Abstract
Background: The aberrant activation of oncogenic signaling such as Ras/MAPK signaling is a frequent event in human cancers. In addition to genetic changes, epigenetic silencing of inhibitors in Ras/MAPK signaling contributes to the activation of Ras/MAPK signaling. Recently, ANXA6 has been shown to interact with Ras-GAP1 and inhibit Ras activation in human breast cancer. However, whether and how it is involved in human cancers remain unknown. Methods: Real-time PCR was used to determine ANXA6 expression in gastric cancer cells and primary gastric carcinomas. Next, we explored the methylation of ANXA6 promoter in cell lines and tumor tissues with methylation-specific PCR and bisulfite genomic sequencing. We also investigated the function of ANXA6 in gastric cancer cells with colony formation assay and western blotting analysis. Results: ANXA6 was down-regulated in gastric cancer cells and primary gastric carcinomas. Ectopic ANXA6 expression inhibited the growth of gastric cancer cells and the activity of Ras/MAPK signaling. Its expression was restored after pharmaceutical demethylation. ANXA6 promoter was methylated in gastric cancer cell lines (6/6) and primary gastric carcinoma tissues (29/156). Interestingly, the knockdown of oncoprotein Yin Yang 1 (YY1) also restored ANXA6 expression and promoted the demethylation of ANXA6 promoter. However, ANXA6 methylation was not associated with clinical parameters such as differentiation, and TNM staging. Neither Kaplan-Meier Curve nor Cox regression analysis revealed a significant role of ANXA methylation to predict the survival of gastric cancer patients. Conclusions: We firstly reported that ANXA6 is epigenetically silenced through promoter methylation in human cancers and YY1 is important to initiate or maintain ANXA6 promoter methylation in gastric cancer cells. ANXA6 functions as a tumor suppressor in gastric cancer cells through the inhibition of Ras/MAPK signaling. ANXA6 methylation is not a prognostic factor for gastric cancer patients.
Keywords: ANXA6, methylation, gastric cancer, Ras signaling
Introduction
Human cancers result from the accumulation of multiple genetic and epigenetic changes that eventually lead to the deregulation of signaling pathways critical to cell proliferation and differentiation such as Ras/MAPK signaling. As the first oncogene identified from human tumors, Ras is one of the most well-known oncogenes aberrantly activated in human carcinogenesis [1-3]. Aberrant activation of Ras signaling often results from point mutations that render Ras constitutively active. Such oncogenic mutations have been frequently found in many human cancers including pancreatic cancer, colorectal cancer and lung cancer [3]. However, Ras mutations are rarely found in some cancers such as gastric cancer, suggesting that other mechanisms such as epigenetic changes may be responsible for aberrant Ras activation in these cancers. Through modulating gene expression, epigenetic changes were found as the alternative of genetic changes contributing to the deregulation of oncogenic signaling [4]. As one of most important events of epigenetic regulatory network, promoter methylation functions as signals for the recruitment of suppressive factors to inhibit gene expression. Many important regulators in various signaling pathways are frequently silenced by promoter methylation to facilitate oncogenesis [5-7].
Ras activity is tightly regulated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs). Ras-GEFs can promote Ras activation in response to the binding of growth factors to their receptors. In contrast, GAPs trigger the hydrolysis of bound GTP into GDP and inactivate Ras. The loss function of Ras-GAPs, attributed to either genetic changes including deletions and point mutations or epigenetic changes such as promoter methylation, has been frequently reported in human cancers. Annexin VI (ANXA6) belongs to a family of calcium-dependent membrane and phospholipid binding proteins with their regulations and functions in human cancers largely unknown [8]. Recently, ANXA6 has been shown to interact with Ras-GAP1 and inhibit Ras activation in human breast cancer [9]. In the current study, we found that ANXA6 was down-regulated through promoter hypermethylation in human gastric cancer, the second leading cause of cancer death worldwide.
Materials and methods
Cells, siRNAs and antibodies
Cells were generally cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and incubated at 5% CO2, 37°C and 95% humidity. For pharmacological demethylation, cells were treated for 72 h with 5 μM 5-aza-2’-deoxycytidine (Aza) (Sigma, St Louis, MO, USA). Aza was replenished every 24 h. An equivalent concentration of the vehicle (DMSO) was used as the control. The cells were then harvested for total RNA and genomic DNA extraction. Double str- and siRNAs were synthesized by Genepharma (Shanghai, China). All antibodies are products from cell signaling technology (Danvers, MA, USA).
Total RNA and genomic DNA extraction
Cells were homogenized in Trizol reagent (Invitrogen). Total RNA and genomic DNA were extracted using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The concentrations of RNA and DNA were quantified by NanoDrop 1000 (Nanodrop, Wilmington, DE, USA).
RT-PCR
The reverse transcription reaction was performed using 1 μg of total RNA with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time PCR was performed using the SYBR Green Master Mix Kit and ABI PRISM 7500 Real-Time PCR System (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for the normalization. The primers used for ANXA6 RT-PCR are CTGGACATAATCACCTCACG (RTPCR-F) and TTGGCATCACAATAGGCAGG (RTPCR-R).
Bisulfite treatment of DNA and methylation analysis
Methylation status of ANXA6 was determined by MSP (methylation specific PCR) and BGS (bisulfite genomic sequencing) using bisulfite modified genomic DNA as the template [10]. Genomic DNA was disulphide-treated with Zymo DNA Modification Kit (Zymo Research, Orange, CA, USA) according to the protocol provided by the manufacturer. MSP was carried out for 40 cycles with annealing temperature at 60°C, as previously described [11]. The primers used are GGTTTCGATTTAGCGAGCGT (MSP-F), CCATCAACCGATAACTAAACG (MSP-R), GGGTTTTGATTTAGTGAGTGT (USP-F) and CCCATCAACCAATAACTAAACA (USP-R). For BGS, PCR was performed using the annealing temperature as 55°C. Primers used for BGS are GTAAGATTTTAAGAGTTAAGGAGT (BGS-F) and GGTCAACCAAACCCTCTACAA (BGS-R).
Immunoblotting
Proteins were resolved by SDS-PAGE and transferred onto Hybond C nitrocellulose membranes (Amersham Life Science, Buckinghamshire, UK). Milk-Blocked membranes were probed with primary antibodies in blocking buffer overnight at 4°C. Finally, membranes were incubated with secondary antibodies conjugated with HRP (horse-radish peroxidase) and signals were visualized with enhanced chemiluminescence (Amersham Life Science).
Cell growth assay
Cell growth was measured by CellTiter 96® AQueous Assay kit (Promega, Madison, WI, USA). Briefly, cells were seeded in 12-well plates and transfected with plasmids or siRNAs for 48 hours. Cells were then harvested and re-seeded into 96-well plates. After 24 hours, the quantity of formazan was measured at 490 nm after one hour incubation with CellTiter 96® AQueous One Solution Reagent following the instructions provided.
Statistical analysis
The difference in the expression of ANXA6 between tumor and adjacent non-tumor tissues was analyzed by the Wilcoxon matched pairs test. The χ2 tests were used to analyze the association of patient characteristics with ANXA6 methylation. The probability of overall survival was calculated with the Kaplan-Meier method and differences between curves were evaluated with the log-rank test. Relative risks of death associated with ANXA6 methylation and other predictor variables were estimated by the univariate Cox proportional hazards model. Multivariate Cox models were also constructed to estimate the relative risk for ANXA6 methylation with adjustments of age, gender, H. pylori infection, Lauren type, differentiation and TNM stage. All analyses were performed using SPSS for Windows, version 14.0. A p value < 0.05 was taken as statistically significant.
Results
ANXA6 is down-regulated in gastric cancer
To explore whether ANXA6 is relevant to gastric carcinogenesis, we determined the expression of ANXA6 in a panel of human gastric carcinoma cell lines. In contrast to its high expression in stomach epithelium, ANXA6 mRNA levels are down-regulated in gastric cancer cell lines (Figure 1A). Moreover, ANXA6 expression levels in primary gastric carcinoma tissues are significantly lower than its expression in adjacent non-tumor stomach tissues (Figure 1B).
Figure 1.
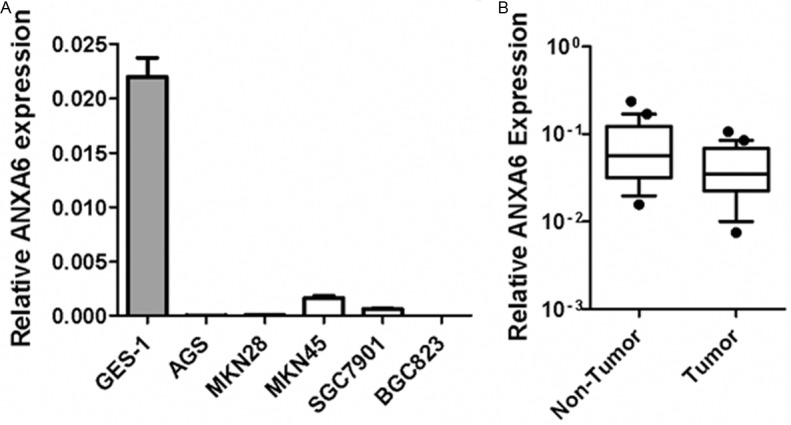
ANXA6 is down-regulated in gastric cancer. A: ANXA6 expression in a panel of gastric cancer cell lines was determined by real-time RT-PCR. B: ANXA6 expression in primary stomach tissues were determined by real-time RT-PCR (Wilcoxon matched pairs t-test, p<0.05).
ANXA6 acts as a tumor suppressor in gastric cancer cells
Next, we explored the biological relevance of ANXA6 downregulation in gastric cancer cells. Transient expression of ANXA6 in MKN28 cells inhibited cell growth (Figure 2A). Similarly, stable ANXA6 expression inhibited the clonogenicity of MKN28 cells (Figure 2B) and attenuated the activation of Ras/MAPK signaling (Figure 2C).
Figure 2.
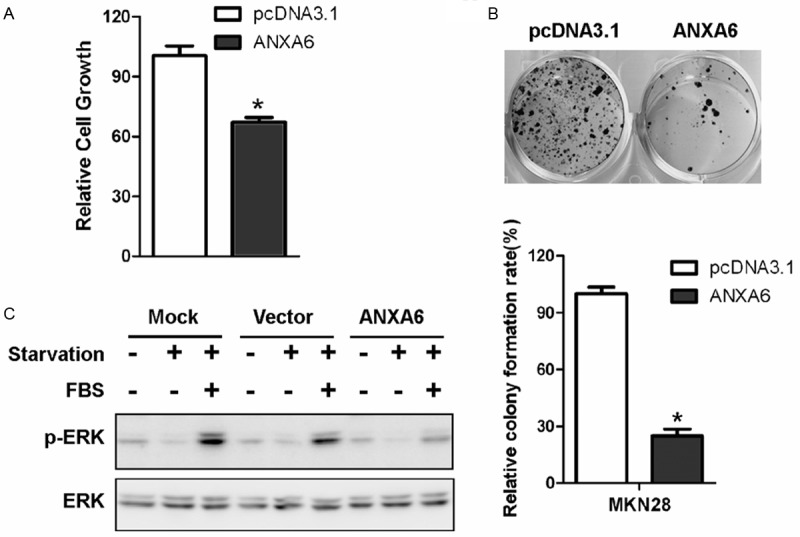
ANXA6 acts as a tumor suppressor in gastric cancer cells. ANXA6 expression inhibits the growth (A) and clonogenicity (B) of MKN28 cells. C: ANXA6 expression inhibited the activation of MAPK signaling in MKN28 cells.
ANXA6 is down-regulated through promoter methylation
Due to the presence of a typical CpG Island (CGI) in the promoter region of ANXA6 gene (Figure 3A), we postulated that methylation of ANXA6 CGI might be responsible for the downregulation of ANXA6 in gastric cancer cells. Indeed, ANXA6 expression was significantly restored in cells treated with demethylation agents like Aza (Figure 3B). Moreover, methylation specific PCR revealed the methylation of ANXA6 CGI in cancer cells (Figure 3C). Bisulfite genomic sequencing further confirmed methylation of ANXA6 CGI (Figure 3D).
Figure 3.
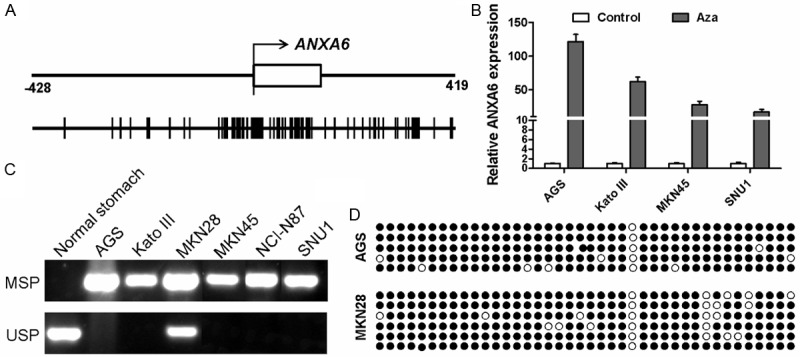
Methylation of ANXA6 promoter CGI mediates ANXA6 downregulation. A: ANXA6 contains a typical CpG Island (CGI) in its promoter region. The number indicates the location of CGI from the transcription start site. One vertical black line indicate one CpG site. B: Expression of ANXA6 before and after Aza treatment were determined by real-time RT-PCR as in Figure 1B. Methylation of ANXA6 promoter in GES-1 and gastric cancer cells were determined by MSP (C) and BGS (D).
Ying Yang 1 is important to the promoter methylation of ANXA6
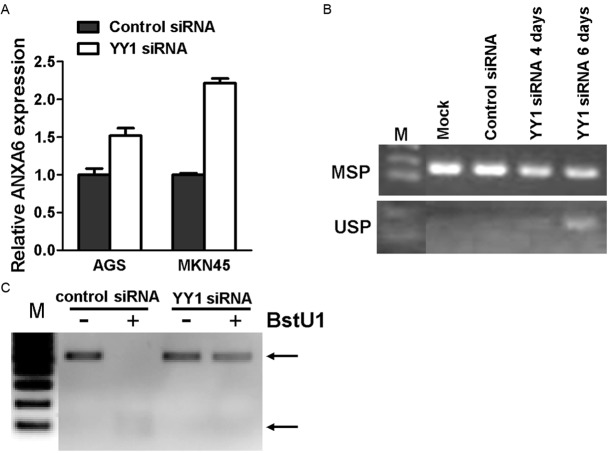
In an effort to explore how the methylation of ANXA6 was maintained in gastric cancer cells, we found several binding consensus of Ying Yang 1 (YY1) within the promoter region of ANXA6 (GCCAT, -2516, -1843 and -1578). Since Ying Yang 1 was well known to play an important role in the initiation and maintenance of DNA methylation, we wonder whether YY1 is relevant to the CGI methylation of ANXA6. The methylation of ANXA6 CGI was reduced in cells treated with YY1 siRNA (Figure 4A and 4B). Consistently, ANXA6 expression was restored after YY1 depletion (Figure 4C), indicating that YY1 promotes CGI methylation and consequent silencing of ANXA6.
Figure 4.
YY1 is important to the methylation of ANXA6 promoter. A: ANXA6 expression in gastric cancer cells before and after YY1 depletion were determined by real-time RT-PCR. B: Methylation of ANXA6 promoter in gastric cancer cells before and after YY1 depletion were analyzed by MSP (B) and Cobra (C).
Methylation of ANXA6 promoter CGI in primary tumors
To further illustrate the clinical relevance of ANXA6 methylation to gastric carcinogenesis, we examined the methylation of ANXA6 CGI in primary gastric carcinoma tissues. ANXA6 promoter CGC was methylated in 29 of 156 cases (18.6%) (Figure 5A). However, the methylation of ANXA6 CGI was not associated with gender, H. pylori infection, TNM staging, Lauren typing and differentiation (Table 1). Kaplan-Meier Curve analysis indicates that the methylation of ANXA6 promoter has no effect on the survival of gastric cancer patients (Figure 5B). In addition, neither univariate Cox regression nor multivariate Cox regression indicates a significant value of ANXA6 methylation in the prognosis evaluation of gastric cancer patients (Table 2).
Figure 5.
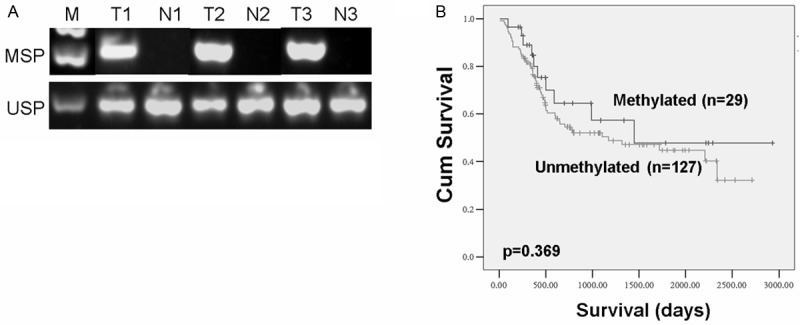
Methylation of ANXA6 promoter CGI in primary tumors. A: Methylation of ANXA6 promoter in primary stomach tissues were analyzed by MSP. T: tumor tissues; N: Adjacent non-tumor tissues. B: The effect of ANXA6 methylation on the survival of gastric cancer patients was analyzed by Kaplan-Meier Curve analysis.
Table 1.
Methylation of ANXA6 in gastric cancer
| Variable | Methylated (n=29) | % | Nonmethylated (n=127) | % | p value |
|---|---|---|---|---|---|
| Mean age, y ± SD | 55.3 ± 12.7 | 58.3 ± 12.4 | 0.393 | ||
| Gender | 0.972 | ||||
| M | 17 | 58.6% | 74 | 58.3% | |
| F | 12 | 41.4% | 53 | 41.7% | |
| H. pylori | 0.541 | ||||
| Positive | 7 | 35.0% | 23 | 28.0% | |
| Negative | 13 | 65.0% | 59 | 72.0% | |
| TNM | 0.470 | ||||
| I | 4 | 17.4% | 22 | 21.2% | |
| II | 6 | 26.1% | 14 | 13.5% | |
| III | 6 | 26.1% | 26 | 25.0% | |
| IV | 7 | 30.4% | 42 | 40.4% | |
| Lauren | 0.836 | ||||
| Intestinal | 25 | 86.2% | 105 | 84.7% | |
| Non-intestinal | 4 | 13.8% | 19 | 15.3% | |
| Differentiation | 0.093 | ||||
| Low | 21 | 84.0% | 70 | 65.4% | |
| Moderate or High | 4 | 16.0% | 37 | 34.6% |
Table 2.
Cox regression analysis of ANXA6 methylation in gastric cancer
| ANXA6 methylation | Univariate | Multivariate | ||
|---|---|---|---|---|
|
| ||||
| RR (95% CI) | p-value | RR (95% CI) | p-value | |
| Yes | 1.36 (0.69-2.66) | 0.371 | 0.94 (0.43-2.02) | 0.864 |
| No | 1.00 | 1.00 | ||
Discussion
Aberrant Ras activation is believed to play a critical role in many if not all cancers. Many genetic and epigenetic alterations can contribute to aberrant Ras activation during the stepwise process of cancer development [2,5,12-15]. Point mutations that render Ras constitutively active have been frequently found in many tumors such as pancreatic carcinoma. However, such oncogenic Ras mutations have rarely been detected in gastric cancer. Many genetic and epigenetic changes as well as environmental factors may contribute to aberrant Ras activation in gastric cancer instead of oncogenic Ras mutations. For example, growth factor receptors like epithelial growth factor receptors (EGFRs) are overexpressed via gene amplification in gastric cancer [16]. In addition, Helicobacter pylori infection, one of the risk factors for gastric cancer, can activate Ras through EGFR transactivation [17]. Recently, we found that promoter methylation mediates the epigenetic silencing of klotho which is a transmembrane protein to affect the interaction of membrane receptors with ligands such as insulin or insulin-like growth factors [18]. In contrast, ezrin which facilitate Ras activation by promoting the interaction of Ras with SOS was upregulated in cancer cells, resulting from the downregulation of microRNA-204 [6]. Herein, we presented another mechanism for aberrant Ras activation in gastric carcinogenesis.
ANXA6 can inhibit Ras activation through its interaction with Ras-GAP1 [9,19]. Recently, ANXA6 was found as a scaffold for protein kinase Cα (PKCα) to promote the inactivation of epidermal growth factor receptor (EGFR) which functions upstream of Ras/MAPK signaling pathway [20,21]. In addition, ANXA6 was found to inhibit cancer cell growth, indicating ANXA6 is a functional tumor suppressor [22]. Similar to RASAL which is a functional Ras-GAP and epigenetically silenced in multiple types of human cancers, ANXA6 was found to be down-regulated in human breast cancer [9]. However, it remains unknown how ANXA6 is down-regulated in breast cancer cells, particularly EGFR-overexpressing and estrogen receptor (ER)-negative cells. There is a typical CGI located in the promoter of ANXA6 and we found for the first time that promoter methylation is responsible for ANXA6 downregulation in human gastric cancer cells. YY1 which is a ubiquitously distributed transcription factor belonging to the GLI-Kruppel class of zinc finger proteins can activate or repress gene expression through directing histone deacetylases and histone acetyltransferases to the promoter. We found that ANXA6 promote contains several binding sites for YY1 and YY1 is important to ANXA6 methylation. Recently, DNA methylation was recognized as a dynamic process due to the existence of active demethylation in human cells such as embryonic stem cells [23]. ANXA6 CGI starts to be demethylated 4 days after YY1 depletion, indicating that YY1 most likely induces passive demethylation through disrupting the initiation or maintenance of DNA methylation. ANXA6 is down-regulated in many human cancers (www.oncomine.org) and it would be interesting to known whether the downregulation of ANXA6 in these cancers is also attributed to YY1-involved promoter methylation.
Promoter methylation was recently recognized as the biomarkers for cancer detection and prognosis prediction in addition to screening or defining novel tumor suppressor genes [24-26]. However, we failed to find any association of ANXA6 with clinical parameters such as overall survival, differentiation and staging. The positive rate of ANXA6 promoter methylation is only 18.6% and there are only 29 patients with ANXA6 promoter methylated in the cohort. This may underscore the clinical relevance of ANXA6 promoter methylation.
Conclusion
In conclusion, we firstly reported that ANXA6 is epigenetically silenced through promoter methmethylation in human cancers and YY1 is important to initiate or maintain ANXA6 promoter methylation in gastric cancer cells. ANXA6 functions as a tumor suppressor in gastric cancer cells through the inhibition of Ras/MAPK signaling. Our results did not support ANXA6 methylation as a prognostic factor for gastric cancer patients.
Acknowledgements
The project was supported by National Natural Science Foundation of China (81071652; 81071963), Qianjiang Scholarship of Zhejiang Province (2011R10061; 2011R10073), 973 project (2012CB526604) and Key Innovation Teams of Science and Technology in Zhejiang Province (2012R10046).
Disclosure of conflict of interest
No conflicts of interest exist for all authors.
References
- 1.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 3.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 4.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin H, Wang X, Ying J, Wong AH, Cui Y, Srivastava G, Shen ZY, Li EM, Zhang Q, Jin J, Kupzig S, Chan AT, Cullen PJ, Tao Q. Epigenetic silencing of a Ca(2+)-regulated Ras GTPase-activating protein RASAL defines a new mechanism of Ras activation in human cancers. Proc Natl Acad Sci U S A. 2007;104:12353–12358. doi: 10.1073/pnas.0700153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam EK, Wang X, Shin VY, Zhang S, Morrison H, Sun J, Ng EK, Yu J, Jin H. A microRNA contribution to aberrant Ras activation in gastric cancer. Am J Transl Res. 2011;3:209–218. [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS, Feng H, Ching AK, Cheung KF, Wong HK, Tong JH, Jin H, Choy KW, Yu J, To KF, Wong N, Huang TH, Sung JJ. EZH2-mediated concordant repression of Wnt antagonists promotes β-catenin-dependent hepatocarcinogenesis. Cancer Res. 2011;71:4028–39. doi: 10.1158/0008-5472.CAN-10-3342. [DOI] [PubMed] [Google Scholar]
- 8.Arcuri C, Giambanco I, Bianchi R, Donato R. Annexin V, annexin VI, S100A1 and S100B in developing and adult avian skeletal muscles. Neuroscience. 2002;109:371–388. doi: 10.1016/s0306-4522(01)00330-x. [DOI] [PubMed] [Google Scholar]
- 9.Vila de Muga S, Timpson P, Cubells L, Evans R, Hayes TE, Rentero C, Hegemann A, Reverter M, Leschner J, Pol A, Tebar F, Daly RJ, Enrich C, Grewal T. Annexin A6 inhibits Ras signalling in breast cancer cells. Oncogene. 2009;28:363–377. doi: 10.1038/onc.2008.386. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Lam EK, Wang X, Zhang J, Cheng YY, Lam YW, Ng EK, Yu J, Chan FK, Jin H, Sung JJ. Promoter hypermethylation mediates downregulation of thiamine receptor SLC19A3 in gastric cancer. Tumour Biol. 2009;30:242–248. doi: 10.1159/000243767. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Lau KK, So LK, Lam YW. CHD5 is down-regulated through promoter hypermethylation in gastric cancer. J Biomed Sci. 2009;16:95. doi: 10.1186/1423-0127-16-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin H, Sperka T, Herrlich P, Morrison H. Tumorigenic transformation by CPI-17 through inhibition of a merlin phosphatase. Nature. 2006;442:576–579. doi: 10.1038/nature04856. [DOI] [PubMed] [Google Scholar]
- 13.Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature. 1992;356:713–715. doi: 10.1038/356713a0. [DOI] [PubMed] [Google Scholar]
- 14.Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 15.Morrison H, Sperka T, Manent J, Giovannini M, Ponta H, Herrlich P. Merlin/neurofibromatosis type 2 suppresses growth by inhibiting the activation of Ras and Rac. Cancer Res. 2007;67:520–527. doi: 10.1158/0008-5472.CAN-06-1608. [DOI] [PubMed] [Google Scholar]
- 16.Kim MA, Lee HS, Lee HE, Jeon YK, Yang HK, Kim WH. EGFR in gastric carcinomas: prognostic significance of protein overexpression and high gene copy number. Histopathology. 2008;52:738–746. doi: 10.1111/j.1365-2559.2008.03021.x. [DOI] [PubMed] [Google Scholar]
- 17.Keates S, Sougioultzis S, Keates AC, Zhao D, Peek RM Jr, Shaw LM, Kelly CP. cag+ Helicobacter pylori induce transactivation of the epidermal growth factor receptor in AGS gastric epithelial cells. J Biol Chem. 2001;276:48127–48134. doi: 10.1074/jbc.M107630200. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Wang X, Wang X, Jie P, Lu H, Zhang S, Lin X, Lam EK, Cui Y, Yu J, Jin H. Klotho is silenced through promoter hypermethylation in gastric cancer. Am J Cancer Res. 2011;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- 19.Grewal T, Evans R, Rentero C, Tebar F, Cubells L, de Diego I, Kirchhoff MF, Hughes WE, Heeren J, Rye KA, Rinninger F, Daly RJ, Pol A, Enrich C. Annexin A6 stimulates the membrane recruitment of p120GAP to modulate Ras and Raf-1 activity. Oncogene. 2005;24:5809–5820. doi: 10.1038/sj.onc.1208743. [DOI] [PubMed] [Google Scholar]
- 20.Koese M, Rentero C, Kota BP, Hoque M, Cairns R, Wood P, Vila de Muga S, Reverter M, Alvarez-Guaita A, Monastyrskaya K, Hughes WE, Swarbrick A, Tebar F, Daly RJ, Enrich C, Grewal T. Annexin A6 is a scaffold for PKCalpha to promote EGFR inactivation. Oncogene. 2013;32:2858–72. doi: 10.1038/onc.2012.303. [DOI] [PubMed] [Google Scholar]
- 21.Grewal T, Koese M, Rentero C, Enrich C. Annexin A6-regulator of the EGFR/Ras signalling pathway and cholesterol homeostasis. Int J Biochem Cell Biol. 2010;42:580–584. doi: 10.1016/j.biocel.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Sakwe AM, Koumangoye R, Guillory B, Ochieng J. Annexin A6 contributes to the invasiveness of breast carcinoma cells by influencing the organization and localization of functional focal adhesions. Exp Cell Res. 2011;317:823–837. doi: 10.1016/j.yexcr.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Chen S, Xue M, Zhong J, Wang X, Gan L, Lam EK, Liu X, Zhang J, Zhou T, Yu J, Jin H, Si J. Homeobox D10 gene, a candidate tumor suppressor, is down-regulated through promoter hypermethylation and associated with gastric carcinogenesis. Mol Med. 2012;18:389–400. doi: 10.2119/molmed.2011.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM, Jin H, To KF, Chan FK, Sung JJ. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640–651. e1. doi: 10.1053/j.gastro.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 26.Yu J, Tao Q, Cheung KF, Jin H, Poon FF, Wang X, Li H, Cheng YY, Rocken C, Ebert MP, Chan AT, Sung JJ. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508–518. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]