biochemical fractionation to separate cytosolic and membrane fractions, as described previously (26). Equal amounts of membrane fraction proteins from biotinylated and non-biotinylated R28 cells were passed through spin columns with NeutrAvidin gels (Pierce). The bound biotinylated proteins were eluted by incubating the beads for 60 min in 1× SDS-PAGE sample buffer with 50 mm DTT.
Real-time Microelectronic Cell Monitoring
The cells were plated on a microplate biosensor platform, and real-time electrical impedance was followed with the RT-CESTM (real-time microelectronic cell sensor system) microelectronic cell sensor system from ACEA Biosciences, a system described previously (29, 38, 39). The microelectronic system measures a dimensionless parameter termed “cell index,” which is a relative change in electrical impedance to represent cell status. R28 cells were seeded at 3000 cells/well on 16-well strips with a 96-well format. Background impedance was measured with medium alone before adding the cells. The cells were allowed to attach for 8 h with medium containing 5% FBS. Then the medium was replaced with serum-free medium without or with the desired concentrations of PEDF. For treatment with peptides, PEDF and peptides were preincubated in 25 μl of medium and rocked at 4 °C for 4 h before addition to the cell cultures at the indicated final concentrations in serum-free medium. Real-time electrical impedance was monitored every hour in each well for several h. Data from two replicates were averaged. The EC50 was calculated using GraphPad Prism5 software.
Terminal dUTP Nick End Labeling (TUNEL) Staining
The TUNEL assay was performed in 24-well plates using an ApopTag fluorescein in situ apoptosis detection kit (Chemicon International) following the manufacturer's protocol. The nuclei were counterstained with Hoechst, and images were taken using a fluorescence microscope. Six different fields were imaged per treatment from three independent wells in each experiment. Cells were counted with ImageJ version 1.42, using fixed threshold values within each experiment to quantify based on intensity. The data are shown as the ratio of TUNEL-positive cells over the total cells per field.
Fl-PEDF Binding to Cells
R28 cells were seeded into 6-well plates and grown to 80% confluence in complete medium. Media were removed, and the cells were washed and incubated in serum-free medium for 1 h at 4 °C prior to treatment. Mixtures of 20 nm Fl-PEDF and increasing concentrations of P1 peptide in serum-free medium were preincubated at 4 °C for 2 h. The mixtures were added to the cell cultures and incubated for 30 min at 4 °C. Media were removed, and the cells were washed 3 times with ice-cold PBS before the addition of 100 μl of radioimmune precipitation assay lysis and extraction buffer (25 mm Tris-HCl (pH 7.6), 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS; Pierce)/well. Cells were scraped from the wells and passed through a 25-gauge × ⅝ inch needle before mixing with 4× SDS sample buffer.
Cell Viability Assays
Cell viability was measured by determining the relative levels of intracellular ATP as a biomarker for live cells using a CellTiter-GloTM (Promega) kit and following the manufacturer's instructions. In this assay, recombinant thermostable luciferase catalyzed the mono-oxygenation of beetle luciferin in the presence of Mg2+, ATP, and molecular oxygen, generating luminescence. Cells in PBS were mixed with the CellGlo reagent, and after 10 min of incubation at room temperature, the solution in each well was transferred into wells in a 96-well OptiPlate. The luminescence intensity corresponding to the ATP levels was measured using an automated plate reader (Envision).
Statistical Analyses
All values were expressed as the mean ± S.D. or S.E. The data were analyzed by the two-tailed unpaired Student's t test. A p value of less than 0.05 was considered statistically significant.
RESULTS
Expression of PEDF-R Fragments Fused to N-end-His6/Xpress Tags
To identify a PEDF binding domain on PEDF-R, we prepared deletion mutant expression vectors designed to express the L4 region (flanked by transmembrane domains 3 and 4) and C-terminal truncated PEDF-R forms containing the patatin-like phospholipase (PLA2) domain with the Ser47 and Asp166 catalytic dyad toward the N terminus (Fig. 1A). Selection of the coding sequences for the constructs was based on the exon map of human PEDF-R (Fig. 1B). Exons 5–7 and part of exon 8 were used for the L4 construct and lacked the patatin-like phospholipase domain (exons 2–4). The PEDF-R4 polypeptide was derived from the first four coding exons of the PEDF-R gene. PEDF-R(D166A), a PEDF-R altered with a single amino acid substitution at the predicted PLA2 active site; PEDF-RΔ203–232, with an internal truncation between His203–Leu232; and PEDF-R4Δ203–232, with a C-terminal truncation at Ile202, were also prepared (Fig. 1B).
Binding of PEDF to Recombinant L4 Polypeptides
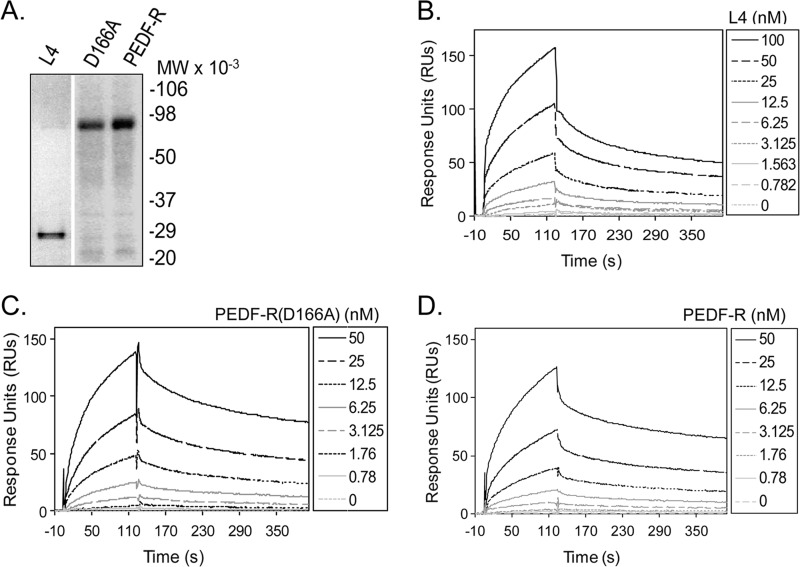
We reasoned that extracellular PEDF molecules may interact with PEDF-R ectodomains to elicit biological responses on target cells and therefore explored binding of PEDF to L4. The migration patterns of purified recombinant His6/Xpress-tagged L4, PEDF-R(D166A), and PEDF-R were as expected from their open reading frames (Fig. 2A). Real-time SPR performed with a recombinant human PEDF surface and the PEDF-R fragments as analytes showed an increase in SPR response units compared with the reference cell without PEDF. The kinetic parameters for the L4·PEDF interactions were determined with injections of recombinant L4 polypeptide at concentrations ranging from 0 to 100 nm. The SPR sensograms with the L4 polypeptide (Fig. 2B) show that binding to PEDF occurred in a concentration-dependent manner, similar to the sensograms with full-length PEDF-R(D166A) and PEDF-R (Fig. 2, C and D). The kinetic parameters revealed rapid and reversible interactions with high association and low dissociation rates. The dissociation constants for two batches of L4 for L4·PEDF interactions (KD) were 8.89 ± 0.1 and 5.97 ± 0.11 nm, which were comparable with those for the PEDF-R·PEDF interactions (KD = 1.98 ± 0.4 and 3.03 ± 0.72 nm) (26) and the PEDF-R(D166A)·PEDF interactions (KD = 2.52 ± 0.04 nm). These results demonstrated that L4, the region between Leu159 and Met325 of PEDF-R, contained structural determinants for binding PEDF and that substitution of a key amino acid within the catalytic dyad did not affect the affinity for PEDF.
FIGURE 2.
Real-time binding of PEDF to extracellular loop L4, PEDF-R(D166A), and PEDF-R. A, N-terminal His6/Xpress-tagged PEDF-R polypeptide fragments were synthesized in an in vitro transcription/translation system. Recombinant proteins were purified by His-tag affinity column chromatography. Proteins were resolved by 10–20% polyacrylamide gel electrophoresis and detected by Western analysis versus anti-Xpress antibodies. A Western blot is shown. Migration positions of molecular weight markers (prestained SDS-PAGE standards) are given to the right. B–D, SPR sensograms were recorded with L4 (B), PEDF-R(D166A) (C), and PEDF-R (D) injected at the indicated concentrations over PEDF immobilized on a CM5 sensor chip. The SPR responses for the reference surface and for the 0 nm PEDF were subtracted from the ones obtained at the various concentrations during the evaluation with BIAevaluation software (y axis) and are shown as a function of time. Kinetic analysis of the interaction was performed, and the data were analyzed using the BIAevaluation software (BIAcore 3000), and the KD value was obtained using the 1:1 Langmuir fitting. The kinetic and thermodynamic values were as follows. B, ka = 1.33 ± 0.05 × 105; kd = 1.18 ± 0.005 × 10−3; KD = 8.89 ± 0.1 nm for L4. C, ka = 4.24 ± 0.10 × 105; kd = 1.07 ± 0.005 × 10−3; KD = 2.52 ± 0.04 nm for PEDF-R(D166A). D, ka = 3.48 ± 0.1 × 105 m−1 s−1; kd = 6.88 ± 0.003 × 10−3 s−1; KD = 1.98 ± 0.4 nm for PEDF-R. A representative experiment for each interaction is shown. Each experiment was performed at least two times. χ2 values for goodness of fit are 7.19 for L4, 8.12 for PEDF-R(D166A), and 2.06 for PEDF-R.
Screening of L4-derived Synthetic Peptides for PEDF Binding
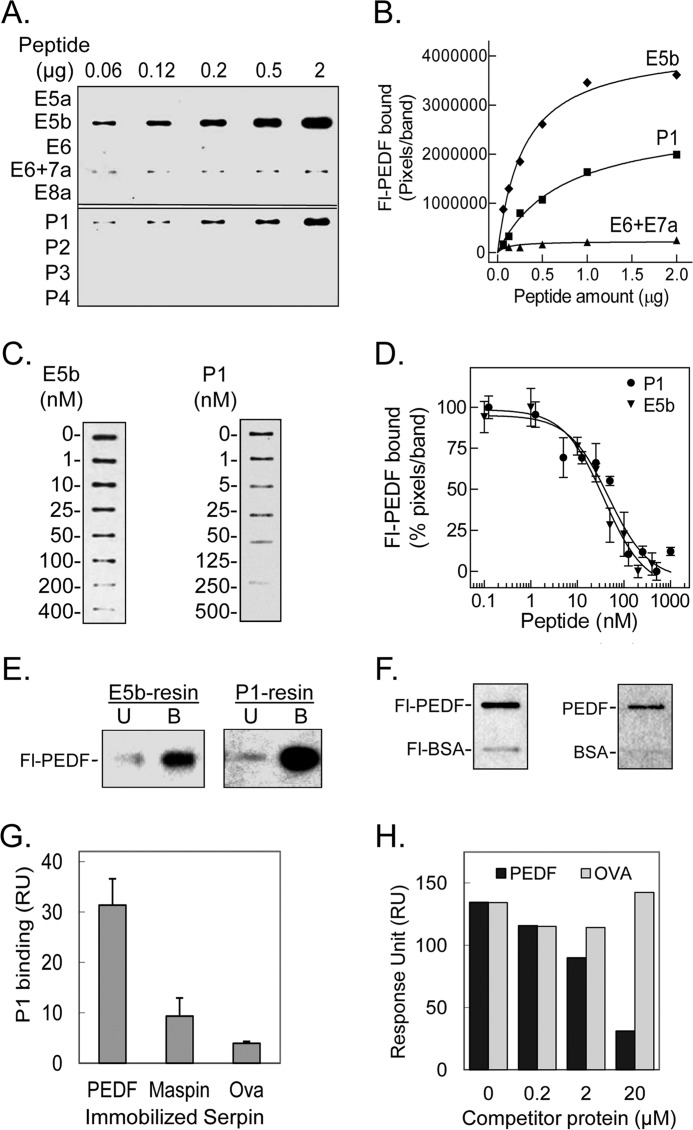
To identify a region in L4 responsible for PEDF binding, peptides spanning the L4 sequence were designed and were chemically synthesized (see Fig. 1C). Screening for PEDF binding was performed by ligand blot with Fl-PEDF as the ligand. Increasing amounts of each peptide (0.06–2 μg) immobilized on a nitrocellulose membrane were probed with 1 nm Fl-PEDF. Binding of Fl-PEDF was clearly detected for slots containing both peptides E5b (Ile193–Leu232) and P1 (Thr210–Leu249), very low with E6 + E7a peptide (Val233–Pro272), and undetectable with the other peptides (Fig. 3A). Quantification of the binding to E5b and P1 peptides showed that the amount of bound Fl-PEDF increased in a dose-dependent fashion (Fig. 3B).
FIGURE 3.
Binding of PEDF to peptides derived from L4 of PEDF-R. A, ligand blot of PEDF-R-derived peptides. Increasing amounts of the indicated peptide were immobilized per slot on a nitrocellulose membrane. After blocking, the ligand Fl-PEDF at 1 nm in 0.1% BSA TBS-T was added to the membrane and incubated for 1.5 h at room temperature. Bound Fl-PEDF was detected by Western blotting versus anti-fluorescein. B, quantification of PEDF bound to PEDF-R-derived peptides. The intensity of the bands in A was densitometrically quantified using UN-SCAN-IT software, and pixels per band were plotted versus the amount of peptide, as indicated. C, competition of Fl-PEDF binding to immobilized peptides with E5b and P1 in solution. E5b or P1 (1 μg) were immobilized on to the nitrocellulose membrane in triplicates per condition. After blocking, the membrane was incubated with a mixture of 1 nm Fl-PEDF and increasing concentrations of E5b or P1 (as indicated) in 0.1% BSA-TBS-T. Bound Fl-PEDF was either detected directly using the fluorescence imager or as described above. One set of slots for each peptide is shown. D, the bands in C were quantified by densitometry. The intensities of the bands in percentage of pixels per band relative to without peptide in solution were determined and plotted as a function of the concentration of peptide in solution. Each point corresponds to the average of triplicates ± S.E. (error bars) per concentration. The data were normalized and analyzed using GraphPad Prism software version 5 with an equation for binding, competitive with one site, fit log IC50. The estimated IC50 values for E5b and P1 were 37.5 ± 1.4 and 47.2 ± 1.3 nm, respectively. E, PEDF and Fl-PEDF (300 ng) were subjected to E5b peptide or P1 peptide affinity chromatography in Tris-buffered saline containing 0.05% Tween 20 (TBS-T). Flow-through containing the unbound material was collected. After extensive washes with binding buffer, bound protein was eluted with SDS-PAGE sample buffer. Western blots of unbound (U) and bound (B) protein versus anti-PEDF are shown. F, PEDF and BSA conjugated and non-conjugated with fluorescein were immobilized on nitrocellulose membranes. E5b in solution at 200 nm was used as the ligand for the blot. Ligand blots immunoreacted with anti-PEDF-R are shown. G, comparison of P1 binding response to PEDF and other serpins. Serpins were immobilized on a sensor chip, and the analyte (P1 at 2 μm) was injected over the surface. Responses to the same amount of P1 injected on immobilized maspin, ovalbumin (Ova), and PEDF surfaces are shown. H, competition of P1 binding to PEDF immobilized on a CM5 sensor chip with PEDF or ovalbumin in solution. Mixtures of P1 (2 μm) with the indicated concentrations of serpins were injected over the surface as analytes. Relative response as a function of time is plotted.
To determine binding of soluble E5b and P1 to PEDF, the Fl-PEDF ligand was mixed with either peptide in solution before testing binding to immobilized E5b or P1, respectively. Increasing concentrations of E5b (1–400 nm) in solution decreased the binding of Fl-PEDF to immobilized E5b (Fig. 3C) with an estimated half-maximum displacement IC50 of 37.5 ± 1.4 nm (Fig. 3D). Similarly, increasing concentrations of P1 (from 1.25 nm to 1.25 μm) in solution decreased the binding of Fl-PEDF to immobilized P1 (Fig. 3C) with an estimated half-maximum displacement IC50 of 47.2 ± 1.3 nm (Fig. 3D). These observations indicated that E5b and P1 specifically bound and displaced Fl-PEDF in solution.
The binding of PEDF to E5b and P1 cross-linked to agarose resins was also assessed. Fl-PEDF was incubated with resins containing about 200 μg/ml bed volume of each peptide. After separating bound from unbound protein by low speed centrifugation, Fl-PEDF proteins were clearly detected in the bound fractions of both peptide resins (Fig. 3E). In other experiments, binding of soluble E5b peptide to Fl-PEDF and Fl-BSA immobilized on nitrocellulose membranes showed binding to the PEDF proteins and insignificant to undetectable binding to the BSA proteins, implying that E5b specifically bound PEDF (Fig. 3F).
We have previously demonstrated that the affinity of PEDF-R for PEDF is significantly higher than that for maspin and even higher than that for ovalbumin (26). To explore if this characteristic of PEDF-R remains in the PEDF-R peptides, we compared the binding of P1 peptide to the three serpins. SPR was performed with P1 peptide (2 μm) as the analyte over a surface with immobilized maspin, ovalbumin, or PEDF. Fig. 3G shows that binding was significantly lower to maspin and ovalbumin than to PEDF, as reported before for the full-length PEDF-R. We also performed competition of binding to a PEDF surface when P1 was co-injected with increasing concentrations of PEDF or ovalbumin (0.2, 2, and 20 μm). PEDF co-injection decreased the binding of P1 to immobilized PEDF, and no such decrease was seen with ovalbumin (Fig. 3H). This shows that PEDF in solution competed for P1 binding to immobilized PEDF, whereas ovalbumin did not. These results imply that, like the full-length PEDF-R, P1 peptide preferred to bind PEDF over other serpins.
Altogether, these results demonstrated that PEDF bound specifically to the region in L4 spanning between Ile193 and Leu249 of human PEDF-R (E5b/P1) that is encoded in part by exons 5 and 6 (see Fig. 1C). Given that E5b and P1 individually bound PEDF and the lack of PEDF binding to peptide E6 (Val233–Gly253), the results implied a PEDF binding region located within the sequence Thr210–Leu232 of exon 5 of PEDF-R.
Phospholipase Activity of PEDF-R Lacking the PEDF-binding Region
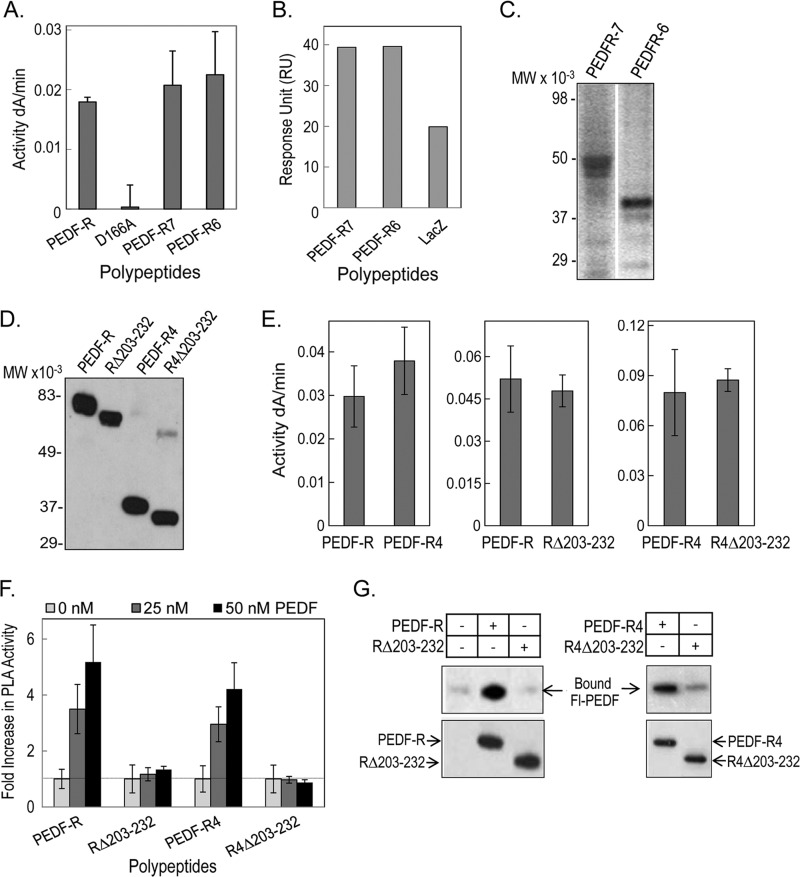
We have previously demonstrated that PEDF-R exhibits PLA2 enzymatic activity that PEDF can enhance (26). Fig. 4A shows that PEDF-R(D166A), which contains substitution D166A at the homologous lipase catalytic dyad, lost phospholipase activity. However, C-terminally truncated polypeptides PEDF-R7 (Met1–Arg351) and PEDF-R6 (Met1–Glu306), encoded by the first seven and six exons of PNPLA2, containing both the patatin-like PLA2 domain and the PEDF-binding region, exhibited PLA2 activity and bound PEDF (Fig. 4, A and B). The polypeptides migrated by SDS-PAGE as expected from the open reading frames of their respective constructs (Fig. 4C).
FIGURE 4.
Phospholipase activity and PEDF binding of truncated PEDF-R and PEDF-R lacking the ligand binding region (His203–Leu232). A, PLA2 activity of recombinant PEDF-R variants. PLA2 activity assays were performed with 1,2-dilinoleoyl-phosphatidylcholine as the substrate, 20 μl of the purified His6/Xpress-tagged PEDF-R variant polypeptides (∼0.6 pmol/reaction), and lipoxygenase as the coupling enzyme in PLA2 reaction buffer (3 mm deoxycholate, 5 mm Tris-Cl, pH 7.5). The appearance of linoleoyl hydroperoxide, the coupled reaction product, was measured spectrophotometrically by increasing absorbance at 234 nm/min for 10 min at room temperature (n = 4). B, comparison of PEDF-R binding response to PEDF. Maximum response units after injection of PEDF-R polypeptides are plotted. PEDF was immobilized on a CM5 sensor chip, and PEDF-R polypeptides were the analytes injected over the surface. C, Western blot of purified protein fractions of PEDF-R7 and PEDF-R6 versus anti-Xpress. A total of 20 μl of each fraction were resolved by SDS-PAGE. D, Western blot of purified recombinant His6/Xpress-tagged PEDF-R, PEDF-RΔ203–232 (RΔ203–232), PEDF-R4, and PEDF-R4Δ203–232 (R4Δ203–232) proteins versus anti-Xpress. A total of 20 μl of each fraction were resolved by SDS-PAGE. E, PLA2 activity assays of recombinant PEDF-R variants were performed as in A. F, effect of PEDF additions on the PLA2 activity of PEDF-R polypeptides. Equivalent amounts of affinity-purified PEDF-R and variant polypeptides (∼0.6 pmol/reaction) were preincubated with increasing amounts of PEDF in PLA2 reaction buffer for 10 min on ice. PLA2 activity was measured as above and represented as -fold change over basal activity without PEDF. Shown is the average -fold change from two independent experiments (n = 4). G, binding assays by His tag pull-down. Soluble fractions of cell-free expression reactions containing His6/Xpress-tagged PEDF-R, RΔ203–232, PEDF-R4, and R4Δ203–232 (∼700 ng each) were mixed with Fl-PEDF protein (250 ng) in PLA2 reaction buffer and incubated for 2 h at 4 °C with gentle rotation. This was followed by the addition of Ni-NTA resin beads (25 μl), and the suspension was incubated for 1 h at 4 °C with gentle rotation. Bound PEDF (pull-down) was extracted with SDS-sample buffer and analyzed by Western blot versus anti-PEDF. Error bars, ±S.D.
Next, we hypothesized that PEDF binding is critical for the enhancement of enzymatic activity of PEDF-R. From our previous findings, we predicted that PEDF-R region 203–232 contains PEDF binding determinants because no binding was detected with peptides E5a (Arg163–Ile202) or E6 (Val233–Gly253). PEDF-R polypeptide without this region PEDF-RΔ203–232 (RΔ203–232) was prepared and assayed for PLA2 activity. This region was also removed from PEDF-R4 (Met1–Leu232) to obtain PEDF-R4Δ203–232 (R4Δ203–232), which extended from the N-terminal end of PEDF-R to the region upstream of the N terminus of P1. The soluble purified recombinant proteins migrated by SDS-PAGE as expected from the open reading frames of their respective constructs (Fig. 4D). The phospholipase activity of the recombinant proteins was measured immediately after purification. Fig. 4E shows that PEDF-R4, RΔ203–232, and R4Δ203–232 displayed basal PLA2 activity similar to that of the full-length PEDF-R. Then the ability of PEDF to stimulate PLA2 activity was assayed in the presence of 25 and 50 nm PEDF. Fig. 4F shows that the increase in PLA2 enzymatic activity of PEDF-R and of PEDF-R4 was 2.5–3-fold with 25 nm PEDF and 4–5-fold with 50 nm PEDF, respectively. However, this increase was not observed for the PLA2 activities of RΔ203–232 and R4Δ203–232 with PEDF additions. Binding of PEDF to PEDF-R and PEDF-R4 lacking the 203–232 region was evaluated. Pull-down of the His6-tagged polypeptides shows that although Fl-PEDF bound to full-length PEDF-R, it failed to bind to the PEDF-RΔ203–232 variant (Fig. 4G). Similarly, Fl-PEDF binding to the PEDF-R4Δ203–232 variant decreased significantly relative Fl-PEDF binding to PEDF-R4 (Fig. 4G). These observations demonstrated that the His203–Leu232 region was critical for PEDF binding and PEDF-R stimulation and that increase of the enzymatic activity depended on ligand binding.
PEDF-R Is Critical for PEDF Retinal Survival Activity
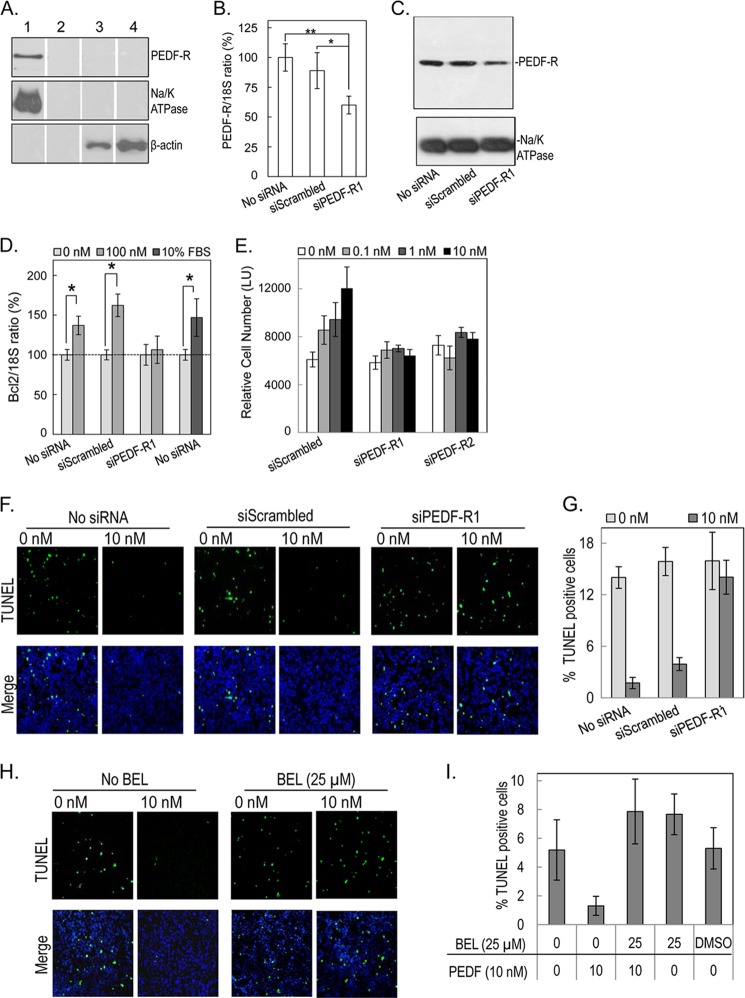
We and others have shown that rat retinal precursor R28 cells respond to PEDF stimuli, express the Pnpla2 gene, and contain active PEDF-R protein in their membrane extracts (26, 35, 40, 41). To determine if PEDF-R is a functional receptor for the activities of PEDF in these cells, we first corroborated the location of the PEDF-R protein to the R28 cell surface. Specific cell surface labeling experiments were performed to label accessible primary amines (e.g. lysine residues) of extracellularly exposed protein regions. Membrane protein extracts from cells labeled with or without membrane-impermeable biotin were passed through the avidin column. Fig. 5A shows that PEDF-R and Na/K ATPase, a marker for plasma membrane, were detected only in membrane fractions from biotinylated cells (lane 1) and not in fractions from non-biotinylated control cells (lane 2) or in their respective cytosolic fractions (lanes 3 and 4). The intracellular marker β-actin was detected only in the cytosolic fractions. The result demonstrated that PEDF-R in R28 is a plasma membrane protein and has an extracellular biotinylation-sensitive region that probably contains lysines available for conjugation with biotin.
FIGURE 5.
Effects of PEDF on PEDF-R deficient retina R28 cells. A, selective biotinylation of cell surface proteins of R28 cells was performed using a membrane-impermeable, thiol-cleavable, and amine-reactive sulfo-N-hydroxysuccinimide-SS-biotin reagent with avidin affinity resin. Biotinylated and non-biotinylated cells were subjected to biochemical fractionation, and the resulting membrane fraction was passed through NeutrAvidinTM gel and analyzed by Western blotting against antibodies to PEDF-R (top), Na/K ATPase (plasma membrane marker) (middle), and β-actin (cytoplasmic marker) (bottom). Samples were as follows: membrane fraction of biotinylated R28 cells (lane 1); non-biotinylated R28 cells (lane 2); cytosolic fraction of biotinylated R28 cells (lane 3); and non-biotinylated R28 cells (lane 4). B, RT-PCR for PEDF-R in R28 cells transfected with siScrambled or siRNA targeting PEDF-R (48 h post-transfection). *, p < 0.05; **, p < 0.005. C, proteins from cells transfected with siRNAs for 48 h were fractionated, and membrane fractions were resolved by SDS-PAGE followed by immunoblotting with anti-PEDF-R. The blot was stripped and immunoreacted with antibodies to Na/K-ATPase as loading control (bottom). D, RT-PCR for Bcl-2 in R28 cells untransfected and transfected with siScrambled or siPEDF-R of B and treated for 6 h with or without PEDF (100 nm) or 10% FBS. Bar graphs depict the mean ± S.D. of data from two independent experiments normalized to the treatment without PEDF (n = 2 each and n = 1 for 10% FBS). *, p < 0.05. E, quantitation of cell numbers using intracellular ATP as a live cell biomarker of cells at 48 h post-transfection with siScrambled or siPEDF-R. Cells were treated with increasing PEDF concentrations as indicated. Relative cell numbers are averages of triplicate wells ± S.D. (error bars). F, representative TUNEL images of untransfected and transfected cells, as indicated. After 24 h, cells in 24-well plates were treated for 48 h with or without 10 nm PEDF (indicated) in triplicate wells. Cells were fixed and processed for TUNEL staining (green) and counterstained with Hoechst dye (blue) for the nucleus. G, quantification of TUNEL-positive cells treated as in F. Images of 6 fields/well of triplicate wells were taken and quantified for TUNEL-positive cells. The plot shows TUNEL-positive cells represented as a percentage of total cells per field for each treatment. H, representative TUNEL images of cells pretreated with bromoenol lactone (BEL) (25 μm) or DMSO (vehicle control) for 1 h before a 48-h treatment with or without 10 nm PEDF (indicated) in triplicate wells. Cells were fixed and processed for TUNEL staining (green) and counterstained with Hoechst dye (blue) for the nucleus. I, quantification of TUNEL-positive cells treated as in H.
Next, we investigated if PEDF-R is required for PEDF-mediated activities in R28 cells. R28 cells deficient in PEDF-R were prepared and used in signaling, viability, and apoptosis assays. The siRNA-mediated knockdown of PEDF-R resulted in a ∼40% decrease of Pnpla2 transcripts with a concomitant decline in PEDF-R protein level in R28 membrane extracts (Figs. 5, B and C). No significant differences were observed with scrambled siRNA.
Murakami et al. (40) have reported that PEDF protects R28 cells from death via up-regulation of the antiapoptotic Bcl-2 gene. Given that R28 cell survival was maximized when cells were treated with 10–100 nm PEDF (Fig. 6), we evaluated Bcl-2 expression in R28 cells treated with 100 nm PEDF. Fig. 5D shows a clear increase in Bcl-2 expression after 6 h of PEDF treatment in untransfected and scrambled siRNA-transfected cells but not in siPEDF-R1-transfected cells. Treatments with 10% FBS were controls for the Bcl-2 increase. Similar data were obtained with another batch of cells where PEDF-R levels were knocked down with siPEDF-R2 (data not shown). We note that Pnpla2 silencing did not affect Bcl-2 expression levels (data not shown). These results demonstrated that PEDF-R was required for the PEDF-mediated up-regulation of Bcl-2 in R28 cells.
FIGURE 6.
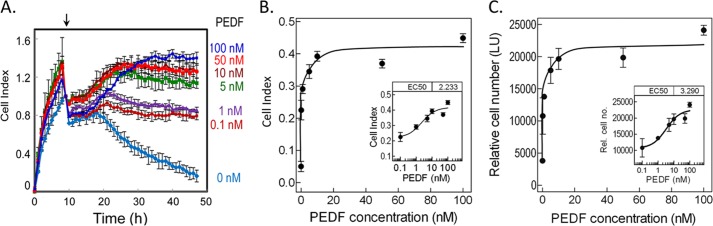
Cell survival effects of PEDF on R28 cells. A, kinetics of PEDF cell survival activity with increasing concentrations of PEDF using a real-time microelectronic cell sensor system. Cells were plated in duplicates, and after 8 h, serum-containing medium was replaced with serum-free medium containing increasing concentrations of PEDF (as indicated). Cell index was monitored each hour for 48 h. Cell index values were averaged. Data were plotted with cell index ± S.D. (error bars) as a function of time. The arrow indicates the time at which PEDF was added. B, the plot of cell index value at the end point as a function of PEDF concentration was obtained using GraphPad Prism with an equation for dose-response-stimulation (log[agonist] versus response). The calculated half-maximal response was EC50 = 2.233 ± 1.873 nm (see inset). Each point is the average of duplicate wells ± S.D. C, relative cell numbers were determined at the end point using intracellular ATP as a live cell biomarker. A plot of relative cell numbers as a function of PEDF concentration was obtained using GraphPad Prism as in B. The calculated half-maximal response was EC50 = 3.29 ± 1.98 nm (see inset). Each point is the average of duplicate wells ± S.D.
We also determined the effects of silencing PEDF-R on the survival activity of PEDF in serum-starved R28 cells. Cells were treated with increasing concentrations of PEDF (0, 0.1, 1, and 10 nm) for 48 h before assaying. Fig. 5E shows that PEDF treatment increased the relative number of live cells in scrambled siRNA-transfected cells but not in siPEDF-R1- or siPEDF-R2-transfected cells.
We also measured apoptosis using TUNEL staining in serum-starved R28 cells treated with or without 10 nm PEDF for 48 h. Fig. 5, F and G, shows that, in untransfected or scrambled siRNA-transfected cells, we observed a significant decrease in the percentage of TUNEL-positive cells with PEDF treatment (∼2–4%) as compared with treatment without PEDF (∼15%). However, no decrease in TUNEL-positive cells (14–16%) was observed in siPEDF-R1-transfected cells, regardless of PEDF treatment. In another experiment with a different batch of cells treated with 10 nm PEDF, there was no decrease in TUNEL-positive cells in siPEDF-R2-transfected cells compared with scrambled siRNA-transfected cells (data not shown). These results demonstrated that PEDF-R was essential for the PEDF-mediated survival and antiapoptotic activities in R28 cells.
Next, we investigated the role of PEDF-R PLA2 activity in mediating the effects of PEDF. As we have previously described (26), a calcium-independent phospholipase A2 inhibitor, bromoenol lactone, inhibits PEDF-R PLA2 enzymatic activity. Here, we performed TUNEL staining in serum-starved R28 cells treated with or without 10 nm PEDF in the presence or absence of the inhibitor (25 μm). Interestingly, in the presence of the inhibitor, we found no change in the number of TUNEL-positive cells with PEDF treatment (∼7.8%) in comparison with cells without PEDF treatment (∼7.6%) (Fig. 5, H and I). Bromoenol lactone or DMSO alone did not affect the TUNEL-positive cells. These observations demonstrate that lipase activity was indeed required for the antiapoptotic activity of PEDF in R28 cells and support the hypothesis of a lipid-mediated signaling response to PEDF.
PEDF-binding Peptides of PEDF-R Block the Retinal Cell Survival Activity of PEDF
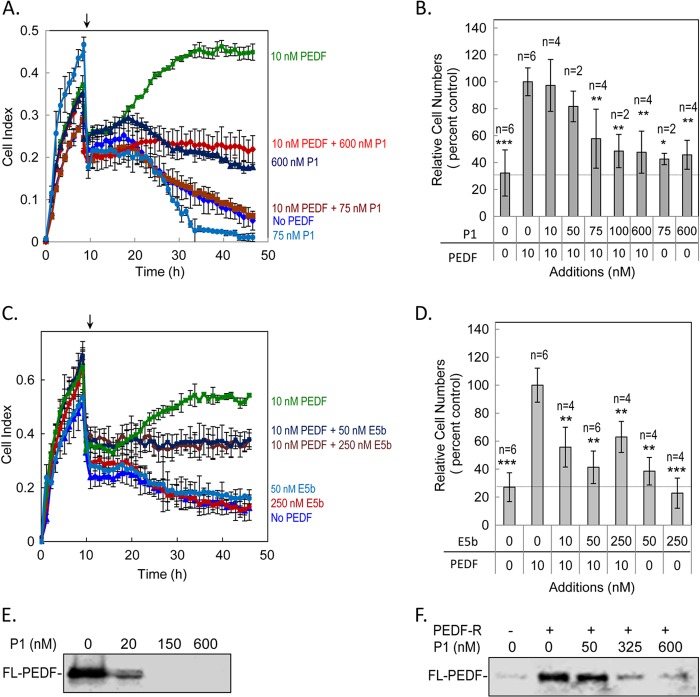
The above findings suggest that a PEDF-binding region in PEDF-R may act as a soluble receptor fragment that, upon binding the ligand, would prevent interactions with cell surface receptors and, in turn, the biological effects of PEDF. To explore this hypothesis, we evaluated the ability of peptides E5b and P1 to block the PEDF-mediated survival activity using a real-time microelectronic system to monitor cells and cell viability at the end point. The microelectronic system measured the changes in cell-based impedance represented as cell index. Fig. 6A shows plots of cell index values for R28 cells monitored in real time and treated with varying PEDF concentrations (0.1–100 nm). During the first hours, cell index increased significantly, consistent with cell attachment to plates in the presence of serum. After removing serum and adding PEDF, the cell index remained constant in all wells for the next 8–10 h. At later times, the cell index decreased in wells without PEDF, whereas it increased in wells with PEDF over time, having a maximum increase between 10 and 100 nm. Cell index increases were in a concentration-dependent fashion. A half-maximum effect calculated at the end point was estimated to be EC50 = 2.23 ± 1.87 nm (Fig. 6B). At the end point, cell viability assays using intracellular ATP as a biomarker of live cells showed that the increase in cell index paralleled the R28 relative cell numbers, with a half-maximum effect (EC50 = 3.29 ± 1.98 nm) (Fig. 6C), which is similar to that obtained with the microelectronic system.
Then serum-starved R28 cells were treated with 10 nm PEDF in the presence of P1 or E5b peptides as competitors for PEDF binding and monitored for cell growth using the real-time microelectronic system. Fig. 7A shows that cell index values increased over time between 20 and 48 h (end point) in the presence of PEDF alone. In contrast, the cell index stayed the same or even decreased during this time period when PEDF was mixed with an excess of P1. Peptide-mediated blocking of PEDF activity was very effective with 75 nm of P1. Peptide P1 by itself at 75 nm and 600 nm was the control. The cell index value at the end point was corroborated to cell viability using intracellular ATP as a biomarker of live cells (Fig. 7B). A dose-dependent decrease in relative cell numbers was observed with PEDF mixed with peptide P1 (10–600 nm). Fig. 7C shows that preincubation of PEDF with E5b peptide also attenuated the PEDF-mediated survival effect, as demonstrated by real-time monitoring. As with P1 treatment, the relative cell numbers at the end point following E5b treatment also corroborated with cell index data (Fig. 7D). Peptide-mediated blocking was effective with 10 nm of E5b (i.e. a 1:1 ligand:peptide molar ratio). The lack of dose-dependent blocking of PEDF action by E5b (10, 50, and 250 nm) in R28 cells suggested that maximal blocking was reached with the concentrations tested. The difference in the number of viable cells with 50 and 250 nm E5b in the presence of PEDF was not statistically significant.
FIGURE 7.
Effect of peptides P1 and E5b on PEDF-mediated survival effects. Kinetics of PEDF survival activity in the presence of the indicated concentrations of peptide P1 (A) or E5b (C) in serum-starved R28 cells using a real-time microelectronic cell sensor system. Wells with PEDF alone were positive control, no addition was negative control, and P1 or E5b alone were internal controls. Each condition was tested in duplicate wells per experiment. Three experiments were performed. The cell index values were averaged. Data were plotted with cell index ± S.D. (error bars) as a function of time. The arrow indicates the time at which PEDF was added. At the end point, cell viability assays were performed with cells that had been treated with PEDF in the presence of peptide P1 (B) or E5b (D) using intracellular ATP as a live cell biomarker. Relative cell numbers from two or three independent experiments were normalized to the 10 nm PEDF condition and plotted for each condition. Each point is the average of the indicated number of wells ± S.D. *, p < 0.05; **, p < 0.005; ***, p < 0.0005 with respect to 10 nm PEDF (100%) treatment. E, Fl-PEDF binding to R28 cells. Cells were treated with Fl-PEDF (20 nm) preincubated for 2 h without or with increasing concentrations of peptide P1 as indicated. Treatments were performed for 1 h at 4 °C, and total cell lysate was collected and resolved by SDS-PAGE. Bound Fl-PEDF was detected directly using the fluorescence imager. F, Fl-PEDF binding to recombinant full-length PEDF-R. Soluble fractions of cell-free expression reactions containing His6/Xpress-tagged PEDF-R were mixed with Fl-PEDF protein (50 nm) preincubated without or with increasing concentrations of peptide P1 as indicated. This was followed by the addition of Ni-NTA resin beads, and bound Fl-PEDF (pull-down) was extracted with SDS-sample buffer, resolved by SDS-PAGE, and detected directly using the fluorescence imager.
The results imply that the peptides acted as soluble receptor fragments to prevent PEDF binding to R28 cells. Binding of Fl-PEDF (20 μm) to R28 cells in the presence of P1 was investigated. Fig. 7E shows that Fl-PEDF bound to R28 cells, and when preincubated with increasing concentrations of P1 (20–600 nm), the bound Fl-PEDF decreased in a dose-dependent manner. P1 also blocked the binding of Fl-PEDF to recombinant PEDF-R in a dose-dependent fashion (Fig. 7F). These results demonstrated that the ligand-binding peptides P1 and E5b inhibited the PEDF survival activity, preventing the binding of PEDF to the cells and PEDF-R. The findings imply that the ligand-binding peptides acted as a soluble receptor fragments to block PEDF biological activity.
DISCUSSION
Here we report the identification and characterization of a PEDF binding region on PEDF-R that is critical for PLA2 enzymatic enhancement and blocks PEDF-mediated cell survival activities. Most importantly, the data provide evidence for the essential role of PEDF-R in antiapoptotic effects via interactions with PEDF. This is the first study that identifies a functional PEDF binding region in PEDF-R and its relevance for ligand activities.
The observation that both the full-length PEDF-R and the L4 polypeptide have similar affinities for PEDF suggests that the L4 ectodomain satisfies most, if not all, of the structural requirements of PEDF-R for ligand binding. In this report, we identify a PEDF binding region within L4 (e.g. E5b (Ile193–Leu232) and P1 (Thr210-Leu249) that has significantly higher affinity for PEDF than other areas of L4. The fact that both E5b and P1 have identical apparent affinities for PEDF suggests that it is the structure of their overlapping region (Leu232–Thr210) that contributes to interactions between the L4 ectodomain and PEDF and comprises the ligand binding domain (LBD) of PEDF-R. Although the binding kinetic values (KD) of the ligand-binding peptides to PEDF cannot be derived from the current data, differences in folding of the short peptide fragments may decrease the PEDF affinity relative to longer polypeptides. Nevertheless, our findings show that the interactions between PEDF and the ligand-binding peptides occur in solution or when either ligand or peptide is immobilized. Additionally, the peptides and the full-length PEDF-R both prefer to bind PEDF over other serpins and bovine serum albumin. The amino acid sequence of the LBD (210TNTSIQFNLRNLYRLSKALFPPEPL232) is a unique and highly conserved PEDF-R fragment not previously known to bind PEDF. In silico data suggest that the LBD can fold to form α-helices, and these results are supported by circular dichroism spectra of P1 (data not shown). Speculation on the nature of the LBD·PEDF complex implies the involvement of partial ionic interactions given that the P1·PEDF and E5b·PEDF complexes are moderately sensitive to increasing ionic strength (data not shown). Although the LBD fragment is positively charged and enriched in leucine residues, the surface surrounding the neurotrophic active region (44-mer) is mainly acidic (42, 43) and well suited to interact with the basic LBD, probably via ionic and additional non-ionic interactions. Although there is little information on the secondary structure of L4, it is anticipated that PEDF would interact similarly with either L4 or the full-length PEDF-R protein.
Our findings have implications for the regulation of the PLA2 activity of PEDF-R. The PLA2 active site of PEDF-R is formed by a homologous Ser47 and Asp166 catalytic dyad within the conserved motifs GXSXG and DX(G/A) (26). These amino acid residues play an essential role in the lipase activity, as evident by the findings that altering Asp166 abolishes PLA2 activity (Fig. 4) and altering either Ser47 or Asp166 abolishes triglyceride lipase activity (44, 45). Furthermore, in the present study, the C-terminal truncated variants of PEDF-R demonstrated PLA2 activity similar to that of full-length PEDF-R. Although we found the shortest domain with PLA2 activity to be Met1–Ile202 (PEDF-R4Δ203–232) (Fig. 4), the reported minimum domain for triglyceride lipase activity is slighter longer (Met1–Leu254) (46). Differences in substrate recognition (phospholipids versus triglycerides) and/or in recombinant protein expression and purification optimization (i.e. we used nanolipoprotein particles to improve the folding of recombinant polypeptides) could explain this discrepancy. Other reports describe the presence of an inhibitory region toward the C-terminal end of PEDF-R (44, 47, 48). This region does not seem to be inhibitory for PLA2 activity because the C-terminal truncated variants of this study exhibit PLA2 activity identical to that of full-length polypeptides. Given that the triglyceride lipase activity of recombinant PEDF-R variants was not assayed, we cannot remark further on what effects these truncations may have on such activity. Nonetheless, our data clearly demonstrate that the C-terminal half of PEDF-R is dispensable for phospholipid substrate accessibility and/or PLA2 activity. It has also been reported that the triglyceride lipase activity can be activated or inhibited by CGI-58 or G0S2, respectively (46, 49, 50), and that PEDF-R binds to these two regulator proteins with a yet unknown binding site (46, 51). It remains to be determined if these proteins can compete with PEDF for the binding to PEDF-R. The fact that PEDF binds and stimulates PEDF-R only when it contains the LBD indicates that PEDF, through interaction with the LBD, can positively regulate the PEDF-R enzyme (Fig. 8). At the molecular level, the interaction with PEDF could induce structural and conformational changes in PEDF-R that enhance the enzymatic catalysis of phospholipid substrates, although this has not yet been verified. Nevertheless, our study clearly demonstrates the requirement of the LBD for enzymatic stimulation of PEDF-R by PEDF.
FIGURE 8.
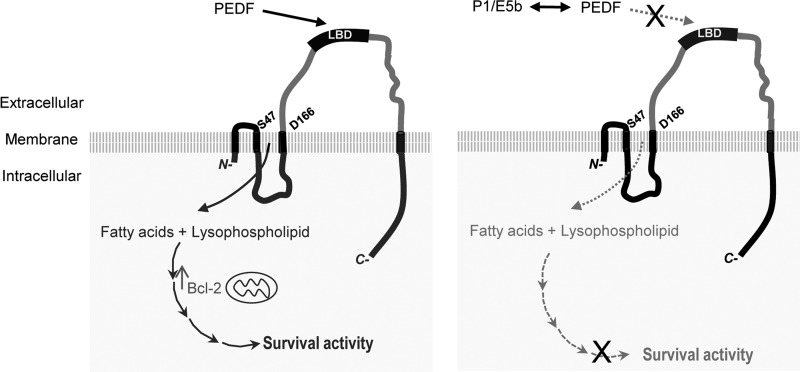
Scheme to illustrate our current understanding of PEDF·PEDF-R interactions. Plasma membranes of retina cells contain transmembrane PEDF-R, which is essential for PEDF-mediated survival and antiapoptotic effects. The amino acids Ser47 and Asp166 of PEDF-R are critical for lipase activity. PEDF binds to the LBD within the L4 ectodomain of PEDF-R to stimulate PLA2 activity. Left, it is proposed that the PLA2 activity would lead to the release of fatty acids and lysophospholipids from phospholipid substrates present in the lipid bilayer of plasma membranes. The products activate yet unknown signaling pathways to up-regulate Bcl-2 and cell survival (solid lines). Right, LBD-containing peptides (e.g. E5b and P1) act as soluble receptors that bind free PEDF to compete with binding to LBD, resulting in blocking of the survival activity of PEDF (dotted lines). ×, blocking.
The results also have implications in cell biology. The location of PEDF-R in plasma membranes of retina cells (Fig. 5A) (26, 35) makes it an excellent target for cell surface interactions with extracellular PEDF to subsequently trigger cell signaling. Comparison of the affinity of the PEDF·PEDF-R interaction (KD = 2.3–3.8 nm) with the efficacy of the PEDF retina survival effects (EC50 = 2.2–3.3 nm) supports this idea. More convincingly, our findings provide evidence for the key role of PEDF-R in mediating the retina survival actions of PEDF, as demonstrated by the attenuation of the survival and antiapoptotic activities of PEDF in Pnpla2-deficient R28 cells. The dependence of PEDF on PEDF-R has been reported for other biological activities, emphasizing the essential role of the PEDF·PEDF-R interaction. For example, human embryonic stem cells deficient in the PNPLA2 gene lacked the PEDF self-renewal activity (52); PNPLA2 knockdown in human melanoma cells resulted in the loss of the ability of PEDF to promote the mesenchymal-like phenotype that prevents metastasis (53); PEDF-mediated activation of PPARγ and repression of IL-8 required PEDF-R in human prostate cancer cells (54), and stimulation of PEDF-R triglyceride lipase activity in mouse muscle and liver is dependent on interactions with PEDF (55). We propose that the PEDF·PEDF-R interaction is critical for PEDF-mediated retina survival activity (Fig. 8). It is anticipated that LBD peptides will prove to be useful for probing the involvement of PEDF-R in other PEDF-mediated cellular effects. For example, P1 blocked PEDF-mediated axon growth on primary retinal ganglion cells,4 supporting the idea that PEDF specifically interacts with PEDF-R to induce neurotrophic effects.
Here, we defined a novel role for PEDF-R in retina survival. Previously, it was described that PEDF increased the Bcl-2 mRNA and Bcl-2 protein levels in both serum-starved R28 cells and in the retinas of Royal College Surgeon rats, a model for photoreceptor degeneration (40). In both systems, the Bcl-2 signal induced nuclear translocation of apoptosis-inducing factor, a caspase-independent apoptosis inducer (40). Although these studies did not report a receptor for PEDF signaling, our findings demonstrate that PEDF-R can mediate these effects, revealing it to be a PEDF receptor in the retina. However, the precise mechanisms by which PEDF-R transmits the PEDF signal between the cell surface and nucleus are not yet known. Our findings that the PEDF·PEDF-R interaction stimulates the phospholipase activity of PEDF-R provide insight into potential signaling mechanisms. The phospholipase A2 activity is of great interest, given that the enzyme is known to release bioactive fatty acids that function as either second messengers or eicosanoid precursors, both of which mediate signal transduction (56). Phospholipase PLA2 can also release bioactive lysophospholipids, which are known to interact with their G protein-coupled receptors to influence cell signaling (57). The retina and CNS contain high levels of ω-3 fatty acids, such as docosahexaenoic acid (DHA) and eicosapentaenoic acid, that are required for proper function and survival of retinal and neuronal cells (58, 59). Given that DHA constitutes more than 50% of the fatty acids in photoreceptor membrane phospholipids (60), it is likely that DHA-containing phospholipids are available substrates for PEDF-R in the retina. Although there is no direct evidence for DHA release from phospholipids by PEDF-R, several circumstantial lines of evidence support it. For example, PEDF significantly enhances the release of neuroprotectin D1, a neuroprotectant derivative of DHA with antiangiogenic, antioxidant, and anti-inflammatory properties in the retina and CNS (61–63). Moreover, PEDF, DHA, and neuroprotectin D1 exert neurotrophic activity by delaying the onset of the apoptotic pathway, including increases in antiapoptotic Bcl-2 protein levels (40, 62, 64, 65). Thus, a plausible upstream mechanism for transmitting PEDF signal in retina cells might involve DHA release by PEDF-R at the cell surface.
In conclusion, this is the first study to identify the LBD as a functional PEDF-binding region within the L4 ectodomain of PEDF-R that satisfies the minimum requirement for ligand binding. The LBD may form part of the receptor/ligand binding interface necessary for enzymatic stimulation of PEDF-R, a protein essential for the survival and antiapoptotic effects of PEDF. Our findings constitute an important advance in elucidating the molecular mechanisms of PEDF action and enhance our understanding of the process by which the neurotrophic PEDF interacts with cells.
Supplementary Material
Acknowledgments
We thank Dr. Gail Siegel (SUNY, Buffalo, NY) for generously providing R28 cells, Natalia Balko and Rachel Chen for technical assistance in the preparation of PEDF-R expression plasmids and recombinant proteins, and Drs. Stanislav Tomarev and Lijin Dong for critical reading of the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health, NEI, Intramural Research Program.

This article contains supplemental Table 1.
Y. Yin, personal communication.
- PEDF
- pigment epithelium-derived factor
- Fl-PEDF
- fluorescein-conjugated PEDF
- PEDF-R
- pigment epithelium-derived factor receptor
- PLA2
- phospholipase A2
- LBD
- ligand binding domain
- SPR
- surface plasmon resonance
- Ni-NTA
- nickel-nitrilotriacetic acid
- DHA
- docosahexaenoic acid
- EC50
- 50% of effective concentration
- IC50
- 50% of inhibitory concentration.
REFERENCES
- 1. Barnstable C. J., Tombran-Tink J. (2004) Neuroprotective and antiangiogenic actions of PEDF in the eye. Molecular targets and therapeutic potential. Prog. Retin. Eye Res. 23, 561–577 [DOI] [PubMed] [Google Scholar]
- 2. Becerra S. P. (2006) Focus on Molecules. Pigment epithelium-derived factor (PEDF). Exp. Eye Res. 82, 739–740 [DOI] [PubMed] [Google Scholar]
- 3. Bouck N. (2002) PEDF. Anti-angiogenic guardian of ocular function. Trends Mol. Med. 8, 330–334 [DOI] [PubMed] [Google Scholar]
- 4. Jablonski M. M., Tombran-Tink J., Mrazek D. A., Iannaccone A. (2000) Pigment epithelium-derived factor supports normal development of photoreceptor neurons and opsin expression after retinal pigment epithelium removal. J. Neurosci. 20, 7149–7157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takita H., Yoneya S., Gehlbach P. L., Duh E. J., Wei L. L., Mori K. (2003) Retinal neuroprotection against ischemic injury mediated by intraocular gene transfer of pigment epithelium-derived factor. Invest. Ophthalmol. Vis. Sci. 44, 4497–4504 [DOI] [PubMed] [Google Scholar]
- 6. Cayouette M., Smith S. B., Becerra S. P., Gravel C. (1999) Pigment epithelium-derived factor delays the death of photoreceptors in mouse models of inherited retinal degenerations. Neurobiol. Dis. 6, 523–532 [DOI] [PubMed] [Google Scholar]
- 7. Becerra S. P., Fariss R. N., Wu Y. Q., Montuenga L. M., Wong P., Pfeffer B. A. (2004) Pigment epithelium-derived factor in the monkey retinal pigment epithelium and interphotoreceptor matrix. Apical secretion and distribution. Exp. Eye Res. 78, 223–234 [DOI] [PubMed] [Google Scholar]
- 8. Tanimoto S., Kanamoto T., Mizukami M., Aoyama H., Kiuchi Y. (2006) Pigment epithelium-derived factor promotes neurite outgrowth of retinal cells. Hiroshima J. Med. Sci. 55, 109–116 [PubMed] [Google Scholar]
- 9. Cao W., Tombran-Tink J., Elias R., Sezate S., Mrazek D., McGinnis J. F. (2001) In vivo protection of photoreceptors from light damage by pigment epithelium-derived factor. Invest. Ophthalmol. Vis. Sci. 42, 1646–1652 [PubMed] [Google Scholar]
- 10. Pang I. H., Zeng H., Fleenor D. L., Clark A. F. (2007) Pigment epithelium-derived factor protects retinal ganglion cells. BMC Neurosci. 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyazaki M., Ikeda Y., Yonemitsu Y., Goto Y., Murakami Y., Yoshida N., Tabata T., Hasegawa M., Tobimatsu S., Sueishi K., Ishibashi T. (2011) Pigment epithelium-derived factor gene therapy targeting retinal ganglion cell injuries. Neuroprotection against loss of function in two animal models. Hum. Gene Ther. 22, 559–565 [DOI] [PubMed] [Google Scholar]
- 12. Ogata N., Wang L., Jo N., Tombran-Tink J., Takahashi K., Mrazek D., Matsumura M. (2001) Pigment epithelium derived factor as a neuroprotective agent against ischemic retinal injury. Curr. Eye Res. 22, 245–252 [DOI] [PubMed] [Google Scholar]
- 13. Unterlauft J. D., Eichler W., Kuhne K., Yang X. M., Yafai Y., Wiedemann P., Reichenbach A., Claudepierre T. (2012) Pigment epithelium-derived factor released by Müller glial cells exerts neuroprotective effects on retinal ganglion cells. Neurochem. Res. 37, 1524–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeCoster M. A., Schabelman E., Tombran-Tink J., Bazan N. G. (1999) Neuroprotection by pigment epithelial-derived factor against glutamate toxicity in developing primary hippocampal neurons. J. Neurosci. Res. 56, 604–610 [DOI] [PubMed] [Google Scholar]
- 15. Bilak M. M., Corse A. M., Bilak S. R., Lehar M., Tombran-Tink J., Kuncl R. W. (1999) Pigment epithelium-derived factor (PEDF) protects motor neurons from chronic glutamate-mediated neurodegeneration. J. Neuropathol. Exp. Neurol. 58, 719–728 [DOI] [PubMed] [Google Scholar]
- 16. Araki T., Taniwaki T., Becerra S. P., Chader G. J., Schwartz J. P. (1998) Pigment epithelium-derived factor (PEDF) differentially protects immature but not mature cerebellar granule cells against apoptotic cell death. J. Neurosci. Res. 53, 7–15 [DOI] [PubMed] [Google Scholar]
- 17. Taniwaki T., Hirashima N., Becerra S. P., Chader G. J., Etcheberrigaray R., Schwartz J. P. (1997) Pigment epithelium-derived factor protects cultured cerebellar granule cells against glutamate-induced neurotoxicity. J. Neurochem. 68, 26–32 [DOI] [PubMed] [Google Scholar]
- 18. Sanchez A., Tripathy D., Yin X., Luo J., Martinez J., Grammas P. (2012) Pigment epithelium-derived factor (PEDF) protects cortical neurons in vitro from oxidant injury by activation of extracellular signal-regulated kinase (ERK) 1/2 and induction of Bcl-2. Neurosci. Res. 72, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dawson D. W., Volpert O. V., Gillis P., Crawford S. E., Xu H., Benedict W., Bouck N. P. (1999) Pigment epithelium-derived factor. A potent inhibitor of angiogenesis. Science 285, 245–248 [DOI] [PubMed] [Google Scholar]
- 20. Mori K., Gehlbach P., Yamamoto S., Duh E., Zack D. J., Li Q., Berns K. I., Raisler B. J., Hauswirth W. W., Campochiaro P. A. (2002) AAV-mediated gene transfer of pigment epithelium-derived factor inhibits choroidal neovascularization. Invest. Ophthalmol. Vis. Sci. 43, 1994–2000 [PubMed] [Google Scholar]
- 21. Alberdi E., Aymerich M. S., Becerra S. P. (1999) Binding of pigment epithelium-derived factor (PEDF) to retinoblastoma cells and cerebellar granule neurons. Evidence for a PEDF receptor. J. Biol. Chem. 274, 31605–31612 [DOI] [PubMed] [Google Scholar]
- 22. Becerra S. P., Notario V. (2013) The effects of PEDF on cancer biology. Mechanisms of action and therapeutic potential. Nat. Rev. 13, 258–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Becerra S. P., Sagasti A., Spinella P., Notario V. (1995) Pigment epithelium-derived factor behaves like a noninhibitory serpin. Neurotrophic activity does not require the serpin reactive loop. J. Biol. Chem. 270, 25992–25999 [DOI] [PubMed] [Google Scholar]
- 24. Filleur S., Volz K., Nelius T., Mirochnik Y., Huang H., Zaichuk T. A., Aymerich M. S., Becerra S. P., Yap R., Veliceasa D., Shroff E. H., Volpert O. V. (2005) Two functional epitopes of pigment epithelial-derived factor block angiogenesis and induce differentiation in prostate cancer. Cancer Res. 65, 5144–5152 [DOI] [PubMed] [Google Scholar]
- 25. Aymerich M. S., Alberdi E. M., Martínez A., Becerra S. P. (2001) Evidence for pigment epithelium-derived factor receptors in the neural retina. Invest. Ophthalmol. Vis. Sci. 42, 3287–3293 [PubMed] [Google Scholar]
- 26. Notari L., Baladron V., Aroca-Aguilar J. D., Balko N., Heredia R., Meyer C., Notario P. M., Saravanamuthu S., Nueda M. L., Sanchez-Sanchez F., Escribano J., Laborda J., Becerra S. P. (2006) Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J. Biol. Chem. 281, 38022–38037 [DOI] [PubMed] [Google Scholar]
- 27. Bernard A., Gao-Li J., Franco C. A., Bouceba T., Huet A., Li Z. (2009) Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor. J. Biol. Chem. 284, 10480–10490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Notari L., Arakaki N., Mueller D., Meier S., Amaral J., Becerra S. P. (2010) Pigment epithelium-derived factor binds to cell-surface F1-ATP synthase. FEBS J. 277, 2192–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deshpande M., Notari L., Subramanian P., Notario V., Becerra S. P. (2012) Inhibition of tumor cell surface ATP synthesis by pigment epithelium-derived factor. Implications for antitumor activity. Int. J. Oncol. 41, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park K., Lee K., Zhang B., Zhou T., He X., Gao G., Murray A. R., Ma J. X. (2011) Identification of a novel inhibitor of the canonical Wnt pathway. Mol. Cell. Biol. 31, 3038–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Etschmaier K., Becker T., Eichmann T. O., Schweinzer C., Scholler M., Tam-Amersdorfer C., Poeckl M., Schuligoi R., Kober A., Chirackal Manavalan A. P., Rechberger G. N., Streith I. E., Zechner R., Zimmermann R., Panzenboeck U. (2011) Adipose triglyceride lipase affects triacylglycerol metabolism at brain barriers. J. Neurochem. 119, 1016–1028 [DOI] [PubMed] [Google Scholar]
- 32. Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. (2004) Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids. Ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279, 47066–47075 [DOI] [PubMed] [Google Scholar]
- 33. Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. (2004) Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 34. Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. (2004) Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279, 48968–48975 [DOI] [PubMed] [Google Scholar]
- 35. Subramanian P., Notario P. M., Becerra S. P. (2010) Pigment epithelium-derived factor receptor (PEDF-R). A plasma membrane-linked phospholipase with PEDF binding affinity. Adv. Exp. Med. Biol. 664, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stratikos E., Alberdi E., Gettins P. G., Becerra S. P. (1996) Recombinant human pigment epithelium-derived factor (PEDF). Characterization of PEDF overexpressed and secreted by eukaryotic cells. Protein Sci. 5, 2575–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiménez-Atiénzar M., Cabanes J., Gandía-Herrero F., Escribano J., García-Carmona F., Pérez-Gilabert M. (2003) Determination of the phospholipase activity of patatin by a continuous spectrophotometric assay. Lipids 38, 677–682 [DOI] [PubMed] [Google Scholar]
- 38. Abassi Y. A., Jackson J. A., Zhu J., O'Connell J., Wang X., Xu X. (2004) Label-free, real-time monitoring of IgE-mediated mast cell activation on microelectronic cell sensor arrays. J. Immunol. Methods 292, 195–205 [DOI] [PubMed] [Google Scholar]
- 39. Solly K., Wang X., Xu X., Strulovici B., Zheng W. (2004) Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev. Technol. 2, 363–372 [DOI] [PubMed] [Google Scholar]
- 40. Murakami Y., Ikeda Y., Yonemitsu Y., Onimaru M., Nakagawa K., Kohno R., Miyazaki M., Hisatomi T., Nakamura M., Yabe T., Hasegawa M., Ishibashi T., Sueishi K. (2008) Inhibition of nuclear translocation of apoptosis-inducing factor is an essential mechanism of the neuroprotective activity of pigment epithelium-derived factor in a rat model of retinal degeneration. Am. J. Pathol. 173, 1326–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Notari L., Miller A., Martínez A., Amaral J., Ju M., Robinson G., Smith L. E., Becerra S. P. (2005) Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9. Implications for downregulation in hypoxia. Invest. Ophthalmol. Vis. Sci. 46, 2736–2747 [DOI] [PubMed] [Google Scholar]
- 42. Simonovic M., Gettins P. G., Volz K. (2001) Crystal structure of human PEDF, a potent anti-angiogenic and neurite growth-promoting factor. Proc. Natl. Acad. Sci. U.S.A. 98, 11131–11135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alberdi E., Hyde C. C., Becerra S. P. (1998) Pigment epithelium-derived factor (PEDF) binds to glycosaminoglycans. Analysis of the binding site. Biochemistry 37, 10643–10652 [DOI] [PubMed] [Google Scholar]
- 44. Duncan R. E., Wang Y., Ahmadian M., Lu J., Sarkadi-Nagy E., Sul H. S. (2010) Characterization of desnutrin functional domains. Critical residues for triacylglycerol hydrolysis in cultured cells. J. Lipid Res. 51, 309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lake A. C., Sun Y., Li J. L., Kim J. E., Johnson J. W., Li D., Revett T., Shih H. H., Liu W., Paulsen J. E., Gimeno R. E. (2005) Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J. Lipid Res. 46, 2477–2487 [DOI] [PubMed] [Google Scholar]
- 46. Cornaciu I., Boeszoermenyi A., Lindermuth H., Nagy H. M., Cerk I. K., Ebner C., Salzburger B., Gruber A., Schweiger M., Zechner R., Lass A., Zimmermann R., Oberer M. (2011) The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One 6, e26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kobayashi K., Inoguchi T., Maeda Y., Nakashima N., Kuwano A., Eto E., Ueno N., Sasaki S., Sawada F., Fujii M., Matoba Y., Sumiyoshi S., Kawate H., Takayanagi R. (2008) The lack of the C-terminal domain of adipose triglyceride lipase causes neutral lipid storage disease through impaired interactions with lipid droplets. J. Clin. Endocrinol. Metab. 93, 2877–2884 [DOI] [PubMed] [Google Scholar]
- 48. Schweiger M., Schoiswohl G., Lass A., Radner F. P., Haemmerle G., Malli R., Graier W., Cornaciu I., Oberer M., Salvayre R., Fischer J., Zechner R., Zimmermann R. (2008) The C-terminal region of human adipose triglyceride lipase affects enzyme activity and lipid droplet binding. J. Biol. Chem. 283, 17211–17220 [DOI] [PubMed] [Google Scholar]
- 49. Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J. G., Gorkiewicz G., Zechner R. (2006) Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 50. Yang X., Lu X., Lombès M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., Liu J. (2010) The G0/G1 switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 11, 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu X., Yang X., Liu J. (2010) Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle 9, 2719–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gonzalez R., Jennings L. L., Knuth M., Orth A. P., Klock H. E., Ou W., Feuerhelm J., Hull M. V., Koesema E., Wang Y., Zhang J., Wu C., Cho C. Y., Su A. I., Batalov S., Chen H., Johnson K., Laffitte B., Nguyen D. G., Snyder E. Y., Schultz P. G., Harris J. L., Lesley S. A. (2010) Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proc. Natl. Acad. Sci. U.S.A. 107, 3552–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ladhani O., Sánchez-Martinez C., Orgaz J. L., Jimenez B., Volpert O. V. (2011) Pigment epithelium-derived factor blocks tumor extravasation by suppressing amoeboid morphology and mesenchymal proteolysis. Neoplasia 13, 633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hirsch J., Johnson C. L., Nelius T., Kennedy R., Riese W., Filleur S. (2011) PEDF inhibits IL8 production in prostate cancer cells through PEDF receptor/phospholipase A2 and regulation of NFκB and PPARγ. Cytokine 55, 202–210 [DOI] [PubMed] [Google Scholar]
- 55. Borg M. L., Andrews Z. B., Duh E. J., Zechner R., Meikle P. J., Watt M. J. (2011) Pigment epithelium-derived factor regulates lipid metabolism via adipose triglyceride lipase. Diabetes 60, 1458–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Balsinde J., Winstead M. V., Dennis E. A. (2002) Phospholipase A2 regulation of arachidonic acid mobilization. FEBS Lett. 531, 2–6 [DOI] [PubMed] [Google Scholar]
- 57. Goetzl E. J., Tigyi G. (2004) Lysophospholipids and their G protein-coupled receptors in biology and diseases. J. Cell. Biochem. 92, 867–868 [DOI] [PubMed] [Google Scholar]
- 58. Kim H. Y., Akbar M., Kim K. Y. (2001) Inhibition of neuronal apoptosis by polyunsaturated fatty acids. J. Mol. Neurosci. 16, 223–227; discussion 279–284 [DOI] [PubMed] [Google Scholar]
- 59. SanGiovanni J. P., Chew E. Y. (2005) The role of ω-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog. Retin. Eye Res. 24, 87–138 [DOI] [PubMed] [Google Scholar]
- 60. Fliesler S. J., Anderson R. E. (1983) Chemistry and metabolism of lipids in the vertebrate retina. Prog. Lipid Res. 22, 79–131 [DOI] [PubMed] [Google Scholar]
- 61. Bazan N. G. (2005) Neuroprotectin D1 (NPD1). A DHA-derived mediator that protects brain and retina against cell injury-induced oxidative stress. Brain Pathol. 15, 159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lukiw W. J., Cui J. G., Marcheselli V. L., Bodker M., Botkjaer A., Gotlinger K., Serhan C. N., Bazan N. G. (2005) A role for docosahexaenoic acid-derived neuroprotectin D1 in neural cell survival and Alzheimer disease. J. Clin. Invest. 115, 2774–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mukherjee P. K., Marcheselli V. L., Serhan C. N., Bazan N. G. (2004) Neuroprotectin D1. A docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 101, 8491–8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rotstein N. P., Aveldaño M. I., Barrantes F. J., Roccamo A. M., Politi L. E. (1997) Apoptosis of retinal photoreceptors during development in vitro. Protective effect of docosahexaenoic acid. J. Neurochem. 69, 504–513 [DOI] [PubMed] [Google Scholar]
- 65. Rotstein N. P., Politi L. E., German O. L., Girotti R. (2003) Protective effect of docosahexaenoic acid on oxidative stress-induced apoptosis of retina photoreceptors. Invest. Ophthalmol. Vis. Sci. 44, 2252–2259 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.