Background: Both eukaryotic and prokaryotic GAPs use dual finger mechanisms to deactivate Rab GTPases.
Results: The Legionella pneumophila GAP LepB employs a novel catalytic network to accelerate GTP hydrolysis.
Conclusion: A non-canonical hydrolytic mechanism underlies the catalytic prowess and specificity of LepB.
Significance: The selective pressure driving evolution of unconventional Rab GAP mechanisms is related to oncogenic substitutions in Ras.
Keywords: GTPase, Host-Pathogen Interactions, Rab, Rab Proteins, X-ray Crystallography, GAP, GTPase Activating Protein, Legionella pneumophila, LepB, Rab1
Abstract
GTPase activating proteins (GAPs) from pathogenic bacteria and eukaryotic host organisms deactivate Rab GTPases by supplying catalytic arginine and glutamine fingers in trans and utilizing the cis-glutamine in the DXXGQ motif of the GTPase for binding rather than catalysis. Here, we report the transition state mimetic structure of the Legionella pneumophila GAP LepB in complex with Rab1 and describe a comprehensive structure-based mutational analysis of potential catalytic and recognition determinants. The results demonstrate that LepB does not simply mimic other GAPs but instead deploys an expected arginine finger in conjunction with a novel glutamic acid finger, which forms a salt bridge with an indispensible switch II arginine that effectively locks the cis-glutamine in the DXXGQ motif of Rab1 in a catalytically competent though unprecedented transition state configuration. Surprisingly, a heretofore universal transition state interaction with the cis-glutamine is supplanted by an elaborate polar network involving critical P-loop and switch I serines. LepB further employs an unusual tandem domain architecture to clamp a switch I tyrosine in an open conformation that facilitates access of the arginine finger to the hydrolytic site. Intriguingly, the critical P-loop serine corresponds to an oncogenic substitution in Ras and replaces a conserved glycine essential for the canonical transition state stereochemistry. In addition to expanding GTP hydrolytic paradigms, these observations reveal the unconventional dual finger and non-canonical catalytic network mechanisms of Rab GAPs as necessary alternative solutions to a major impediment imposed by substitution of the conserved P-loop glycine.
Introduction
The ability of the intracellular bacterial pathogen Legionella pneumophila to evade lysosomal degradation and replicate within diverse phagocytic cell types, including amoebae and human lung macrophages (1), depends on manipulation of conserved host proteins involved in membrane trafficking and other fundamental processes (2, 3). L. pneumophila uses an essential type IV Dot/Icm secretion system to inject at least 275 effector proteins (4). After phagocytosis, the Legionella-containing vacuole (LCV)4 fuses with vesicles derived from the endoplasmic reticulum (ER) and matures into a rough ER-like replicative organelle (5, 6).
The molecular mechanisms underlying interaction of L. pneumophila with the early secretory pathway have been illuminated by studies based on the observation that the Rab1 GTPase, which is essential for fusion of ER-derived vesicles with the cis-Golgi, is recruited to the LCV shortly after phagocytosis and cycles off as the LCV matures (7, 8). Rab1 recruitment depends on the effector DrrA/SidM, a Rab1 guanine nucleotide exchange factor (GEF) targeted to the LCV through interaction with phosphatidylinositol 4-phosphate (9–11). The effector LepB accumulates on the LCV as DrrA and Rab1 cycle off and has been shown to function as a GTPase activating protein (GAP) for Rab1 (12). Another effector, LidA, binds several Rab GTPases including Rab1 with low nm to pm affinity and may sequester Rab proteins or tether ER-derived vesicles with the LCV to facilitate SNARE-mediated fusion (9). Interestingly, DrrA also promotes non-canonical SNARE pairing of plasma membrane syntaxins on the LCV with Sec22b on ER-derived vesicles (13, 14) and AMPylates a tyrosine residue in the switch II region of Rab1 (15). Recent studies have identified an effector that phosphocholinates Rab1 on an adjacent serine (16) as well as effectors with Rab1 de-AMPylation and de-phosphocholination activities (17, 18). These modifications impair or abrogate interactions with DrrA, LepB, and some host proteins but not LidA (19, 20).
Several studies have delineated the structural bases for Rab1 activation, AMPylation, and interaction with LidA (15, 21–23). LepB is not homologous to known GAPs and, consequently, the structural basis for Rab1 deactivation has been unclear. TBC (Tre, Bub2, Cdc16) domains in host Rab GAPs supply catalytic arginine and glutamine fingers in trans and utilize the cis-glutamine in the DXXGQ motif of Rab GTPases for binding rather than catalysis (24). An analogous dual trans finger mechanism was identified in the Rab1 GAPs VirA and EspG from Shigella flexneri and enteropathogenic Escherichia coli, respectively (25). A recently reported crystallographic study of the LepB GAP core in complex with Rab1-GDP and the ground state mimetic beryllium fluoride (BeF3) revealed that LepB supplies an arginine finger but not a glutamine finger and instead utilizes the cis-glutamine in the Rab1 DXXGQ motif (26). This structural observation, which was supported by mutation of the putative arginine finger and cis-glutamine, suggested that LepB utilizes a Ras/Rho GAP-like catalytic mechanism rather than the dual finger mechanism described for TBC domains (24, 27) and VirA/EspG (25).
Although considerable attention has been focused on the role of the trans/cis-arginine and glutamine as principal catalytic residues in GAP-GTPase reactions, computational as well as experimental studies suggest that the overall electrostatic environment, conformational transitions between the ground and active states, and other factors including steric hindrance in the hydrolytic site also play important roles (28–30). Beyond identification of expected catalytic arginine and glutamine residues based on the ground state complex (26), the structural mechanism for LepB-mediated acceleration of GTP hydrolysis has not been investigated in detail.
Here, we report the crystal structure of the LepB GAP core in complex with Rab1-GDP and the transition state mimetic aluminum fluoride (AlF3), which is interpreted in the context of a family-wide profile of Rab substrate specificity and an extensive structure-based mutational analysis of potential catalytic determinants. The structure and supporting mutational analyses reveal an unprecedented catalytic network consisting of six distinct polar/charged residues that are critical for GAP activity. This non-canonical catalytic mechanism accounts for the unusually high catalytic prowess of the LepB GAP core, contributes to its exquisite substrate-specificity, and provides insight into the selective pressure driving evolution of unconventional hydrolytic mechanisms in Rab GAPs.
EXPERIMENTAL PROCEDURES
Constructs
Constructs amplified with Vent polymerase were digested with BamHI-hf/SalI-hf and ligated into a modified pET15b vector incorporating an N-terminal His6 tag (human Rab1A), a modified pET28b vector incorporating a His6-SUMO tag (LepB), or pGEX-4T1/pGEX-6P1 (Rab GTPases). Mutations were generated using the QuikChange II XL kit (Stratagene). Constructs were verified by sequencing.
Expression and Purification
BL21(DE3)-RPIL cells transformed with LepB and Rab expression plasmids were cultured for 16 h at 21 °C in 2×YT after induction with 50 μm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 0.2–0.4. Cell pellets resuspended in 50 mm Tris, pH 8.0, 0.2 mg/ml lysozyme (Sigma), 10 μg/ml protease free DNase I (Worthington) were incubated on ice for 40 min and lysed by sonication. Lysates were supplemented with 0.5% Triton X-100 and centrifuged at 33,000 × g for 1 h at 4 °C. His6 constructs were initially purified using nickel-nitrilotriacetic acid-Sepharose HP (GE Healthcare) equilibrated with 50 mm Tris, pH 8.0, 100 mm NaCl, washed with buffer containing 500 mm NaCl and 15 mm imidazole, and eluted with 300 mm imidazole. To remove His6 SUMO, fusion proteins were digested with 0.12 mg/ml His6-SUMO protease for 12 h at 4 °C and passed through a nickel-nitrilotriacetic acid Sepharose HP column. GST fusions were initially purified using glutathione-Sepharose (GE Healthcare) equilibrated and washed with 50 mm Tris, pH 8.0, 100 mm NaCl and eluted with 10 mm reduced glutathione. Constructs were further purified by anion exchange chromatography on a HiTrap HPQ column (GE Healthcare), equilibrated, washed with 10 mm Tris, pH 8.0, and eluted with a gradient of 0–1 m NaCl followed by gel filtration on a Superdex-75 column (GE Healthcare) in 10 mm Tris, pH 8.0, 100 mm NaC1. All buffers were supplemented with 0.1% 2-mercaptoethanol and, for Rab GTPases, 2 mm MgCl2.
GAP Assays and Stopped Flow Experiments
Rab GTPases were incubated for 1–3 h at 25 °C in 20 mm Tris, pH 8.0, 150 mm NaCl, 5 mm EDTA, and 1 mm DTT with a 10-fold molar excess of GTP that was subsequently removed using a D-Salt column (Pierce). The kinetics of phosphate release after intrinsic and GAP-accelerated GTP hydrolysis were continuously monitored using a fluorescent phosphate sensor consisting of the E. coli phosphate-binding protein (PBP) labeled with 7-diethylamino-3-[N-(2-maleimidoethyl)carbamoyl]coumarin (MDCC) on a cysteine residue introduced in a loop proximal to the binding site as described (31). Reactions were initiated by mixing 4 μm His6/GST-Rab-GTP with 10 mm MgCl2, 5 μm PBP-MDCC phosphate sensor, and varying concentrations of His6-SUMO-LepB326–623 or His6-TBC1D201–350 in 20 mm Tris, pH 7.5, 150 mm NaCl. GAP assays were conducted in 96-well half-area microplates (Corning) using a Safire microplate spectrometer (TECAN) with emission and excitation wavelengths of 425 and 457 nm, respectively. Stopped flow measurements were performed using a SX20 instrument (Applied Photophysics) with emission detected through a 455-nm-long pass filter after excitation at 425 nm. For intrinsic tryptophan fluorescence experiments, samples were excited at 297 nm, and the emission was detected at 340 nm (microplate assays) or through a 320-nm long pass filter (stopped flow). Time courses were fit with an exponential model,
where I(t), I0, and I∞ are the fluorescence intensities at the indicated times. The turnover number (kcat) and Michaelis constant (Km) were obtained by fitting the resulting values of kobs with the hyperbolic model,
Catalytic efficiencies (kcat/Km) were obtained from the slopes of linear fits with,
Rab Specificity Profile
GST-Rab GTPases were loaded with GTP as described above, and the kinetics of GTP hydrolysis were monitored in microplate format at 0, 10, 50, and 250 nm His6-SUMO-LepB326–623. Initial velocities (vo) obtained by linear regression were converted to concentration per unit time using a conversion factor determined by titration of PBP-MDCC with a phosphate standard. Catalytic efficiencies (kcat/Km) were estimated from the slopes of linear fits with
Crystallization, Data Collection, and Structure Determination
LepB326–623 and His6-Rab1A4–177 were transformed into BL834(DE3) and expressed in defined MOPS medium (Molecular Dimensions) supplemented with 40 mg/ml selenomethionine. Purified LepB constructs were incubated in a 1:1 ratio with His6-Rab14–177 for 12 h at 4 °C in 10 mm Tris, pH 8.0, 2 mm MgCl2, 2 mm AlCl3, and 20 mm NaF. Initial crystallization conditions were identified by sparse matrix screening in 96-well sitting drop IntelliPlates (Art Robbins) formatted with equal 200-nl volumes of 10 mg/ml complex and crystallization solution using a Gryphon crystallization robot (Art Robbins). For diffraction experiments, 10 mg/ml SeMET-substituted complex was crystallized in hanging drops with 12% PEG 4000, 0.1 m Hepes, pH 7.2, 0.2 m potassium acetate, 2 mm AlCl3, 20 mm NaF, and 10 mm 2-mercaptoethanol. Crystals were transferred to a cryo-stabilizer solution consisting of 20% PEG 4000, 100 mm Hepes, pH 7.2, 0.2 m potassium acetate, 2 mm AlCl3, 20 mm NaF, 10 mm 2-mercaptoethanol, 20% glycerol, and flash-frozen in liquid nitrogen. The crystals are in the space group I4122 with cell dimensions a = b = 139.4 Å, c = 384.5 Å, a mosaicity of 0.5, and a solvent content of 58%. A two-wavelength anomalous diffraction (MAD) data set was collected at the selenium edge using the NSLS X29 beamline at Brookhaven National Laboratory and processed with HKL2000 (32).
The structure was solved by MAD using sites and density modified phases calculated by AutoSol in Phenix (33). Density modification included solvent flipping and averaging over two of the three complexes in the asymmetric unit. The resulting σA-weighted experimental map was used for automated model building with Buccaneer (34). Models complete to 80–90% were obtained for two complexes along with a partial model in multiple fragments for the third complex. A complete model for the third complex (chains E and F) was subsequently obtained by molecular replacement using Phaser (35) with search models consisting of the individual LepB (chain A) and Rab1 (chain B) coordinates. The trace was completed, and the models were refined without non-crystallographic symmetry restraints by iterating manual model building in Coot (36) with simulated annealing in Phenix (33) and positional refinement by Refmac5 (37). Buccaneer, Phaser, and Refmac5 were used as implemented in CCP4 (38). Structural figures were generated with PyMOL.
The third complex is located adjacent to an extended solvent channel and is poorly ordered as indicated by the low quality of the electron density in the experimental and difference maps, high overall B-factors, and torsion angle outliers in the final refined model. Lower symmetry space groups were also considered by systematically locating the contents of the asymmetric unit by molecular replacement with Phaser for all possible space groups including P1. The structures were refined by simulated annealing in Phenix followed by Refmac5 is CCP4. In no instance did we find a solution that resolved the observed disorder or resulted in a lower free R. Thus, at least at the resolution of the data, there is no evidence to support a lower symmetry space group even though refinement in lower symmetry space groups is possible with similar but not better results.
Statistical Analysis
Values and errors represent mean ± S.D. for 2–4 independent measurements.
RESULTS
The LepB GAP Core Has Unusually High Catalytic Activity and Specificity
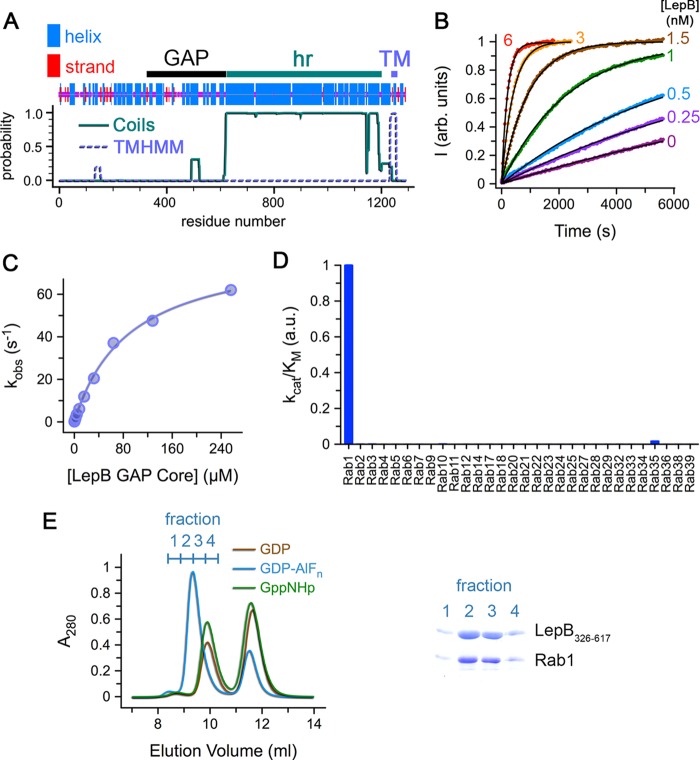
LepB consists of an N-terminal region with a high density of predicted helical structure despite lacking homology with other proteins followed by heptad repeats and a transmembrane domain (Fig. 1A). A His6 construct of the full-length protein was expressed in a soluble form and purified. Limited proteolysis with Glu-C yielded a stable intermediate of ∼33 kDa. N-terminal sequencing revealed a mixture consisting of the His6 tag and a sequence (LKPLTLLM) matching residues 326–334. Based on the apparent molecular mass of the Glu-C fragment, constructs were generated corresponding to residues 326–617, 326–623, and 326–667. The shorter constructs purified as monomers, whereas the longer construct, which includes ∼40 residues from the heptad repeat region, purified as a dimer.
FIGURE 1.
Identification and characterization of the LepB GAP core. A, the region corresponding to the LepB GAP core identified by limited proteolysis with Glu-C is depicted above the predictions of consensus secondary structure (47), heptad repeats (hr) with Coils (48), and transmembrane (TM) helices with TMHMM (49). B, shown are time courses for Rab1 GTP hydrolysis at the indicated concentrations of the LepB GAP core. C, shown is stopped flow analysis of the kinetics for Rab1 GTP hydrolysis as a function of the concentration of the LepB GAP core. D, shown is a profile of the catalytic efficiency (kcat/Km) of the LepB GAP core for GST-fusions of 30 Rab GTPases. E, shown is gel filtration chromatography after incubation of the LepB GAP core with Rab1-GDP, Rab1-GDP-AlF3, or Rab1-GppNHp in a 1:2 stoichiometric ratio. On the right is an SDS-polyacrylamide gel of the fractions corresponding to the first peak (elution volume 8.5–10.5 ml; 0.5-ml fractions). a.u., arbitrary units.
As shown in Fig. 1B, LepB326–623 has a very high catalytic efficiency for Rab1 (kcat/Km = 9.5 × 105 m−1 s−1), which is similar to that of LepB326–667 (kcat/Km = 1.4 × 106 m−1 s−1), >10-fold higher than well characterized TBC domains such as Gyp1p and TBC1D20, and ∼3-fold higher than VirA (Table 1). Detailed kinetic analysis by stopped flow revealed a high kcat of 84 s−1 and a Km of 90 μm (Fig. 1C). These values of kcat/Km, kcat, and Km differ by <3-fold from those reported recently for similar LepB and Rab1 constructs (19, 26, 39). A somewhat higher kcat/Km of 3.6 × 106 m−1 s−1 was reported for the longer LepB299–618 construct (26) compared with the value of 5.5 × 105 m−1 s−1 previously reported for the shorter LepB325–618 construct (19). These moderate variations can be attributed to differences in assay conditions and constructs.
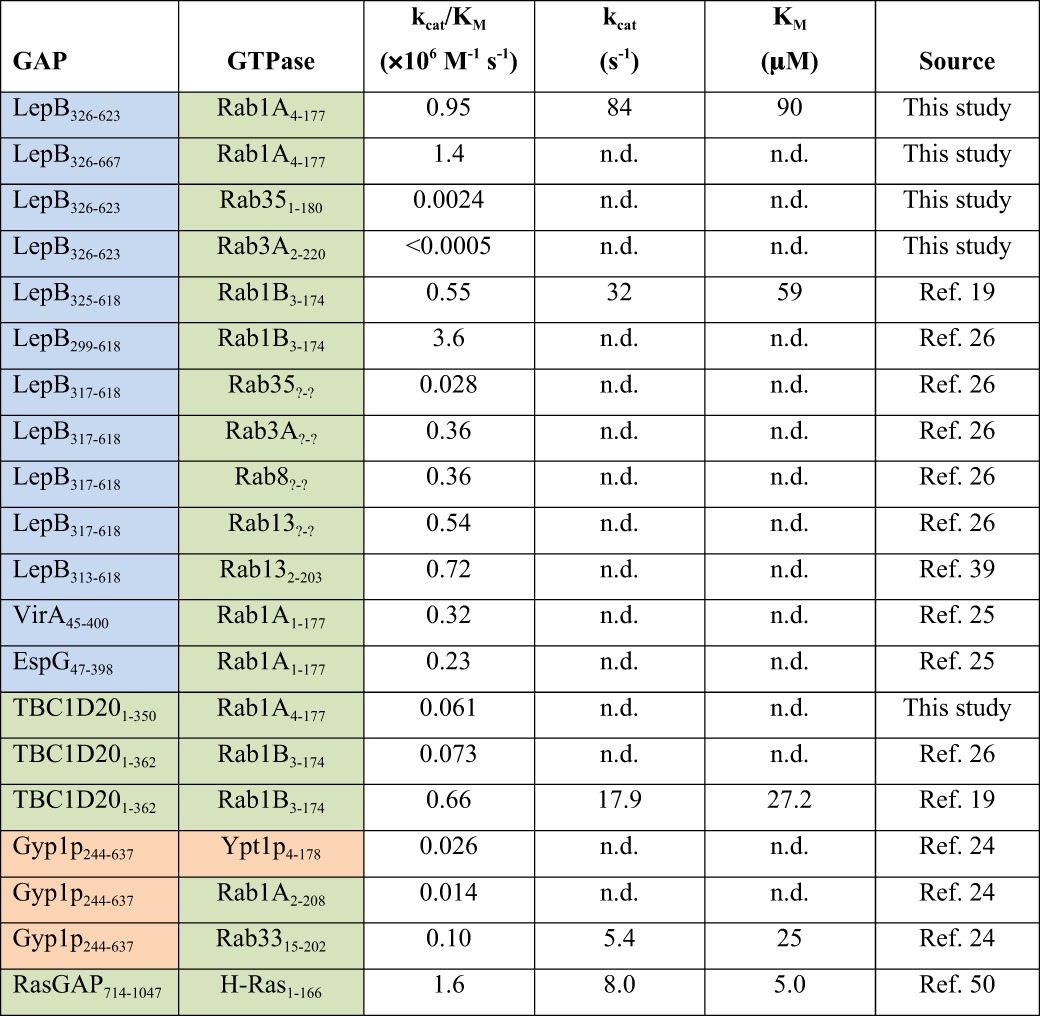
TABLE 1.
Kinetic parameters for the catalytic cores of LepB and other GAPs
n.d., not determined; colors indicate taxonomic origin: bacterial (blue), mammalian (green), yeast (orange).
Because some TBC domains exhibit complex Rab specificities that do not correlate with phylogenetic relationships (40), we profiled the catalytic efficiency of LepB326–623 for 30 Rab GTPases (Fig. 1D). In contrast to Gyp1p and VirA, which have GAP activity for multiple Rabs (24), LepB326–623 is remarkably selective for Rab1. Indeed, kcat/Km is 400-fold lower for Rab35 (the most similar Rab1 paralog) and >2000-fold lower for other Rabs. Similarly high specificity for Rab1 was recently reported for LepB313–618 (39). The LepB GAP core also formed a stable complex with Rab1-GDP and aluminum fluoride (AlF3) that could be isolated by gel filtration (Fig. 1E).
Based on intrinsic tryptophan fluorescence, LepB was reported to have substantial catalytic efficiencies for Rab3A, Rab8, and Rab13 that are only 10-fold lower than for Rab1 and thus comparable to the most active TBC domain GAPs (26). However, in the profile of catalytic efficiency described here (Fig. 1D), which uses a fluorescent sensor to detect phosphate release, we observed weak activity for Rab3A that was at least 2000-fold lower than for Rab1. Using intrinsic tryptophan fluorescence in both stopped flow and microplate experiments (Fig. 2), we readily detected a strong signal for Rab1 that yielded a catalytic efficiency equivalent to that measured by phosphate detection. For Rab3A, on the other hand, we observed an increase in intrinsic tryptophan fluorescence in the microplate experiments that clearly coincided with intrinsic GTP hydrolysis but was not accelerated by LepB. In stopped flow, we observed a small but reproducible transient that depended on Mg2+ but not LepB. This small transient was also observed for Rab3A loaded with GppNHp and evidently reflects conformational changes after Mg2+ addition rather than GTP hydrolysis. Because production of inorganic phosphate is a hallmark of GTP hydrolytic reactions and because we detected only marginal acceleration of the intrinsic hydrolytic rate constant by either method (kcat/Km <500 m−1 s−1), we conclude that Rab3A is a very weak LepB substrate compared with Rab1. Although Rab8 and Rab13 were not included in our profile, neither Rab3A nor Rab13 was detected as substrates in an independently determined profile of LepB313–618 that did not include Rab8 (39). Whether Rab8 is a bona fide substrate remains unclear.
FIGURE 2.
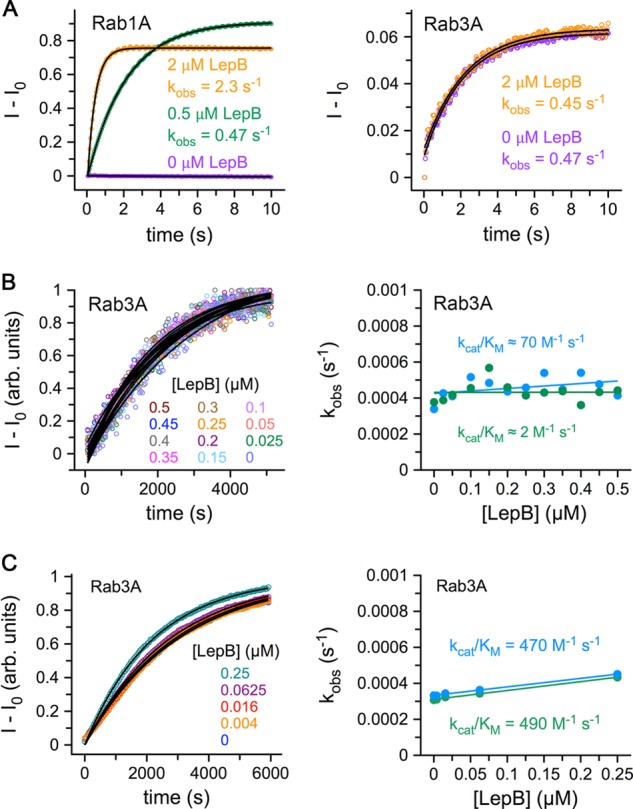
Hydrolytic activities of Rab1A and Rab3A in the presence and absence of LepB. A, stopped flow experiments with excitation at 297 nm and detection through a 320-nm long pass filter are shown. Note that the small transient signal for Rab3A (right panel) requires Mg2+ and is also observed for the GppNHp-loaded protein (data not shown), indicating that it cannot be due to GTP hydrolysis. B, shown are microplate experiments with excitation at 297 nm and detection at 340 nm. C, shown are microplate experiments with phosphate release detected using the MDCC-phosphate-binding protein.
Structure of the LepB GAP Core in a Transition State Mimetic Complex with Rab1
Crystals diffracting to 3.2 Å were obtained for His6-Rab1A4–177-GDP-AlF3 in complex with LepB326–623. The structure was solved by multiwavelength anomalous diffraction (“Experimental Procedures,” Table 2, and Fig. 3). The asymmetric unit contains three independently refined complexes, two of which are well ordered and have similar tertiary structures (root mean square deviations of 0.26 Å after superposition) with no significant differences in the molecular interfaces or hydrolytic site stereochemistry (Fig. 3). The figures, analyses, and presentation are based on chains A (LepB) and B (Rab1) but also apply to chains C (LepB) and D (Rab1). Chains E (LepB) and F (Rab1), on the other hand, are poorly ordered and were, therefore, excluded from subsequent consideration.
TABLE 2.
Data collection and refinement statistics
Numbers in parentheses represent the values for the highest resolution shell.
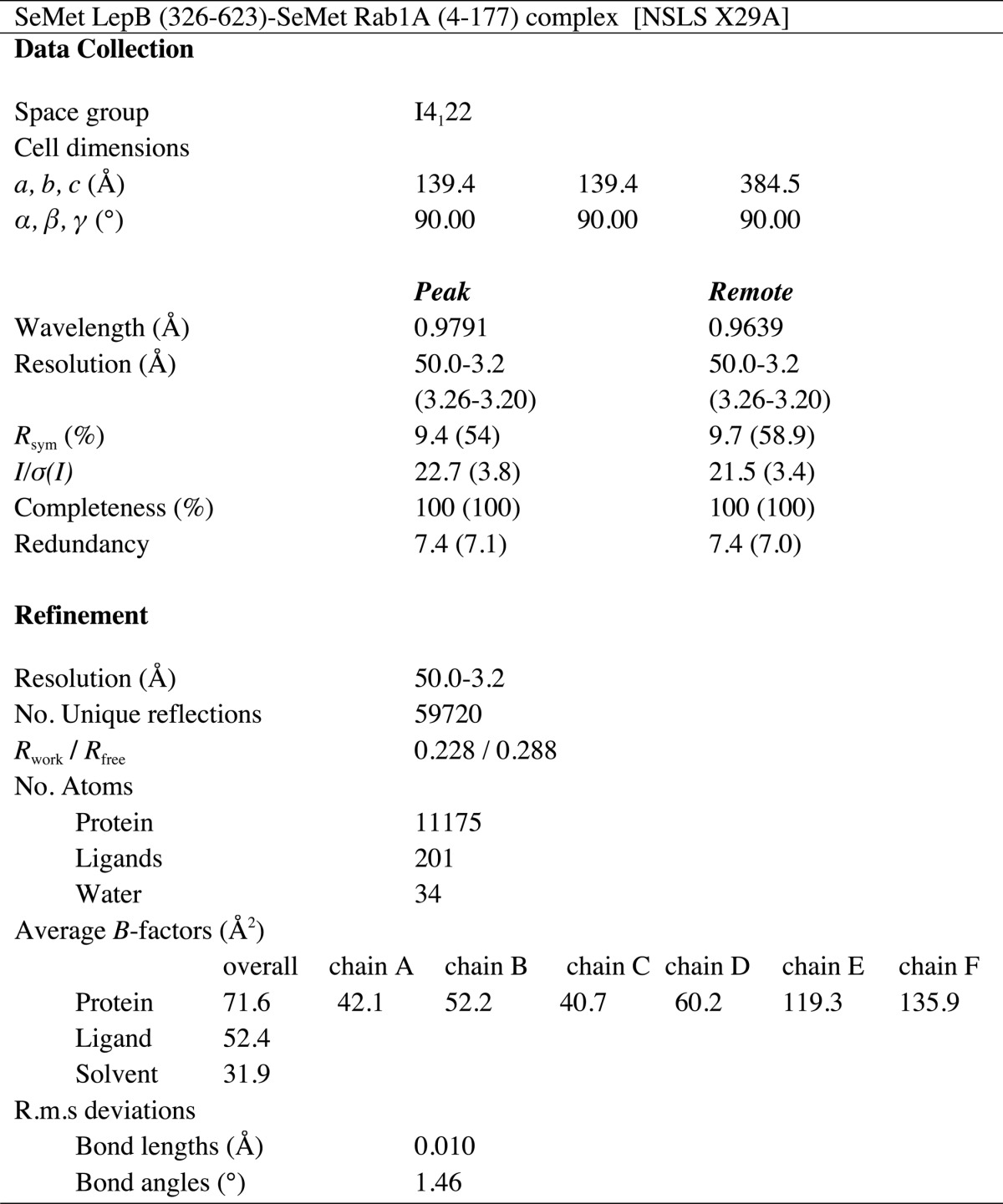
FIGURE 3.
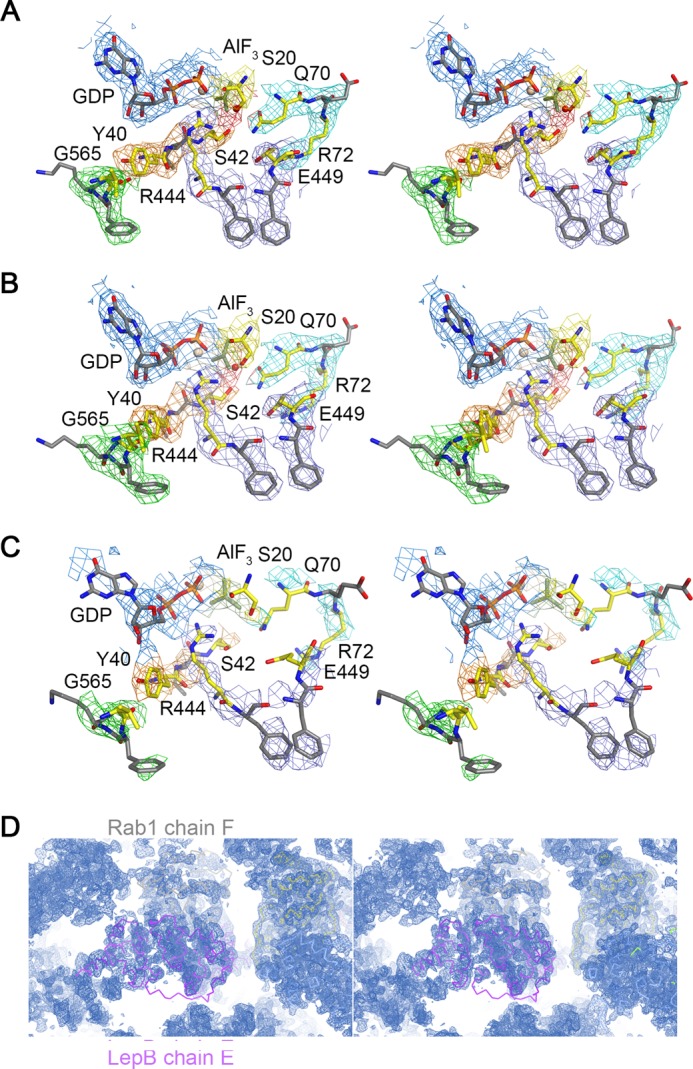
Stereoviews of electron density contoured at 1.0 σ from a σA-weighted 2wFo − DFc map calculated with MAD phases improved by solvent flipping and NCS averaging. A, shown are chains A and B. B, shown are chains C and D. C, shown are chains E and F. The final refined model is also shown. D, shown is the extended solvent channel in the vicinity of chains E and F.
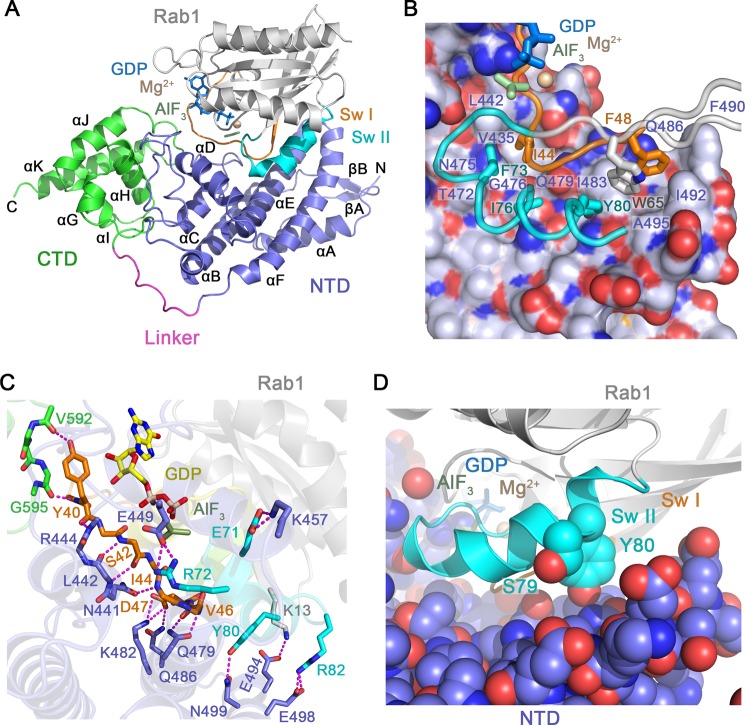
The overall structure of the LepB GAP core is similar to that in the ground state Rab1-GDP-BeF3 complex (26) and consists of two domains (residues 326–524 and 536–621) connected by a polyproline linker (Fig. 4A). The N-terminal domain has a mostly helical secondary structure, with an elongated although distorted three helix bundle (αB, αE, and αF) that scaffolds an N-terminal α/β subdomain (βA, βB, and αA) and a central subdomain composed of two helices (αC and αD) connected by large yet well defined loops. Near the αE/αF loop, the three-helix bundle splays apart to accommodate a substantial hydrophobic core at the interface with the N-terminal subdomain. Docking of the central subdomain with the opposite, less distorted end of the three helix bundle generates a pronounced V-shaped groove corresponding to the core of the Rab1 binding site. The C-terminal domain consists of five helices (αG-αK) with a right-handed super-helical arrangement and, as described below, plays a crucial role with respect to the catalytic efficiency of the GAP core.
FIGURE 4.
Structural basis for Rab1 recognition by LepB. A, shown is the overall view of the LepB catalytic core in complex with Rab1-GDP and AlF3. CTD, C-terminal domain; NTD, N-terminal domain. B, shown is the non-polar interface between LepB and the switch/interswitch regions of Rab1. LepB is rendered as spheres beneath a semi-transparent surface with carbon, nitrogen, and oxygen atoms colored light slate, blue, and red, respectively. Rab1 is rendered as tubes and colored as in panel A. C, shown is are polar interactions between LepB and Rab1, defined using a 3.4 Å distance cutoff with appropriate stereochemistry. D, shown is the location of AMPylated (Tyr-80) and phosphocholinated (Ser-79) residues.
The LepB transition state complex with Rab1-GDP-AlF3 buries a surface area of 2500 Å2 within an interface centered on the N-terminal domain (Fig. 4B). Near the splayed end of the three helix bundle, the invariant hydrophobic triad at the switch/interswitch junction of Rab1 (switch I Phe-48, interswitch Trp-65, and switch II Tyr-80) docks in a hydrophobic groove lined by residues from αE (Ile-483, Gln-486, and Phe-490), the αE/αF loop (Ile-492), and αF (Ala-495). Other hydrophobic switch I (Ile-44) and switch II residues (Phe-73 and Ile-76) are buried in an adjacent hydrophobic groove defined by residues from αE (Gly-476, Thr-472, Asn-475, and Gln-479) and the αC/αD loop (Val-435 and Leu-442). The hydrophobic core of the interface is flanked by an extensive network of polar/ionic interactions (Fig. 4C). At the nucleotide proximal end of the interface, switch I Tyr-40 is clamped by the backbone of residues in the αB/αC loop of the N-terminal domain (His-415 to Gly-416) and αJ/αK loop of the C-terminal domain (Gly-595 to Arg-596) and further secured through polar interactions with the backbone of Arg-444 in the N-terminal domain and Val-592 and Gly-595 in the C-terminal domain (see also Fig. 6A). Consistent with the ground state structure (26), the mode of interaction with the switch II region explains why GAP activity is abrogated by AMPylation of Tyr-80 but only mildly impaired by phosphocholination of Ser-79 (19, 20). Indeed, Tyr-80 is located directly in the interface, whereas Ser-79 is exposed at a proximal location (Fig. 4D).
FIGURE 6.
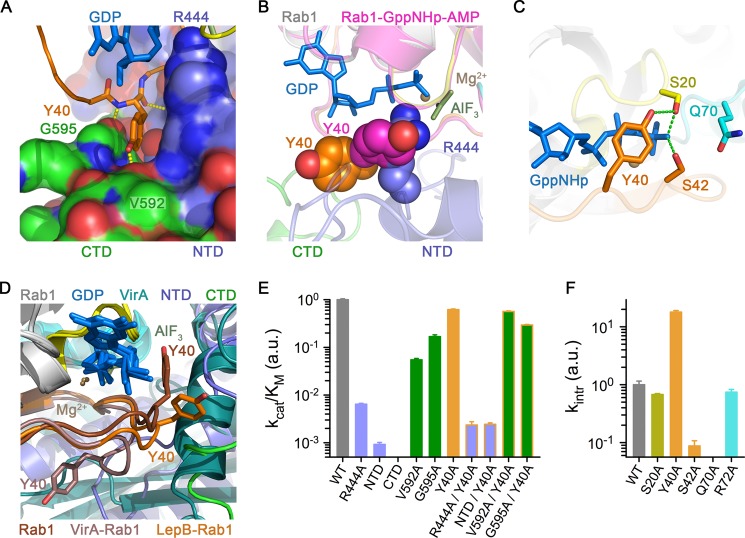
Role of the tandem domain architecture and clamp interaction. A, shown are polar and non-polar interactions between Tyr-40 in the switch I region of Rab1 and the interface of the N-terminal and C-terminal domains of the LepB GAP core. CTD, C-terminal domain; NTD, N-terminal domain. B, shown is a comparison of Rab1 from the LepB complex with AMPylated Rab1-GTP (PDB ID 3NKV) after superposition. C, shown are polar interactions involving the Ser-20, Tyr-40, and Ser-42 in the closed conformation of AMPylated Rab1-GTP. D, shown is a comparison of Tyr-40 conformations in AMPylated Rab1-GTP alone and in the complexes with LepB and VirA (PDB ID 4FMB) after superposition of the Rab1 molecules. E, shown is catalytic efficiency of LepB and/or Rab1 mutations involving residues or domains implicated in the clamp interaction with Tyr-40. F, shown are rate constants for intrinsic GTP hydrolysis by Rab1 mutants.
The Hydrolytic Site Architecture in the Transition State Mimetic Complex Reveals an Unprecedented Non-canonical Polar Network
Inspection of the experimental maps revealed clear density for GDP, Mg2+, and aluminum fluoride, which was modeled as planar AlF3 (Fig. 3). Although not clearly resolved at 3.2 Å resolution, the invariant waters coordinated by Al3+ and Mg2+ could be modeled in density at the expected locations. There was also clear density for an obvious arginine finger (Arg-444 from the αC/αD loop), which appears to mediate canonical polar interactions with the α/β phosphates and one of the equatorial fluoride ions. Unlike TBC domains and VirA/EspG, but consistent with the ground state complex (26), LepB does not supply a trans-glutamine finger (Fig. 5, A and B). Instead, the cis-glutamine in the Rab1 DXXGQ motif (Gln-70) is oriented toward the hydrolytic site, where it appears to mediate a canonical polar interaction with the axial (hydrolytic) water without forming the heretofore invariant hydrogen bond with the equatorial fluoride (Fig. 5A). Intriguingly, however, the loss of this key interaction appears to be compensated by an elaborate network of polar residues, in which Ser-20 in the P-loop contacts the equatorial fluoride that typically interacts with the cis/trans-glutamine, whereas Ser-42 in switch I contacts the same equatorial fluoride, the axial water, and the cis-glutamine. Moreover, alternative rotamer conformations for Gln-70 are excluded by Glu-449, which appears to mediate a polar interaction with Gln-70 while extending to form a salt bridge with Arg-72 in the switch II region. Although the resolution of the structure precludes a detailed analysis of the stereochemistry, it is noteworthy that the same polar network and pattern of putative hydrogen bonding interactions was observed in both of the well ordered, independently refined complexes (Fig. 3).
FIGURE 5.
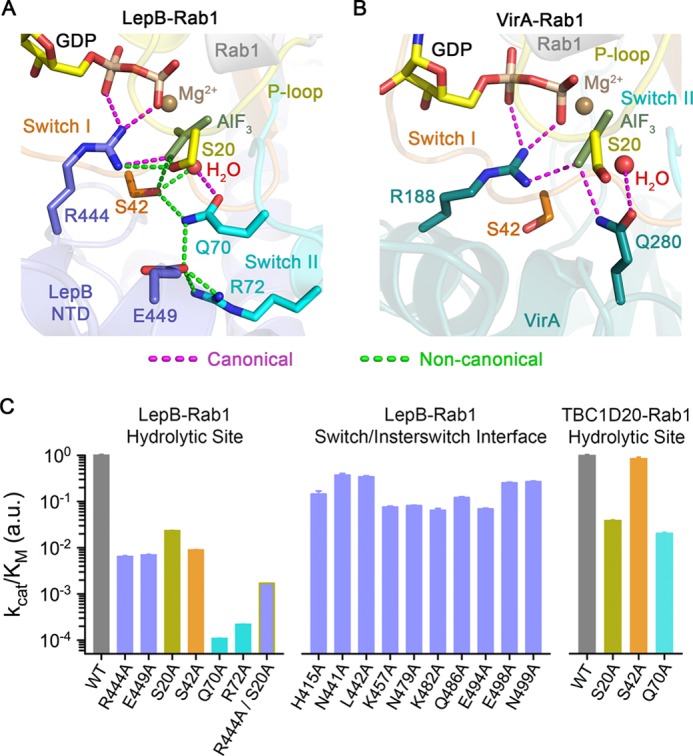
Non-canonical structural mechanism for acceleration of GTP hydrolysis. A, shown is the view of the GTP hydrolytic site in the LepB-Rab1 complex with canonical and non-canonical polar interactions, colored as indicated. B, shown is the view of the GTP hydrolytic site in the VirA-Rab1 complex (PDB ID 4FMB). C, shown is catalytic efficiency of mutations involving LepB and/or Rab1 residues in the non-canonical polar network (left), LepB residues in the polar/non-polar interface with the switch and interswitch regions (middle), and Rab1 residues in the non-canonical polar network with respect to the TBC domain of TBC1D20 (right). Bars are colored according to the location of the mutated residues using the color scheme in Fig. 2A. a.u., arbitrary units.
Residues in the Non-canonical Polar Network Are Critical Determinants of GAP Activity
To examine the functional importance of the putative non-canonical hydrolytic network, the implicated residues were replaced with alanine. All of the mutants described here expressed as soluble monomeric proteins at wild-type levels and were purified to homogeneity. Large decreases in kcat/Km of 100 and 10,000-fold were observed for R444A and Q70A, respectively (Fig. 5C). The large reduction for the Q70A substitution contrasts with the modest effect of the equivalent mutation in Rab substrates for the Gyp1p TBC domain (24) or the moderate effect on TBC1D20 catalysis. These results are in good agreement with the effects of the R444A and Q70A mutations described in the context of the ground state complex (26).
Remarkably, although consistent with the putative non-canonical polar network observed in the transition state structure, the S20A, S42A, R72A, and E449A substitutions reduced kcat/Km to an extent comparable to or greater than mutation of the arginine finger, with effects ranging from 50- to 5000-fold (Fig. 5C). Moreover, the Q70A mutation reduced the intrinsic rate of hydrolysis to an undetectable level, whereas an 11-fold reduction was observed for S42A (see Fig. 6F). Interestingly, the intrinsic rate of S20A is similar to wild-type Rab1, suggesting that the large effect of this mutation on LepB catalysis may reflect loss of the non-canonical polar interaction with Arg-444. Consistent with this interpretation, the R444A mutation diminished kcat/Km by 10-fold for S20A compared with 100-fold for wild-type Rab1. In contrast, catalytic efficiency was reduced 10-fold or less for mutation of 10 other LepB residues that mediate polar interactions with Rab1, supporting a role for these residues in binding rather than catalysis.
Taken together, these observations confirm the trans/cis disposition of the arginine finger/catalytic glutamine in the context of the transition state-mimetic complex and further identify a non-canonical catalytic network that includes four additional residues that are critical for GAP activity and appropriately distributed within the hydrolytic site to contribute to transition state stabilization via direct interactions with an equatorial γ-phosphate oxygen and axial hydrolytic water (Ser-20 and Ser-42) or otherwise restrict the cis-glutamine to a catalytically competent configuration (Glu-449 and Arg-72). The non-canonical intramolecular interactions involving Gln-70 and Ser-42 also appear to be relevant for the intrinsic reaction.
Role of the Tandem Domain Architecture and Tyr-40 Clamp Interaction
As shown in Fig. 6A, Tyr-40 in the switch I region of Rab1 is clamped in a distinctive crevice between the N-terminal domain and C-terminal domain of LepB, where it is secured through polar interactions. In the ground state, Tyr-40 adopts either an “open” conformation trapped by crystal contacts or a “closed” conformation that blocks insertion of the arginine finger (Fig. 6B). The corresponding tyrosine in the yeast ortholog Ypt1p adopts an identical closed conformation (41), strongly suggesting that the closed conformation predominates in the absence of artifactual crystal contacts as demonstrated for the equivalent Tyr-32 in Ras (42). A notable difference in Rab1 is the hydrogen bond between Tyr-40 and Ser-20 (Fig. 6C) rather than a direct or water-mediated contact with the γ-phosphate as in Ras (43). As a consequence of the clamp interaction in the LepB complex or a distinct yet analogous interaction in VirA/EspG, Tyr-40 is maintained in an open conformation compatible with arginine finger insertion (Fig. 6D).
Three different approaches were used to assess the role of the C-terminal domain and clamp interaction. First, Tyr-40 was replaced by alanine, which is predicted to generate a constitutively open Rab1 variant. Consistent with this expectation and the possibility that the Tyr-40 side chain does not contribute to GAP affinity per se, the Y40A mutation had little effect on kcat/Km (Fig. 6E). Interestingly, however, the intrinsic hydrolytic rate was elevated 20-fold (Fig. 6F), strongly suggesting that the closed conformation is not only favored in solution but contributes substantially to ground state stability and thus represents a barrier that must be overcome by GAPs. As a second approach, constructs corresponding to the N- and C-terminal domains were generated. Both domains expressed in a soluble form at levels comparable to the intact catalytic core and were readily purified to homogeneity as uniform monomers with no evidence of breakdown products or aggregation. The kcat/Km for the N-terminal domain was 1000-fold lower than the intact catalytic core, whereas no activity was detected for the C-terminal domain (Fig. 6E). This dramatic difference, however, cannot be due entirely to loss of contacts with the side chain of Tyr-40, as kcat/Km for the N-terminal domain is 300-fold lower with respect to the constitutively open Y40A mutant. The third approach, which involved mutation of LepB residues proximal to the clamp interface, provides the most conclusive evidence that the clamp interaction plays an important role. Indeed, the V592A substitution reduced kcat/Km 20-fold for wild-type Rab1 but had negligible effect with respect to the Y40A mutant. Likewise, the G595A mutation resulted in a 5-fold decrease that is partially restored by the Y40A mutation, consistent with small steric perturbations involving both the backbone and side chain of Tyr-40. These observations clearly demonstrate that the tandem domain architecture is critical for GAP activity and suggest that the clamp interaction destabilizes the ground state in addition to facilitating access of the arginine finger in the transition state.
DISCUSSION
This study reveals how a structurally unique GAP core of a bacterial effector attains extraordinary catalytic output and substrate specificity by employing an atypical tandem domain architecture in combination with a non-canonical transition state polar network involving six distinct “catalytic” residues. Despite similarity with respect to the trans/cis disposition of the arginine finger/catalytic glutamine, as recently deduced from the structure of the ground state complex (26), it is clear that there are profound differences between the conventional structural mechanism for acceleration of GTP hydrolysis observed in other GAPs and that described here for LepB based on the structure of the transition state complex and systematic mutational analysis of polar residues in the LepB-Rab1 interface.
The LepB GAP core occurs in the context of a large, 1294-amino acid protein that also includes an N-terminal domain of unknown structure and function, a long sequence of heptad repeats, and a putative C-terminal transmembrane domain that is likely important for localization to the LCV. Notably, the catalytic efficiency of the LepB GAP core is ∼100-fold greater than that reported earlier for the full-length protein (12). It is possible that the catalytic activity is autoinhibited by elements outside the GAP core; however, the lower apparent activity of the full-length protein could be due to aggregation mediated by the putative transmembrane domain. Additional experiments are required to distinguish these possibilities and determine the function of the N-terminal domain and heptad repeats.
Given the length of the heptad repeat region and the observation that a construct containing ∼40 residues of the heptad repeats proximal to the GAP core purifies as a dimer on gel filtration, we suspect that the heptad repeats may form an extended parallel coiled coil that could project as far as 100 nm into the cytoplasm. The presence of likely hinge regions within and after the putative coiled coil would in principle allow the GAP core to deactivate Rab1 associated with the LCV as well as Rab1 on proximal ER-derived vesicles. Moreover, the exceptional catalytic prowess of the LepB GAP core is consistent with the comparably high Rab1 exchange activity of DrrA as well as the picomolar binding affinity of LidA (21, 23, 44). It is likely that these remarkably high catalytic activities/binding affinities evolved to allow L. pneumophila to compete effectively with host factors for control of Rab1 on the LCV.
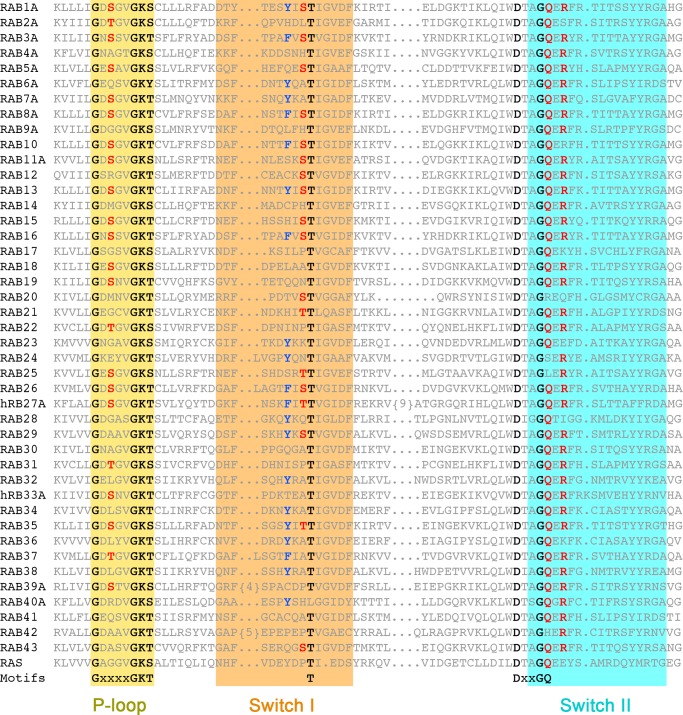
The exquisite specificity of LepB for Rab1 contrasts with the non-phylogenetic specificities of VirA and at least some TBC domains. By supplying both the catalytic arginine and glutamine in trans from orientations that avoid contact with residues in the GTPase (Fig. 7), the dual finger mechanism may be prone to promiscuity. Conversely, the non-canonical trans/cis polar network utilized by LepB is expected to enhance specificity, as the serines in the non-canonical polar network are only partially conserved (Fig. 8). Moreover, many Rab GTPases replace Tyr-40 with residues that may not be compatible with the tyrosine-shaped groove at the interface of the N- and C-terminal domains. Thus, the high specificity of LepB for Rab1 can be attributed to determinants directly related to the catalytic mechanism in addition to effector-like recognition determinants within or proximal to the interface with the switch/interswitch regions (45).
FIGURE 7.
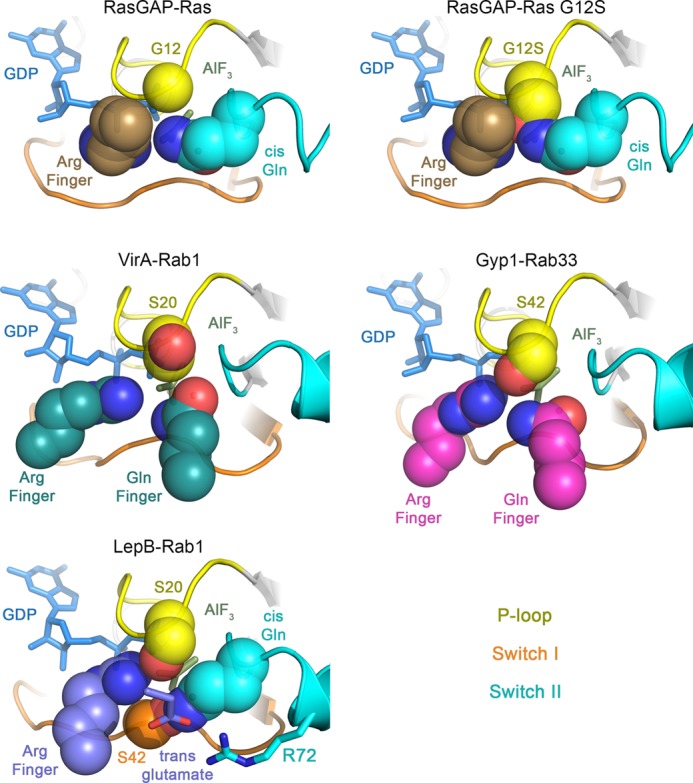
Comparison of hydrolytic site configurations in GAP-GTPase transition state mimetic complexes. Note the steric clash of the P-loop serine with the cis-Gln and arginine finger in the RasGAP complex with Ras G12S. The arginine and glutamine fingers in VirA and Gyp1 avoid conflict with the P-loop serine by inserting from compatible orientations. LepB avoids conflict by inserting the arginine finger from a compatible orientation and supplying a trans-glutamate to redirect the cis-glutamine into a non-canonical polar network involving Ser-20, Ser-42, and Arg-72.
FIGURE 8.
Annotated alignment of the P-loop and switch regions of Rab GTPases. Conserved residues in GTPase motifs are highlighted in black, residues in the non-canonical polar network are in red, and the switch I tyrosine is in blue.
The observation that the heretofore universal transition state stereochemistry of glutamine-dependent hydrolytic reactions can be fundamentally reconfigured expands current paradigms for both the intrinsic and GAP accelerated mechanisms. Moreover, the independent evolution of non-canonical trans/cis and dual trans finger mechanisms in structurally unrelated GAPs from widely divergent eukaryotic and prokaryotic organisms implies a selective pressure common to Rab GTPases but absent in GTPases with conventional cis-glutamine-mediated interactions. Intriguingly, the P-loop serine is invariably replaced by glycine in Ras, Rho family GTPases, and Gα subunits, which collectively define the hallmark cis-glutamine paradigm but rarely in Rab GTPases. Moreover, oncogenic mutations involving the corresponding Gly-12 in Ras strongly impair intrinsic as well as GAP accelerated hydrolysis due to steric conflicts with both the cis-glutamine and trans-arginine (46). The same fundamental conflict with the cis-glutamine applies to nearly all Rab GTPases (30). TBC domains and VirA circumvent this critical impediment by supplying dual trans fingers that insert in orientations compatible with the conventional transition state interactions, whereas LepB exploits the P-loop serine in the context of a non-canonical catalytic network (Fig. 7). These observations implicate substitution of the P-loop glycine as the primary selective pressure driving evolution of alternative mechanisms in Rab GAPs.
While this manuscript was under review, Yu et al. (39) reported a Rab specificity profile for LepB as well as the crystal structure of LepB from Legionella drancourtii in complex with Rab1-GDP and aluminum fluoride. The specificity profile and structure are similar to those described here.
Acknowledgments
We greatly appreciate the assistance of Alexei Soares and the NSLS X29/X12B beamline staff with MAD data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants GM056324 (to D. G. L.) and AI041699 (C. R. R.). This work was also supported by a Jane Coffin Childs postdoctoral fellowship (to R. E. C.).
The atomic coordinates and structure factors (code 4IRU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- LCV
- Legionella-containing vacuole
- ER
- endoplasmic reticulum
- GAP
- GTPase activating protein
- TBC
- Tre, Bub2, Cdc16
- SUMO
- small ubiquitin-like modifier
- PBP
- phosphate-binding protein
- MDCC
- 7-diethylamino-3-[N-(2-maleimidoethyl)carbamoyl]coumarin
- MAD
- multiwavelength anomalous diffraction.
REFERENCES
- 1. Rowbotham T. J. (1980) Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33, 1179–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Isberg R. R., O'Connor T. J., Heidtman M. (2009) The Legionella pneumophila replication vacuole. Making a cosy niche inside host cells. Nat. Rev. Microbiol. 7, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Al-Quadan T., Price C. T., Abu Kwaik Y. (2012) Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 20, 299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu W., Banga S., Tan Y., Zheng C., Stephenson R., Gately J., Luo Z. Q. (2011) Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE 6, e17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Horwitz M. A., Silverstein S. C. (1980) Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Invest. 66, 441–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kagan J. C., Roy C. R. (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4, 945–954 [DOI] [PubMed] [Google Scholar]
- 7. Derré I., Isberg R. R. (2004) Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 72, 3048–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kagan J. C., Stein M. P., Pypaert M., Roy C. R. (2004) Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J. Exp. Med. 199, 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Machner M. P., Isberg R. R. (2006) Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell 11, 47–56 [DOI] [PubMed] [Google Scholar]
- 10. Murata T., Delprato A., Ingmundson A., Toomre D. K., Lambright D. G., Roy C. R. (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat. Cell Biol. 8, 971–977 [DOI] [PubMed] [Google Scholar]
- 11. Brombacher E., Urwyler S., Ragaz C., Weber S. S., Kami K., Overduin M., Hilbi H. (2009) Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J. Biol. Chem. 284, 4846–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ingmundson A., Delprato A., Lambright D. G., Roy C. R. (2007) Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450, 365–369 [DOI] [PubMed] [Google Scholar]
- 13. Arasaki K., Roy C. R. (2010) Legionella pneumophila promotes functional interactions between plasma membrane syntaxins and Sec22b. Traffic 11, 587–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arasaki K., Toomre D. K., Roy C. R. (2012) The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe 11, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Müller M. P., Peters H., Blümer J., Blankenfeldt W., Goody R. S., Itzen A. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 [DOI] [PubMed] [Google Scholar]
- 16. Mukherjee S., Liu X., Arasaki K., McDonough J., Galán J. E., Roy C. R. (2011) Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477, 103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan Y., Luo Z. Q. (2011) Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475, 506–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goody P. R., Heller K., Oesterlin L. K., Müller M. P., Itzen A., Goody R. S. (2012) Reversible phosphocholination of Rab proteins by Legionella pneumophila effector proteins. EMBO J. 31, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Müller M. P., Shkumatov A. V., Oesterlin L. K., Schoebel S., Goody P. R., Goody R. S., Itzen A. (2012) Characterization of enzymes from Legionella pneumophila involved in reversible adenylylation of Rab1 protein. J. Biol. Chem. 287, 35036–35046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neunuebel M. R., Mohammadi S., Jarnik M., Machner M. P. (2012) Legionella pneumophila LidA affects nucleotide binding and activity of the host GTPase Rab1. J. Bacteriol. 194, 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schoebel S., Oesterlin L. K., Blankenfeldt W., Goody R. S., Itzen A. (2009) RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol. Cell 36, 1060–1072 [DOI] [PubMed] [Google Scholar]
- 22. Suh H. Y., Lee D. W., Lee K. H., Ku B., Choi S. J., Woo J. S., Kim Y. G., Oh B. H. (2010) Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J. 29, 496–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng W., Yin K., Lu D., Li B., Zhu D., Chen Y., Zhang H., Xu S., Chai J., Gu L. (2012) Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS Pathog. 8, e1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pan X., Eathiraj S., Munson M., Lambright D. G. (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442, 303–306 [DOI] [PubMed] [Google Scholar]
- 25. Dong N., Zhu Y., Lu Q., Hu L., Zheng Y., Shao F. (2012) Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell 150, 1029–1041 [DOI] [PubMed] [Google Scholar]
- 26. Mihai Gazdag E., Streller A., Haneburger I., Hilbi H., Vetter I. R., Goody R. S., Itzen A. (2013) Mechanism of Rab1b deactivation by the Legionella pneumophila GAP LepB. EMBO Rep. 14, 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gavriljuk K., Gazdag E. M., Itzen A., Kötting C., Goody R. S., Gerwert K. (2012) Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 109, 21348–21353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klähn M., Rosta E., Warshel A. (2006) On the mechanism of hydrolysis of phosphate monoesters dianions in solutions and proteins. J. Am. Chem. Soc. 128, 15310–15323 [DOI] [PubMed] [Google Scholar]
- 29. Zhu G., Liu J., Terzyan S., Zhai P., Li G., Zhang X. C. (2003) High resolution crystal structures of human Rab5a and five mutants with substitutions in the catalytically important phosphate-binding loop. J. Biol. Chem. 278, 2452–2460 [DOI] [PubMed] [Google Scholar]
- 30. Anand B., Majumdar S., Prakash B. (2013) Structural basis unifying diverse GTP hydrolysis mechanisms. Biochemistry 52, 1122–1130 [DOI] [PubMed] [Google Scholar]
- 31. Brune M., Hunter J. L., Corrie J. E., Webb M. R. (1994) Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry 33, 8262–8271 [DOI] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 35. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 39. Yu Q., Hu L., Yao Q., Zhu Y., Dong N., Wang D. C., Shao F. (2013) Structural analyses of Legionella LepB reveal a new GAP fold that catalytically mimics eukaryotic RasGAP. Cell Res. 23, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barr F., Lambright D. G. (2010) Rab GEFs and GAPs. Curr. Opin. Cell Biol. 22, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eathiraj S., Mishra A., Prekeris R., Lambright D. G. (2006) Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364, 121–135 [DOI] [PubMed] [Google Scholar]
- 42. Rudack T., Xia F., Schlitter J., Kötting C., Gerwert K. (2012) Ras and GTPase activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations. Proc. Natl. Acad. Sci. U.S.A. 109, 15295–15300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosnizeck I. C., Graf T., Spoerner M., Tränkle J., Filchtinski D., Herrmann C., Gremer L., Vetter I. R., Wittinghofer A., König B., Kalbitzer H. R. (2010) Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes. Angew Chem. Int. Ed. Engl. 49, 3830–3833 [DOI] [PubMed] [Google Scholar]
- 44. Schoebel S., Cichy A. L., Goody R. S., Itzen A. (2011) Protein LidA from Legionella is a Rab GTPase supereffector. Proc. Natl. Acad. Sci. U.S.A. 108, 17945–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee M. T., Mishra A., Lambright D. G. (2009) Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 10, 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) The Ras-RasGAP complex. Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 47. Deléage G., Blanchet C., Geourjon C. (1997) Protein structure prediction. Implications for the biologist. Biochimie 79, 681–686 [DOI] [PubMed] [Google Scholar]
- 48. Lupas A., Van Dyke M., Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 49. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 50. Ahmadian M. R., Hoffmann U., Goody R. S., Wittinghofer A. (1997) Individual rate constants for the interaction of Ras proteins with GTPase activating proteins determined by fluorescence spectroscopy. Biochemistry 36, 4535–4541 [DOI] [PubMed] [Google Scholar]