Background: The role of the MmpL11 transporter in mycobacteria is not understood.
Results: Mycobacterium smegmatis mmpL11 mutants accumulate mycolic acid precursors and fail to transport monomeromycolyl diacylglycerol and mycolate ester wax to the bacterial surface.
Conclusion: MmpL11 contributes to mycobacterial cell wall biosynthesis.
Significance: MmpL11 plays a conserved role in mycobacterial cell wall biogenesis that is important for M. tuberculosis virulence.
Keywords: Biofilm, Membrane lipids, Membrane proteins, Mycobacteria, Mycobacterium, MmpL, mycolic acid
Abstract
A growing body of evidence indicates that MmpL (mycobacterial membrane protein large) transporters are dedicated to cell wall biosynthesis and transport mycobacterial lipids. How MmpL transporters function and the identities of their substrates have not been fully elucidated. We report the characterization of Mycobacterium smegmatis MmpL11. We showed previously that M. smegmatis lacking MmpL11 has reduced membrane permeability that results in resistance to host antimicrobial peptides. We report herein the further characterization of the M. smegmatis mmpL11 mutant and identification of the MmpL11 substrates. We found that biofilm formation by the M. smegmatis mmpL11 mutant was distinct from that by wild-type M. smegmatis. Analysis of cell wall lipids revealed that the mmpL11 mutant failed to export the mycolic acid-containing lipids monomeromycolyl diacylglycerol and mycolate ester wax to the bacterial surface. In addition, analysis of total lipids indicated that the mycolic acid-containing precursor molecule mycolyl phospholipid accumulated in the mmpL11 mutant compared with wild-type mycobacteria. MmpL11 is encoded at a chromosomal locus that is conserved across pathogenic and nonpathogenic mycobacteria. Phenotypes of the M. smegmatis mmpL11 mutant are complemented by the expression of M. smegmatis or M. tuberculosis MmpL11, suggesting that MmpL11 plays a conserved role in mycobacterial cell wall biogenesis.
Introduction
Tuberculosis infections are one of the leading causes of death due to infectious disease. The World Health Organization estimates that Mycobacterium tuberculosis infects one-third of the world population, and 8 million new cases of tuberculosis are reported annually (1). The mycobacterial cell wall plays a crucial role in mycobacterial intrinsic resistance to external stresses and antibiotics. Cell wall lipids contribute to mycobacterial biofilm formation and have immunomodulatory properties that are essential to the infectious strategy of pathogenic mycobacteria. The composition and architecture of the mycobacterial cell wall are unique. The outer membrane contains an inner leaflet of very long chain mycolic acids, covalently bound to the arabinogalactan-peptidoglycan layer, and an outer leaflet composed of noncovalently associated lipids such as trehalose 6,6′-dimycolate (TDM),2 glycopeptidolipids (GPLs), phthiocerol dimycocerosate (PDIM), and sulfolipids (2, 3). Mycolic acids are β-hydroxyl fatty acids with an α-alkyl side chain and require the fatty acid synthase systems FAS-I and FAS-II and the polyketide synthase Pks13 for synthesis (4). The resulting mycolic acid is transferred as trehalose monomycolate (TMM) to the outer leaflet of the bacterium, where it is a precursor for mycolyl arabinogalactan and TDM. Although mycolic acid side chains differ in length and oxygenation between mycobacterial species (5), heterologous expression of core enzymes demonstrates that their biosynthesis is largely conserved between M. tuberculosis and the fast-growing nonpathogenic species M. smegmatis (6–8).
Microbial biofilms are defined as communities of microorganisms that range from surface-attached colonies to well developed pellicles formed at the air-liquid interface (9). Bacteria within biofilm communities are typically associated with a complex architecture of extracellular material that contains secreted molecules such as polysaccharides, lipids, proteins, and DNA. This matrix provides a physical barrier to environmental stresses and allows for the emergence of a drug-tolerant phenotype. As such, the ability to form biofilms is associated with virulence in a number of bacterial pathogens. Although the presence and role of biofilms during M. tuberculosis infections remain unclear, biofilm formation by M. smegmatis and other environmental species has been established. The extracellular matrix associated with mycobacterial biofilms is notably rich in lipids. Free mycolic acids are associated with the formation of biofilms by M. smegmatis and M. tuberculosis (10, 11). In M. smegmatis, free mycolic acids are liberated from TDM by the hydrolase MSMEG_1529 and are linked to drug resistance in vitro (10, 12). Mutants lacking MSMEG_1529 or that are defective in mycolic acid biosynthesis have impaired biofilm formation. In M. smegmatis and Mycobacterium avium, GPLs also contribute to biofilm formation (13, 14). Reduced biofilm formation has been demonstrated in M. smegmatis lsr2 mutants, and this phenotype was attributed to a lack of mycolyl diacylglycerol (15).
The mycobacterial cell wall is a rich area for research, and biochemical and genetic approaches are being employed to further elucidate the mechanism of cell wall biosynthesis. In mycobacteria, MmpL (mycobacterial membrane protein large) proteins appear to be dedicated to the export of cell wall lipid constituents. MmpL3 was recently demonstrated to be the transporter responsible for delivery of TMM to the mycobacterial surface (16). MmpL3 is predicted to be essential in M. tuberculosis, and its essentiality was demonstrated in M. smegmatis. This is perhaps unsurprising because TDM biosynthesis and incorporation into the mycobacterial cell wall are essential for mycobacterial viability (17, 18). M. smegmatis mutants lacking the MmpL4a and MmpL4b (originally named TmtpB and TmtpC) transporters do not have GPLs on their surface. These mutants, along with mutants lacking the accessory protein MmpS4, have altered colony morphology and reduced sliding motility and biofilm formation (13, 19). In M. tuberculosis, MmpL7 and MmpL8 play respective roles in PDIM and sulfolipid export to the outer leaflet of the cell wall and are required for full virulence in a mouse model (20–24). Although substrates for M. tuberculosis MmpL4, MmpL5, and MmpL11 have not yet been described, data suggest that they also contribute to M. tuberculosis virulence (22, 25). Two independent studies found that an M. tuberculosis mmpL11 mutant is attenuated in a mouse model of infection; however, neither group determined the substrate of MmpL11 or defined the mechanism by which their mutant was attenuated (22, 25).
Our efforts have focused on characterization of the MmpL11 transporter. We showed that the loss of MmpL11 reduces the membrane permeability of M. smegmatis (26). In this work, we demonstrate that the M. smegmatis mmpL11 mutants had differences in cell wall lipid composition compared with wild-type mycobacteria. Specifically, mutant bacteria were unable to transport monomeromycolyl diacylglycerol (MMDAG) and mycolate ester wax to the bacterial surface. In addition, the mycolic acid precursor molecule mycolyl phospholipid (MycPL) accumulated in the absence of MmpL11. MmpL11 is encoded at the MmpL3/MmpL11 chromosomal locus that is conserved across mycobacteria, including Mycobacterium leprae. Expression of the M. tuberculosis MmpL11 protein complemented the M. smegmatis mmpL11 mutant phenotypes. Our data are consistent with a model in which MmpL11 plays a conserved role in mycobacterial cell wall biology and can function across species.
EXPERIMENTAL PROCEDURES
Maintenance of Bacterial Cultures and Cells
M. smegmatis mc2155 was obtained from American Type Culture Collection. Mycobacterial strains were maintained in Middlebrook 7H9 liquid medium (Difco) or on Middlebrook 7H11 agar plates (Difco) supplemented with 10% albumin/dextrose/saline. When required, cultures were incubated with the antibiotics kanamycin (25 μg/ml) and hygromycin (75 μg/ml). Planktonic M. smegmatis strains were grown in 7H9 liquid medium containing oleic acid/albumin/dextrose/catalase supplements and the detergent Tween 80 with shaking.
For biofilm growth, M. smegmatis was grown in polystyrene Petri dishes at 30 °C in modified Sauton's medium without Tween 80. Sauton's medium contained 0.5 g/liter K2HPO4, 0.5 g/liter MgSO4, 4.0 g/liter l-asparagine, 0.05 g/liter ferric ammonium citrate, 4.76% glycerol, and 1.0 mg/liter ZnSO4 at a final pH of 7.0. Pellicle formation in borosilicate glass tubes was observed when M. smegmatis was grown in 7H9 medium without shaking at 37 °C for several days. To enumerate bacteria grown under biofilm-inducing conditions, Tween 80 was added to a final concentration of 0.5% to promote dissociation of biofilm-associated bacteria. The pellicle was mechanically disrupted, and the bacteria were completely dispersed by syringing through a tuberculin syringe. Serial dilutions were performed in PBS containing 0.1% Tween 80 and plated in Middlebrook 7H10 medium.
Mycobacterial Strains and Construction of Complementation Plasmids
The isolation of the M. smegmatis mmpL11 mutant GP02 was described previously (26). For complementation, the M. smegmatis mmpL11 gene was amplified using primers MSMmpL11NdeI (CATATGATGCGCTTGAGCAGCAC) and MSMmpL11H3 (AAGCTTCGCCTCCTCCAGCATTGC) and cloned into pVV16 (27) in frame with a C-terminal His6 affinity tag. For heterologous complementation with M. tuberculosis mmpL11, the gene was amplified from M. tuberculosis CDC1551 genomic DNA using primers RvMmpL11NdeI (ATCATATGATGCGCTTGAGCCGCAACCTG) and RvMmpL11H3 (ATAAGCTTCCTCGCCTCCTCCAACATCGC) and cloned into pVV16 in frame with a C-terminal His6 affinity tag. All plasmid constructs were sequence-verified.
Scanning Electron Microscopy
M. smegmatis biofilms were cultured in a 24-well plate as described above. At the time of harvest, the medium below the pellicle was removed with a tuberculin syringe and replaced with 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer at pH 7.4 (Electron Microscopy Sciences). A 5-mm polystyrene coverslip was used to capture a portion of the pellicle that had been in contact with the fixative. Samples were dried and subsequently gold-sputtered (PELCO 91000 sputter coater) and imaged using a Sirion XL30 scanning electron microscope (FEI, Portland, OR) at the Portland State University Center for Electron Microscopy and Nanofabrication.
Lipid Isolation and Analysis
To harvest total lipids, cultures were harvested by centrifugation and resuspended in chloroform/methanol (2:1, v/v). Following extraction, total lipids were dried under N2 gas. Apolar lipids were harvested as described (11). Briefly, total lipids were resuspended in 5 ml of methanol and 0.3% NaCl (10:1, v/v) and 2.5 ml of petroleum ether. Samples were rocked for 30 min at room temperature; after centrifugation, the upper layer containing the apolar lipids was retained and dried under N2 gas.
Surface lipids were extracted from M. smegmatis biofilms as described previously (20). Briefly, biofilms grown in polystyrene dishes were harvested, resuspended in 5 ml of hexane, and then sonicated for 5 min. After centrifugation at 3000 rpm for 5 min, the hexane-extracted lipids were dried under N2 gas.
The above total, apolar, and surface lipids were resuspended in chloroform/methanol (2:1, v/v), and equivalent amounts were spotted onto TLC plates. TDM, MycPL, and TMM were resolved by TLC in a chloroform/methanol/ammonium hydroxide (80:20:2, v/v/v) solvent system (28). Free mycolates were resolved by TLC in a chloroform/methanol/distilled H2O (90:10:1, v/v/v) solvent system. Meromycolyl diacylglycerol and triacylglycerol were resolved by TLC in a toluene/acetone (99:1, v/v) solvent system. The TLC plates were visualized by spraying with 5% molybdophosphoric acid in ethanol and charred. Lipids were also purified from silica plates (Analtech) using standard preparative TLC procedures.
Radiolabeled lipids and mycolyl arabinogalactan were isolated as described (29). Briefly, M. smegmatis strains were grown to mid-log phase and then radiolabeled with 1 μCi/ml [14C]sodium acetate ([1,2-14C]acetic acid sodium salt, specific activity of 100–120 mCi/mmol, American Radiolabeled Chemicals). At each time point indicated, the cells were pelleted, resuspended in chloroform/methanol (2:1, v/v), and sonicated. TDM, MycPL, and TMM were resolved by TLC using a chloroform/methanol/ammonium hydroxide (80:20:2) solvent system. Mycolyl arabinogalactan was isolated from the delipidated cells and analyzed as mycolic acid methyl esters (MAMEs) as described previously (22). The MAMEs were separated by TLC by running the plate five times using a petroleum ether/diethyl ether (9:1, v/v) solvent system. To detect radiolabeled lipids, TLC plates were exposed to a phosphor screen (GE Healthcare) and visualized using a STORM imager (GE Healthcare). Densitometry on the lipid profiles was performed using NIH ImageJ software. The mean intensities of TDM and mycolyl arabinogalactan were normalized to the total signal from each sample, and the means ± S.D. from three independent experiments were graphed. This normalization ensured that each sample was controlled internally rather than in comparison with other samples and compensated for the reduced radiolabel incorporation that we routinely observed for the mmpL11 mutant.
[14C]Acetate Uptake Assay
M. smegmatis strains were grown to mid-log phase and then radiolabeled with 1 μCi/ml [14C]sodium acetate. These conditions were chosen to mimic the conditions used for lipid incorporation. At the indicated time points, the cells were pelleted, washed twice with PBS, and counted in a liquid scintillation counter.
Mass Spectrometry
Both electrospray ionization (ESI) high-resolution (R = 100,000 at m/z 400) and low-energy collisionally activated dissociation tandem mass spectra were acquired with a Thermo Scientific LTQ Orbitrap Velos mass spectrometer with an Xcalibur operating system. Total lipid extracts were fractionated using a CHROMABOND Sep-Pak amino column (Macherey-Nagel, Duren, Germany) as described previously (15, 16). Fractions were loop-injected onto the ESI source, where the skimmer was set at ground potential, the electrospray needle was set at 4.5 kV, and the temperature of the heated capillary was 275 °C. The automatic gain control of the ion trap was set to 5 × 107, with a maximum injection time of 400 ms. Helium was used as the buffer with collision gas at a pressure of 1 × 10−3 millibars (0.75 millitorrs). The MSn experiments were carried out with a relative collision energy ranging from 30 to 40% and with an activation q value at 0.25. The activation time was set at 10 ms. Mass spectra were accumulated in the profile mode. The MALDI-TOF spectra of the total lipid and fractionated lipid extracts were acquired in the reflector mode using a Voyager-DE STR mass spectrometer (Applied Biosystems) equipped with a 337-nm nitrogen laser and delayed extraction. The final mass spectra were from an average of 5–10 spectra, in which each spectrum was a collection from 200 laser shots. 2,5-Dihydroxybenzoic acid was used as the matrix.
RESULTS
M. smegmatis Mutant mmpL11 Forms Altered Biofilms
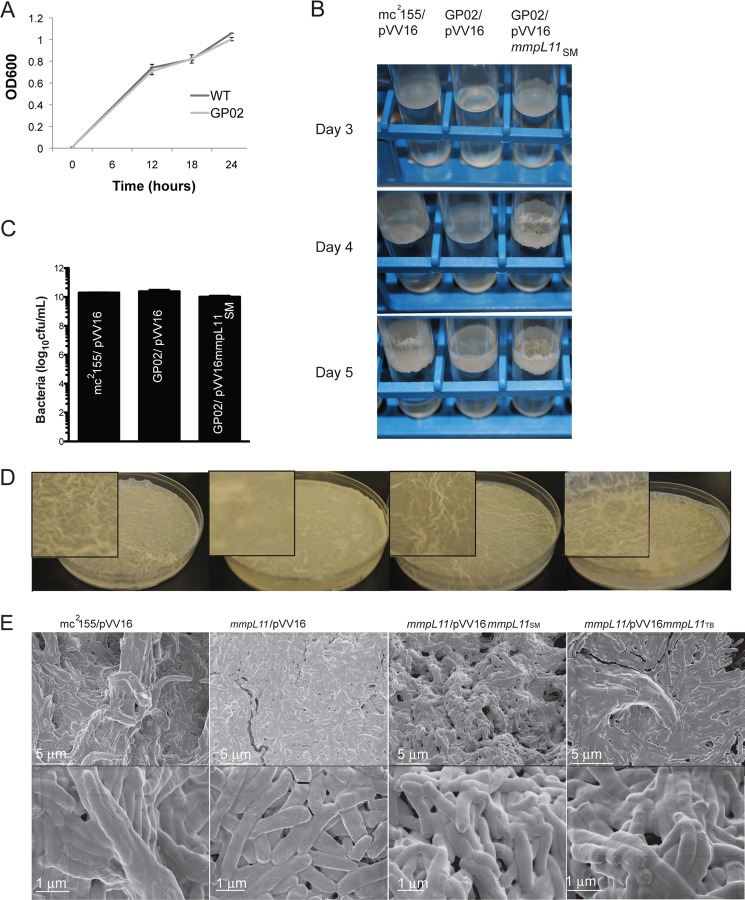
We demonstrated previously that the M. smegmatis mmpL11 transposon mutant GP02 is hyper-resistant to host antimicrobial peptides and has decreased membrane permeability relative to wild-type M. smegmatis (26). Our further characterization of the mutant indicated that there was no difference in the growth rate of wild-type M. smegmatis and GP02 grown planktonically in 7H9 medium containing the detergent Tween 80 to prevent clumping (Fig. 1A). However, we noted differences when the two strains were cultivated in standing culture in medium lacking Tween 80. Surfactants and detergents are known inhibitors of biofilm formation (30, 31). Wild-type M. smegmatis grown under biofilm-inducing conditions formed a pellicle or film at the air-liquid interface 3 days post-inoculation that developed further into a reticulated biofilm (Fig. 1B). The mmpL11 mutant GP02 had delayed pellicle formation relative to wild-type M. smegmatis upon standing culture in borosilicate glass tubes. At later time points, the mmpL11 mutant defect in biofilm formation was seen as a reduced ability of the pellicle to attach and extend upwards along the glass tube (Fig. 1B, middle and lower).
FIGURE 1.
The M. smegmatis mmpL11 mutant grows like the wild type in planktonic cultures but demonstrates impaired biofilm formation. A, growth of wild-type M. smegmatis and the mmpL11 mutant GP02 in complete 7H9 medium containing albumin/dextrose/saline supplements and 0.05% Tween 80 was followed over time. B, pellicle formation by the wild type (mc2155/pVV16), the mmpL11 mutant (GP02/pVV16), and the complemented mutant (GP02/pVV16mmpL11SM). Bacteria were subcultured in glass tubes and allowed to grow at 37 °C for 5 days without shaking. At day 3, a thin film could be observed across the surface of the medium in the wild-type and complemented strains (left). This developed into a textured biofilm over the next 2 days. Attachment of bacteria to the glass tube and growth away from the surface could be observed from the side (right). C, bacteria in pellicle cultures depicted in A were enumerated. Biofilms were disrupted mechanically via syringing, and serial dilutions were plated. The mean ± S.D. of three independent experiments is shown. There was no significant difference in cfu/ml between strains. D, biofilm formation by the wild type (mc2155/pVV16), the mmpL11 mutant (GP02/pVV16), the complemented mutant (GP02/pVV16mmpL11SM), and the heterologously complemented strain (GP02/pVV16mmpL11TB). Equivalent numbers of bacteria were subcultured in liquid biofilm medium (Sauton's medium lacking Tween 80). Plates were incubated for 4 days at 30 °C. E, scanning electron microscopy of biofilms formed by wild-type, mmpL11 mutant, and complemented strains.
The defect in biofilm formation is better appreciated when cultured under biofilm-inducing conditions in polystyrene plates. The mmpL11 mutant GP02 formed a less textured, or reticulated, biofilm compared with wild-type and complemented strains (Fig. 1D). We also noted that the pellicle formed by the mmpL11 mutant was easily disturbed upon handling either the standing cultures in glass tubes or polystyrene plates. In contrast, the pellicle formed by wild-type or complemented strains remained intact.
To ensure that reduced biofilm formation did not result from reduced bacterial numbers in the mutant biofilm cultures, biofilms were mechanically disrupted, and the number of bacteria were determined by plating serial dilutions. This enumeration, which included planktonic bacteria at the bottom of the culture tube and biofilm-associated bacteria, indicated that there was no significant difference in the number of bacteria present in each culture (Fig. 1C). Therefore, replication of the mmpL11 mutant is not impaired in 7H9 medium lacking Tween 80.
The GP02/pVV16SM complemented mutant formed a robust biofilm that initiated sooner than compared with the wild type (Fig. 1B, upper). We believe that this phenotype likely resulted from our complementation strategy, in which mmpL11 is expressed constitutively from the hsp60 promoter of the pVV16 plasmid. Quantification of mmpL11 expression by quantititavie RT-PCR indicated that there was 8-fold more mmpL11 transcript in the GP02/pVV16SM complemented strain relative to wild-type M. smegmatis (mc2155/pVV16) (data not shown).
Scanning electron microscopy analysis of M. smegmatis biofilms revealed reduced extracellular material surrounding the mmpL11 mutant bacteria compared with the wild-type and complemented strains (Fig. 1E). Consistent with the inability of GP02 to form a textured biofilm, the mmpL11 mutant samples appeared flatter and less structured than wild-type biofilms. At the higher magnification, distinct mmpL11 mutant bacteria were discernible, whereas the wild-type and complemented bacteria were enveloped in extracellular material and closely associated with one another.
Development of mycobacterial biofilms is promoted in the presence of iron and copper (31, 32). At least 1 μm ferrous sulfate is required for M. smegmatis biofilm formation, and our standard biofilm culture medium contains 178 μm iron. Addition of 100 μm copper sulfate to the biofilm culture medium did not restore wild-type biofilm formation of the mmpL11 mutant (data not shown). These results indicate that the inability of the mmpL11 mutant to form biofilms is not a result of metal ion deprivation.
MmpL11 Transporter Function Is Likely Conserved across Nonpathogenic and Pathogenic Mycobacteria
MmpL11 is present in a conserved chromosomal locus in nonpathogenic and pathogenic mycobacteria, and the M. smegmatis protein MmpL11 shares 69% identity with the M. tuberculosis protein MmpL11. To determine whether MmpL11 function is conserved among mycobacteria, we assessed whether the M. tuberculosis mmpL11 gene could complement the M. smegmatis mmpL11 mutant. M. tuberculosis MmpL11 restored both textured biofilm formation (Fig. 1D) and levels of extracellular matrix material to those of the wild type (Fig. 1E), consistent with conserved MmpL11 function.
The Cell Wall Lipid Composition Is Altered in the M. smegmatis mmpL11 Mutant
The reduced cell wall permeability and impaired biofilm formation suggested that the mmpL11 mutant had altered cell wall lipid composition compared with wild-type M. smegmatis. GPLs and free mycolic acids are required for M. smegmatis biofilm formation (10, 11, 13, 33). Impaired biofilm formation by the mmpL11 mutant GP02 suggested that these surface-associated lipids might be absent. Therefore, lipids from biofilm-grown cultures of M. smegmatis mc2155 and the mmpL11 mutant GP02 were examined by TLC. GPLs were present in total lipid samples extracted from the wild type and the mmpL11 mutant (Fig. 2A). To examine free mycolates, apolar lipids from biofilm cultures were examined as described previously (11). No significant differences were observed in the presence of free mycolates between the wild-type, mmpL11 mutant, and complemented strains (Fig. 2B). MS analysis also confirmed that there were no differences in the composition of GPLs or free mycolates (supplemental Figs. S1 and S2). To identify other potential differences in lipid composition, total and surface-exposed lipids were extracted and analyzed by TLC using a range of solvent polarities. We noted the absence of apolar lipids A–C in the hexane-extracted lipids of the mmpL11 mutant compared with wild-type M. smegmatis and the complemented strain using the toluene/acetone (99:1, v/v) solvent system (Fig. 2C).
FIGURE 2.
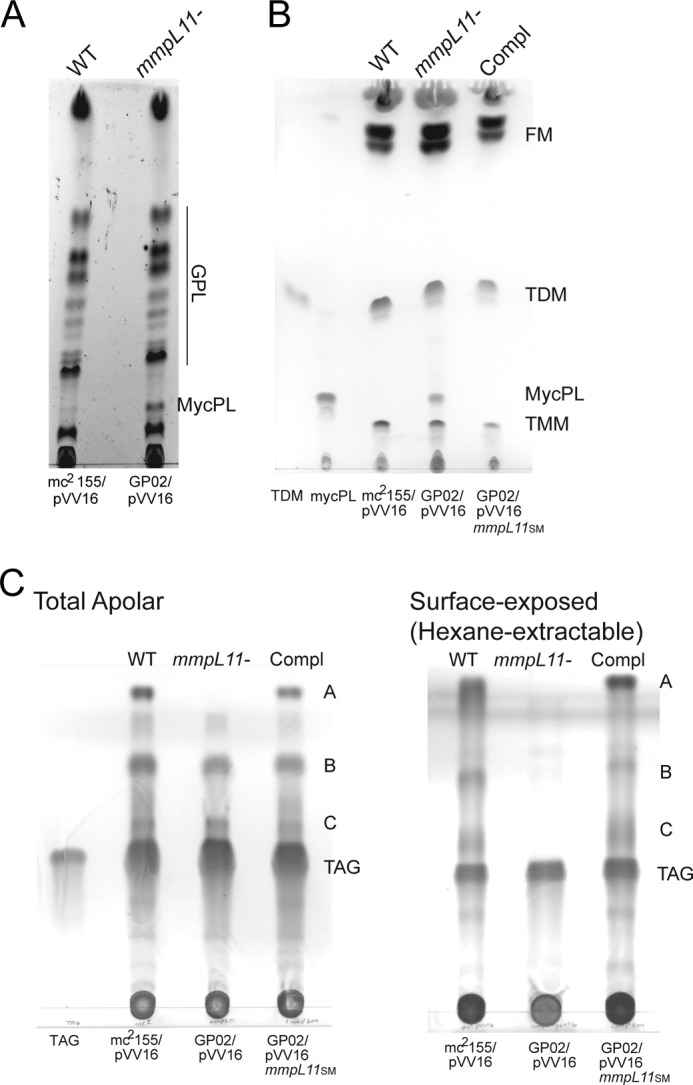
The M. smegmatis mmpL11 mutant has altered surface-exposed lipids. A, GPLs were extracted from M. smegmatis isolates and resolved by TLC in chloroform/methanol/distilled H2O (90:10:1, v/v/v). TLC plates were sprayed with ethanolic sulfuric acid and charred to visualize lipids. B, to visualize free mycolates (FM), apolar lipids were harvested from M. smegmatis biofilms and then resolved by TLC in chloroform/methanol/distilled H2O (90:10:1, v/v/v). The TDM standard (Sigma) and a preparative TLC-purified MycPL are indicated. C, total apolar lipids (left) and hexane-extracted surface lipids (right) were resolved by TLC in toluene/acetone (99:1, v/v). Triacylglycerol (TAG) is indicated. TLC plates in B and C were sprayed with molybdophosphoric acid and charred to visualize lipids. Compl, complemented strain.
Following preparative TLC purification of these lipids, structural analysis with high-resolution multiple-stage MS identified lipids B and C as mycolate ester wax and MMDAG, respectively (Fig. 3 and supplemental Figs. S3 and S4) (34). However, we were unable to obtain structural information on the apolar lipid that ran at the solvent front (lipid A) using ESI/MS, MALDI-TOF-MS, or GC/MS. The data presented in Fig. 2C support a model in which MmpL11 transports MMDAG and mycolate ester wax. In the absence of MmpL11, MMDAG and mycolate ester wax were synthesized but were not transported to the surface, where they would be readily extracted by hexane treatment. Instead, these lipids were retained in the bacterium and were present only in the total lipid extract.
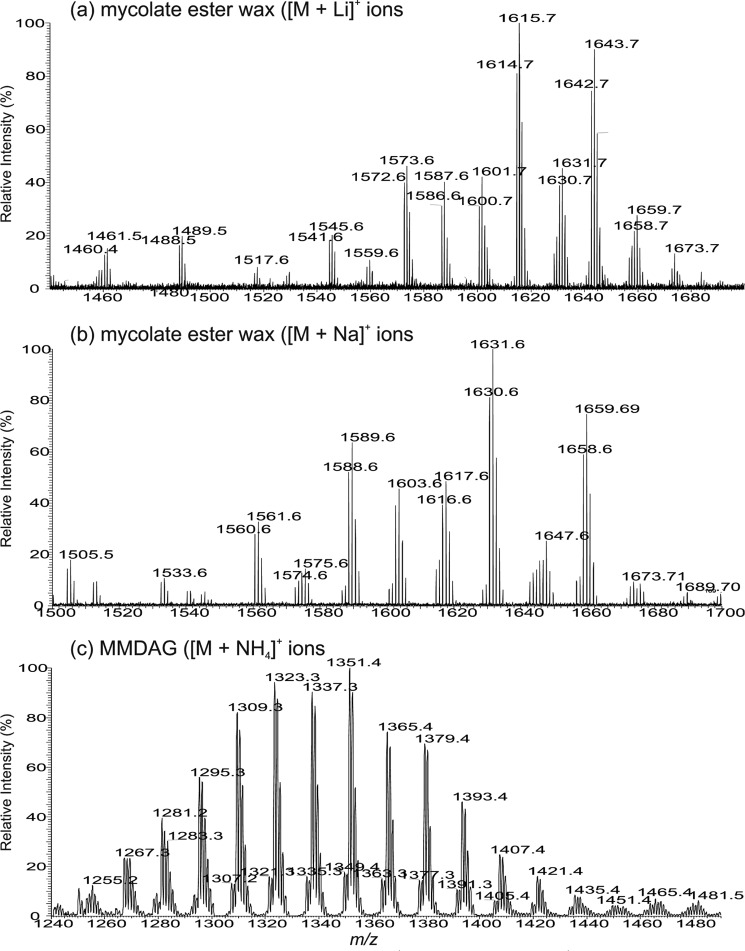
FIGURE 3.
The ESI high-resolution mass spectra of MMDAG and mycolate ester wax. Shown are the ESI high-resolution mass spectra of the [M + Li]+ ions (a) and the [M + Na]+ ions (b) of apolar lipid B, which represents mycolate ester wax, and the ESI mass spectrum of the [M + NH4]+ ions of apolar lipid C (c), which represents MMDAG. The structures of apolar lipid B are pentatriacontatrienyl mycolate esters, the structure of which was characterized by multiple-stage MS as [M + Li]+ ions; the corresponding [M + Na]+ ions (a) were reported previously by Chen et al. (15), but were misassigned as mycolyl diacylglycerol.
In addition, closer examination of the TLC analysis depicted in Fig. 2 (A and B) revealed a relative accumulation of MycPL in the mmpL11 mutant compared with the wild-type and complemented strains. Besra et al. (28) identified MycPL as a mycolic acid-containing precursor molecule in M. smegmatis and demonstrated that the mycolic acid from MycPL incorporates into TMM and TDM using in vitro assays with purified cell extracts. The identity of MycPL was confirmed by MS analysis following preparative TLC purification. On the basis of our data, we suggest a working model in which MmpL11 is the dedicated transporter for MMDAG and mycolate ester wax. Furthermore, the accumulation of MycPL when MmpL11 is absent suggests that this mycolic acid-containing precursor molecule is also a biosynthetic intermediate for MMDAG and mycolate ester wax.
We also compared the lipid profiles of wild-type and mmpL11 mutant strains grown planktonically. When mycolic acid-containing species were examined, we observed a relative accumulation of MycPL in the mutant compared with the wild type (Fig. 4A). We quantified the relative levels of TMM, MycPL, and TDM at four time points of bacterial growth. The relative levels of MycPL were consistently higher in mid-logarithmic and late-logarithmic cultures of the mmpL11 mutant relative to the wild type (supplemental Fig. S5). In contrast, we did not observe significant levels of MMDAG or mycolate ester wax in lipids extracted from planktonic bacteria (Fig. 4B). These results are consistent with the biosynthesis and/or accumulation of these lipid classes in late stationary phase or dormancy-like conditions (35, 36).
FIGURE 4.

The M. smegmatis mmpL11 mutant accumulates MycPL relative to the wild type during planktonic growth. A, total lipids were extracted from mid-logarithmic phase planktonic cultures of the wild type (mc2155/pVV16) and mmpL11 mutant (GP02/pVV16) and then resolved by TLC in chloroform/methanol/ammonium hydroxide (80:20:2, v/v/v). TLC plates were sprayed with ethanolic sulfuric acid and charred to visualize lipids. B, total lipids were extracted from planktonic cultures at the indicated A600 and resolved by TLC in toluene/acetone (99:1, v/v). Triacylglycerol (TAG) is indicated. TLC plates were sprayed with molybdophosphoric acid and charred to visualize lipids (left) or exposed to a phosphor screen and visualized using a STORM imager (right).
Results from MS analysis of Sep-Pak column-fractionated lipids from wild-type mc2155/pVV16, GP02/pVV16, and GP02/pVV16mmpL11 using MALDI-TOF-MS or ESI/MS analysis indicated that TDM, TMM, GPLs, phosphatidylinositol mannosides, cardiolipin, phospholipids, and MycPL were present in the mmpL11 mutant at levels comparable with those in wild-type bacteria. MS analysis also indicated that there were no significant changes in the lipid profiles and compositions between the mmpL11 mutant and wild type (supplemental Figs. S1 and S2).
The Absence of MmpL11 Does Not Impact TMM Transport
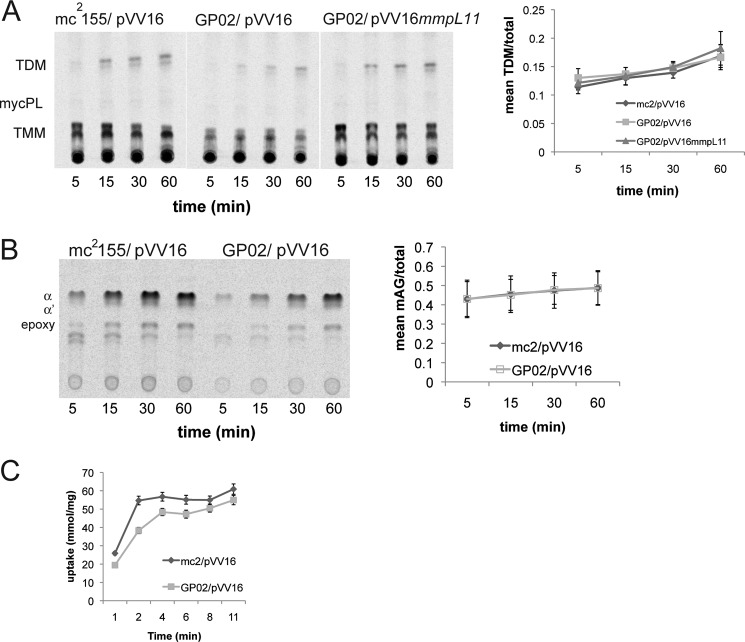
MmpL11 and MmpL3 appear to be closely related phylogenetically (22), and they share 26% amino acid identity in M. smegmatis. Our data support a model in which MmpL11 is the dedicated transporter for MMDAG and mycolate ester wax, whereas work by Jackson and co-workers (16) demonstrates that MmpL3 transports TMM to the bacterial surface. MycPL is likely an intermediate in the biosynthesis of all these mycolic acid-containing lipid substrates. Relative accumulation of the mycolic acid precursor molecule MycPL and TMM in GP02 raised the possibility that TMM transport was impacted in the absence of MmpL11. To rule out reduced TMM export by the mmpL11 mutant, we used [14C]acetate to selectively label mycolic acid-containing lipids. Once TMM is present outside the plasma membrane, it is a substrate for the mycolyltransferases of the Ag85 complex, which transfers one mycolate residue from TMM either to another molecule of TMM, yielding TDM, or to arabinogalactan, yielding mycolyl arabinogalactan. Therefore, TDM or mycolyl arabinogalactan biosynthesis can be used to assess external TMM levels. We pulsed logarithmic phase cultures of wild-type M. smegmatis and the mmpL11 mutant GP02 with [14C]acetate for 5, 15, 30, and 60 min to label mycolic acids. To assess the rate of TDM biosynthesis, we analyzed total extractable lipids by TLC. The rate of TDM biosynthesis by the mmpL11 mutant GP02 was not significantly different from that by wild-type mc2155 (Fig. 5A). This result is in contrast to the reduced TDM biosynthesis observed upon decreased MmpL3 levels (16). Covalently bound cell wall mycolyl arabinogalactan was also isolated, and the mycolic acid side chains were released as MAMEs. Quantification of MAMEs in the wild type and mmpL11 mutant indicated that there was no difference in the rate of mycolyl arabinogalactan biosynthesis or mycolic acid composition (Fig. 5B).
FIGURE 5.
The rates of TDM and mycolyl arabinogalactan biosynthesis in the M. smegmatis mmpL11 mutant are similar to those in the wild type. A, logarithmic phase cultures of wild-type M. smegmatis (mc2155/pVV16), the mmpL11 mutant (GP02/pVV16), and the complemented mutant (GP02/pVV16mmpL11) were pulsed with [14C]acetate for 5, 15, 30, and 60 min to label mycolic acids. A. To examine TDM levels, lipids were extracted and resolved by TLC in chloroform/methanol/ammonium hydroxide (80:20:2, v/v/v). A representative image is shown. Densitometry was performed to determine the intensity of the TDM spots, and the mean ± S.D. of three independent experiments was plotted over time. B, MAMEs of mycolyl arabinogalactan (mAG) were isolated from wild-type M. smegmatis (mc2155/pVV16) and the mmpL11 mutant (GP02/pVV16) and then resolved by TLC in petroleum ether/diethyl ether (9:1, v/v; 5 times). α, α′, and epoxy refer to the three mycolic acid forms present in M. smegmatis. A representative image is shown. Densitometry was performed to determine the intensity of mycolyl arabinogalactan, and the mean ± S.D. of three independent experiments was plotted over time. C, uptake of acetate by wild-type M. smegmatis (mc2155/pVV16) and the mmpL11 mutant (GP02/pVV16) at 37 °C. Analysis of the initial time points indicated uptake rates of 25.9 and 19.4 mmol/mg/min, respectively, with correlation coefficients of 1.0.
Our data suggest that TMM transport, measured as biosynthesis of the cell wall components TDM and mycolyl arabinogalactan, is not greatly impacted by the absence of MmpL11. These results are consistent with the primary function of MmpL3 as the TMM transporter (16) and that of MmpL11 as the MMDAG and mycolate ester wax transporter.
In the course of our radiolabel experiments, we routinely observed reduced incorporation of radiolabel by the mmpL11 mutant GP02. This is consistent with our previous observation obtained using an ethidium bromide uptake assay that the reduced membrane permeability of the mmpL11 mutant is reduced compared with that of the wild type (26). We examined the uptake kinetics of [14C]acetate by intact cells and found that the rate of acetate uptake by the mmpL11 mutant was reduced by 25% relative to the wild type (Fig. 5C). These data provide further evidence that the presence of MmpL11 impacts the cell wall permeability of M. smegmatis.
DISCUSSION
In this work, we have further characterized the role of MmpL11 in M. smegmatis and demonstrated that it contributes to the biogenesis of the mycobacterial cell wall and biofilm formation. In addition, heterologous complementation of the M. smegmatis mmpL11 mutant with the M. tuberculosis mmpL11 gene suggests that the function of MmpL11 transporters in cell wall biosynthesis is conserved between nonpathogenic and pathogenic mycobacteria. On the basis of the observation that the M. smegmatis mmpL11 mutant had altered cell wall permeability and biofilm formation, we hypothesized that there were differences in cell wall lipid composition. Indeed, when cell wall lipids were examined by TLC, we observed that the M. smegmatis mmpL11 mutant accumulated MycPL and lacked MMDAG and mycolate ester wax in the surface lipid profiles, indicating that the reduced biofilm formation is likely attributable to the absence of one or both of these lipid families on the cell wall surface. Biofilm formation in mycobacteria is multifactorial, and the roles of GPLs and free mycolates in biofilm formation by M. smegmatis have been demonstrated (10, 11, 13, 33). Although the role of extracellular lipids in mycobacterial biofilm formation has been established, a recent genetic screen demonstrated that M. tuberculosis biofilm formation also requires a number of additional factors that are not limited to cell wall lipid biosynthesis or transport (37). It is unknown whether homologs of these proteins also play a role in M. smegmatis biofilm formation.
Our finding that the reduced biofilm formation of the M. smegmatis mmpL11 mutant is associated with the absence of mycolate ester wax and MMDAG is consistent with an earlier report that the absence of apolar lipids in an M. smegmatis lsr2 mutant reduces biofilm formation (15). However, our thorough structural analysis of the apolar lipid, lipid species B (Fig. 2C), using high-resolution ESI with multiple-stage MS on the similar ions previously identified as mycolyl diacylglycerol confirmed that the lipid actually belongs to the mycolate ester wax family and consists of a pentatriacontatrienol (35:3) linked to the mycolic acid via an ester bond to form a pentatriacontatrienyl mycolate ester (Fig. 3). Interestingly, the high-resolution ESI mass spectrum profile of this new mycolate ester wax (Fig. 3) is nearly identical to that of the mycolic acid (supplemental Figs. S1 and S2, panel f) isolated from M. smegmatis mc2155, consistent with the notion that the apolar lipids represent mycolate esters. The presence of mycolic acid substituents in the apolar lipids (spots 1 and 2 in Fig. 4 of Chen et al. (15)) was confirmed using alkaline deacylation of the purified lipids, followed by methylation and TLC analysis of fatty acid methyl ester products. The presence of mycolic acid substituents is therefore in agreement with the identification of mycolate ester wax in our analysis.
Understanding the molecular mechanisms underlying the biogenesis of the mycobacterial cell wall not only elucidates the basic biology of pathogenic mycobacteria but also identifies potential targets for antimicrobials. Studies on mycolic acids have been particularly insightful, as these lipids are essential to mycobacterial viability and contribute to M. tuberculosis pathogenicity. In addition, mycolic acid biosynthesis is a target of important anti-tuberculosis drugs. The small molecule inhibitors AU1235, BM212, and SQ109, which target MmpL3 function, were identified recently as potential novel therapeutics for mycobacteria (16, 38, 39). MmpL3 is essential, and block of the TMM transporter MmpL3 results in reduced synthesis of TDM and mycolyl arabinogalactan (16). MmpL3 plays a crucial role in mycobacterial cell wall biosynthesis by transporting TMM to the outer membrane, and our data strongly support a role for MmpL11 in cell wall biosynthesis, specifically in the transport of the mycolic acid-containing glycoconjugate MMDAG and a ester wax mycolate. That we did not see differences in the rate of TDM or mycolyl arabinogalactan biosynthesis indicates that the MMDAG and mycolate ester wax are not intermediates in the biosynthesis of either of these lipids. However, at this time, we cannot rule out that MMDAG and mycolate ester wax are intermediates in another synthesis pathway. Our model for the roles of MmpL3 and MmpL11 is depicted in Fig. 6. MmpL3 and MmpL11 are closely related phylogenetically and share 25% protein identity in M. tuberculosis. Therefore, it is compelling that both proteins appear to transport lipids with mycolic acid functional groups.
FIGURE 6.
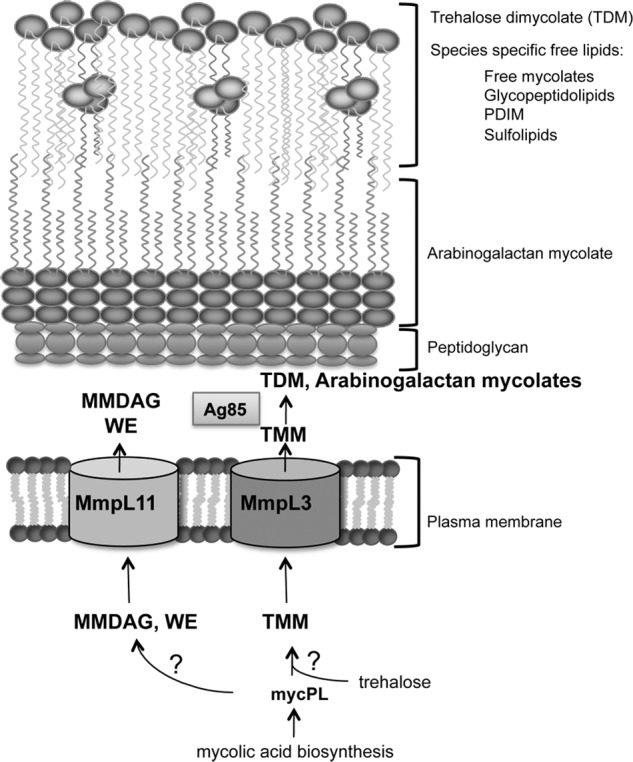
Model depicting MmpL3 and MmpL11 function in mycobacterial cell wall biogenesis. Data from our group and others support a model in which MmpL3 and MmpL11 are conserved mycobacterial cell wall lipid transporters. Mycolic acid biosynthesis yields the intermediate MycPL. The mycolic acid group of MycPL incorporates into TMM, which is transported by MmpL3, and into MMDAG and mycolate ester wax (WE), which are transported by MmpL11. The enzymes responsible for the biosynthesis of these lipid species from MycPL remain unknown.
We also noted the absence of an apolar lipid (lipid A) in both the surface and total lipids extracted from the mmpL11 mutant GP02 (Fig. 2). The identity of this lipid is also of interest to us. Experiments to define the structure of this lipid and to delineate its biosynthetic pathway are in progress.
A recent report attributed heme transport to MmpL3 and MmpL11, and the genetic locus that encodes MmpL3 and MmpL11 also encodes a protein with heme-binding activity (40). Although transport of heme or heme complexes may be one function of these transporters, our data and those of others suggest that MmpL3 and MmpL11 are lipid exporters with a primary function in cell wall biosynthesis. Reduced uptake of heme or heme complexes by the mmpL11 mutant is consistent with the changes in cell wall physiology that we described here and in previous work. The mmpL11 mutant possesses reduced membrane permeability compared with the wild type using an ethidium bromide uptake assay (26). Our present study indicates reduced uptake and incorporation of radiolabeled acetate by the mmpL11 mutant compared with wild-type bacteria (Fig. 5C). Reduced membrane permeability must be taken into consideration because this phenotype likely impacts the overall ability of small molecules including nutrients to access the bacterial cytoplasm. Similarly, M. smegmatis mspA mutants that lack the major porin protein had reduced membrane permeability as measured by uptake of ethidium bromide, radiolabeled glucose, and antibiotics. Strains lacking MspA alone or in combination with other porin proteins grow slower in both rich and minimal media (41, 42).
Further investigation of MmpL transporter function will expand our understanding of how this class of proteins contributes to mycobacterial cell wall biosynthesis. It is proposed that MmpL proteins function in complex with biosynthetic enzymes to promote coordinated synthesis and transport of lipid substrates. For example, M. tuberculosis mmpL7 mutants accumulate PDIM intracellularly, and MmpL7 directly interacts with the polyketide synthase PpsE, which is involved in PDIM biosynthesis (21, 24, 43). If this model holds true for MmpL11, we predict that MmpL11 interacts with the enzymes responsible for catalyzing the formation of MMDAG and mycolate ester wax from MycPL and other substrates. Our current efforts are focused on identifying these biosynthetic enzymes and testing the coupled biosynthesis/transport model.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Awards AI079399 and AI087840 (to G. E. P.). The Washington University Mass Spectrometry Facility is supported by National Institutes of Health Grants P41-GM103422, P60-DK20579, P30-DK56341, and P01-HL57278.

This article contains supplemental Figs. S1–S5.
- TDM
- trehalose 6,6′-dimycolate
- GPL
- glycopeptidolipid
- PDIM
- phthiocerol dimycocerosate
- TMM
- trehalose monomycolate
- MMDAG
- monomeromycolyl diacylglycerol
- MycPL
- mycolyl phospholipid
- MAME
- mycolic acid methyl ester
- ESI
- electrospray ionization.
REFERENCES
- 1. World Health Organization (2010) Tuberculosis, Fact Sheet 104, World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Brennan P. J., Nikaido H. (1995) The envelope of mycobacteria. Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann C., Leis A., Niederweis M., Plitzko J. M., Engelhardt H. (2008) Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takayama K., Wang C., Besra G. S. (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18, 81–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Minnikin D. E., Minnikin S. M., Parlett J. H., Goodfellow M., Magnusson M. (1984) Mycolic acid patterns of some species of Mycobacterium. Arch. Microbiol. 139, 225–231 [DOI] [PubMed] [Google Scholar]
- 6. Bhatt A., Brown A. K., Singh A., Minnikin D. E., Besra G. S. (2008) Loss of a mycobacterial gene encoding a reductase leads to an altered cell wall containing β-oxo-mycolic acid analogs and accumulation of ketones. Chem. Biol. 15, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown A. K., Bhatt A., Singh A., Saparia E., Evans A. F., Besra G. S. (2007) Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153, 4166–4173 [DOI] [PubMed] [Google Scholar]
- 8. Parish T., Roberts G., Laval F., Schaeffer M., Daffé M., Duncan K. (2007) Functional complementation of the essential gene fabG1 of Mycobacterium tuberculosis by Mycobacterium smegmatis fabG but not Escherichia coli fabG. J. Bacteriol. 189, 3721–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Toole G., Kaplan H. B., Kolter R. (2000) Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 [DOI] [PubMed] [Google Scholar]
- 10. Ojha A., Anand M., Bhatt A., Kremer L., Jacobs W. R., Jr., Hatfull G. F. (2005) GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123, 861–873 [DOI] [PubMed] [Google Scholar]
- 11. Ojha A. K., Baughn A. D., Sambandan D., Hsu T., Trivelli X., Guerardel Y., Alahari A., Kremer L., Jacobs W. R., Jr., Hatfull G. F. (2008) Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69, 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ojha A. K., Trivelli X., Guerardel Y., Kremer L., Hatfull G. F. (2010) Enzymatic hydrolysis of trehalose dimycolate releases free mycolic acids during mycobacterial growth in biofilms. J. Biol. Chem. 285, 17380–17389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Recht J., Martínez A., Torello S., Kolter R. (2000) Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182, 4348–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamazaki Y., Danelishvili L., Wu M., Macnab M., Bermudez L. E. (2006) Mycobacterium avium genes associated with the ability to form a biofilm. Appl. Environ. Microbiol. 72, 819–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J. M., German G. J., Alexander D. C., Ren H., Tan T., Liu J. (2006) Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J. Bacteriol. 188, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grzegorzewicz A. E., Pham H., Gundi V. A. K. B., Scherman M. S., North E. J., Hess T., Jones V., Gruppo V., Born S. E. M., Korduláková J., Chavadi S. S., Morisseau C., Lenaerts A. J., Lee R. E., McNeil M. R., Jackson M. (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 18. Bhatt A., Kremer L., Dai A. Z., Sacchettini J. C., Jacobs W. R. (2005) Conditional depletion of KasA, a key enzyme of mycolic acid biosynthesis, leads to mycobacterial cell lysis. J. Bacteriol. 187, 7596–7606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deshayes C., Bach H., Euphrasie D., Attarian R., Coureuil M., Sougakoff W., Laval F., Av-Gay Y., Daffé M., Etienne G., Reyrat J.-M. (2010) MmpS4 promotes glycopeptidolipid biosynthesis and export in Mycobacterium smegmatis. Mol. Microbiol. 78, 989–1003 [DOI] [PubMed] [Google Scholar]
- 20. Converse S. E., Mougous J. D., Leavell M. D., Leary J. A., Bertozzi C. R., Cox J. S. (2003) MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. U.S.A. 100, 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox J. S., Chen B., McNeil M., Jacobs W. R., Jr. (1999) Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 [DOI] [PubMed] [Google Scholar]
- 22. Domenech P., Reed M. B., Barry C. E. (2005) Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Domenech P. (2004) The role of MmpL8 in sulfatide biogenesis and virulence of Mycobacterium tuberculosis. J. Biol. Chem. 279, 21257–21265 [DOI] [PubMed] [Google Scholar]
- 24. Jain M., Cox J. S. (2005) Interaction between polyketide synthase and transporter suggests coupled synthesis and export of virulence lipid in M. tuberculosis. PLoS Pathog. 1, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lamichhane G., Tyagi S., Bishai W. R. (2005) Designer arrays for defined mutant analysis to detect genes essential for survival of Mycobacterium tuberculosis in mouse lungs. Infect. Immun. 73, 2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Purdy G. E., Niederweis M., Russell D. G. (2009) Decreased outer membrane permeability protects mycobacteria from killing by ubiquitin-derived peptides. Mol. Microbiol. 73, 844–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jackson M., Crick D. C., Brennan P. J. (2000) Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 275, 30092–30099 [DOI] [PubMed] [Google Scholar]
- 28. Besra G. S., Sievert T., Lee R. E., Slayden R. A., Brennan P. J., Takayama K. (1994) Identification of the apparent carrier in mycolic acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 91, 12735–12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mikusová K., Slayden R. A., Besra G. S., Brennan P. J. (1995) Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob. Agents Chemother. 39, 2484–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mireles J. R., 2nd, Toguchi A., Harshey R. M. (2001) Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183, 5848–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen K. T., Piastro K., Gray T. A., Derbyshire K. M. (2010) Mycobacterial biofilms facilitate horizontal DNA transfer between strains of Mycobacterium smegmatis. J. Bacteriol. 192, 5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ojha A., Hatfull G. F. (2007) The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth. Mol. Microbiol. 66, 468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Recht J., Kolter R. (2001) Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183, 5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Purdy G. E., Pacheco S., Turk J., Hsu F. F. (2013) Characterization of mycobacterial triacylglycerols and monomeromycolyl diacylglycerols from Mycobacterium smegmatis biofilm by electrospray ionization multiple-stage and high-resolution mass spectrometry. Anal. Bioanal. Chem. 10.1007/s00216-013-7179-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kremer L., de Chastellier C., Dobson G., Gibson K. J. C., Bifani P., Balor S., Gorvel J.-P., Locht C., Minnikin D. E., Besra G. S. (2005) Identification and structural characterization of an unusual mycobacterial monomeromycolyl-diacylglycerol. Mol. Microbiol. 57, 1113–1126 [DOI] [PubMed] [Google Scholar]
- 36. Deb C., Lee C.-M., Dubey V. S., Daniel J., Abomoelak B., Sirakova T. D., Pawar S., Rogers L., Kolattukudy P. E. (2009) A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS ONE 4, e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pang J. M., Layre E., Sweet L., Sherrid A., Moody D. B., Ojha A., Sherman D. R. (2012) The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J. Bacteriol. 194, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. La Rosa V., Poce G., Canseco J. O., Buroni S., Pasca M. R., Biava M., Raju R. M., Porretta G. C., Alfonso S., Battilocchio C., Javid B., Sorrentino F., Ioerger T. R., Sacchettini J. C., Manetti F., Botta M., De Logu A., Rubin E. J., De Rossi E. (2012) MmpL3 is the cellular target of the antitubercular pyrrole derivative BM212. Antimicrob. Agents Chemother. 56, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tahlan K., Wilson R., Kastrinsky D. B., Arora K., Nair V., Fischer E., Barnes S. W., Walker J. R., Alland D., Barry C. E., 3rd, Boshoff H. I. (2012) SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 56, 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tullius M. V., Harmston C. A., Owens C. P., Chim N., Morse R. P., McMath L. M., Iniguez A., Kimmey J. M., Sawaya M. R., Whitelegge J. P., Horwitz M. A., Goulding C. W. (2011) Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U.S.A. 108, 5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stephan J., Mailaender C., Etienne G., Daffé M., Niederweis M. (2004) Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48, 4163–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stephan J., Bender J., Wolschendorf F., Hoffmann C., Roth E., Mailänder C., Engelhardt H., Niederweis M. (2005) The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol. Microbiol. 58, 714–730 [DOI] [PubMed] [Google Scholar]
- 43. Camacho L. R., Constant P., Raynaud C., Lanéelle M. A., Triccas J. A., Gicquel B., Daffé M., Guilhot C. (2001) Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276, 19845–19854 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.