Abstract
In vertebrates, the RAD51 protein is required for genetic recombination, DNA repair, and cellular proliferation. Five paralogs of RAD51, known as RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3, have been identified and also shown to be required for recombination and genome stability. At the present time, however, very little is known about their biochemical properties or precise biological functions. As a first step toward understanding the roles of the RAD51 paralogs in recombination, the human RAD51C and XRCC3 proteins were overexpressed and purified from baculovirus-infected insect cells. The two proteins copurify as a complex, a property that reflects their endogenous association observed in HeLa cells. Purified RAD51C–XRCC3 complex binds single-stranded, but not duplex DNA, to form protein–DNA networks that have been visualized by electron microscopy.
Homologous recombination plays an important role in the repair of DNA double-strand breaks (DSBs) caused by ionizing radiation or from the breakdown of stalled replication forks. Accurate DSB repair, using the sister chromatid as a template, is necessary for the maintenance of genome stability, and defects in this process can lead to the introduction of mutations, chromosomal translocations, apoptosis, and cancer.
The RAD51 protein promotes recombination by catalyzing the invasion of the broken ends of the DSB into the intact sister chromatid. RAD51 is a structural and functional homolog of Escherichia coli RecA and forms helical nucleoprotein filaments in which the DNA lies extended and underwound. Filaments form preferentially on tailed duplex DNA substrates that mimic the resected DSBs thought to be present at chromosomal break sites (1, 2). Strand invasion by RAD51 is stimulated by RAD52, RAD54, and RP-A, resulting in the formation of a heteroduplex joint (2–11). Yeast that are defective in RAD51 exhibit reduced levels of recombination and are sensitive to ionizing radiation, but the cells remain viable. In contrast, disruption of RAD51 in the mouse is lethal (12, 13). Moreover, inactivation of a RAD51 transgene in chicken cells leads to chromosome fragmentation followed by cell death (14). These observations emphasize the essential role that RAD51 and recombinational repair play in normal cellular proliferation. Such an extreme phenotype, however, has not been observed after disruption of RAD52 (15, 16) or RAD54 (17, 18).
Although recombination proteins such as RAD51, RAD52, and RAD54 have been well conserved from yeast to vertebrates, it is not clear whether there are vertebrate homologs of yeast Rad55 and Rad57. Defects in RAD55 and RAD57 result in radiation sensitivity, a phenotype that can be partially complemented by overexpression of Rad51 (19). Biochemical studies of Rad55 and Rad57 have shown that the two proteins form a heterodimer that interacts with Rad51 and stimulates Rad51-mediated pairing reactions (20). It is therefore thought that Rad55/57 play an accessory role in strand invasion, possibly displacing RP-A during nucleoprotein filament assembly by RAD51. The repair-defective phenotype of rad55/57 mutants is elevated at reduced temperatures (21), a property that is often associated with proteins that are composed of multiple subunits or are participants in large multiprotein complexes (22).
Rad55 and Rad57 are known to share limited amino acid similarity with Rad51 and may have been derived by duplication of the ancestral gene encoding Rad51. However, apart from their conserved ATP binding motifs and the ability of Rad55/57 to catalyze ATP hydrolysis (20), they are clearly divergent in function from Rad51. Direct homologs of the yeast RAD55 and RAD57 genes have not been identified in vertebrates. However, five genes that bear a distant resemblance to RAD51 have been identified, and, like Rad55 and Rad57, their products have been classified as members of the RAD51 family (23). The first members of this class, encoded by the XRCC2 and XRCC3 genes (24, 25), were identified by genetic complementation of the repair-deficient irs1 and irs1SF rodent cell lines (26, 27). Furthermore, three distinct genes designated RAD51B (also known as RAD51L1, hREC2, or R51H2), RAD51C (a.k.a. RAD51L2), and RAD51D (a.k.a. RAD51L3 or R51H3) were identified by database analyses on the basis of their sequence homology to RAD51 (28–32). All five RAD51 paralogs share limited (≈20–30%) amino acid sequence identity with RAD51, much of which is concentrated around the two Walker ATP binding sites (23, 33). Multiple protein alignments of the RAD51 family members suggest that RAD51D and XRCC3 are closest to yeast Rad57, whereas XRCC2 is more homologous to yeast Rad55 (23, 34).
The RAD51 family members are required for normal levels of recombination and DSB repair. Whereas cells defective in XRCC2 (irs1) and XRCC3 (irs1SF) are moderately sensitive to x-rays or γ-radiation (≈2 fold), they display an extreme sensitivity (60- to 100-fold) to DNA cross-linking agents such as cisplatin, nitrogen mustard, or mitomycin C (25, 35). The mutant cell lines also exhibit a high incidence of spontaneous and mutagen-induced chromosomal aberrations (36) and show defects in chromosome segregation (37). Moreover, both irs1 and irs1SF show a significant (100- and 25-fold, respectively) decrease in the frequency of DSB repair by homologous recombination (38, 39).
In recent studies, the XRCC2 gene was targeted in the mouse, and disruptions were found to confer an embryonic lethal phenotype (40). XRCC2−/− blastocysts showed a genetic instability phenotype, with high levels of chromosomal aberrations and a sensitivity to γ-rays. They also exhibited developmental defects in the nervous system, indicating a potential role for XRCC2 in the prevention of apoptosis. A similar embryonic lethal phenotype was observed after disruption of RAD51D (41), the product of which physically associates with XRCC2 (42, 43).
The important roles that the RAD51 paralogs play in the maintenance of genome stability during proliferation is also emphasized by the embryonic lethality that is found after targeted disruption of the RAD51B gene (44). Indeed, the embryonic lethal phenotype of the RAD51B disruption is comparable to that of RAD51−/− itself, a phenotype that is more severe than that of the RAD51D knock out. Interestingly, mutations in RAD51B have been found to be associated with uterine leiomyomas, highlighting a role in tumorigenesis (45, 46). To date, however, there are no cell lines with known mutations in RAD51C. Nevertheless, the 17q23 region containing RAD51C was found to be amplified in primary breast tumors and in tumors containing BRCA2 mutations. Amplification of 17q23 has also been observed in pancreatic adenocarcinomas, bladder carcinomas, neuroblastomas, and hepatocellular carcinomas (47, 48).
Each of the paralogs has now been disrupted in chicken DT40 B lymphocytes. All knockout lines were found to be viable, an observation that distinguishes them from the RAD51−/− mutant cell line (14). However, RAD51B−/−, RAD51C−/−, RAD51D−/−, XRCC2−/−, and XRCC3−/− cells all exhibit a reduced growth rate and show chromosome instability (49, 50). The mutants accumulate spontaneous chromosomal breaks, presumably as a consequence of stalled or broken replication forks. As observed with rodent cell lines defective in XRCC2 and XRCC3, the mutant DT40 lines were highly sensitive to DNA cross-linking agents such as mitomycin C. The sensitivity to cross-linking agents was partially rescued by overexpression of RAD51. In contrast, however, the gene targeting deficiencies associated with mutations in RAD51B, XRCC2, or XRCC3 could not be rescued in this way.
Because of the similarities between the RAD51 paralogs, and the related phenotypes associated with their disruption, it is tempting to suggest that the five proteins have similar functions or act in concert during recombination and repair. In this regard, the RAD51 paralogs display numerous interactions: for example, associations between RAD51 and XRCC3, XRCC3 and RAD51C, RAD51C and RAD51B, RAD51C and RAD51D, and RAD51D and XRCC2 have been detected by yeast two-hybrid analyses (43); XRCC3 and RAD51 proteins coimmunoprecipitate (25); and the XRCC2 and RAD51D proteins interact in glutathione S-transferase (GST) pull down experiments (42). If these interactions occur simultaneously, then it is likely that the RAD51 paralogs function with RAD51 and other recombination proteins such as RAD52, RAD54 and BRCA2 (51). Indeed, it is possible that the proteins interact and are constituents of a large protein complex or “recombinosome” that assembles at sites of DNA damage to initiate repair.
Despite their importance, the precise function of each RAD51 paralog remains unclear at the present time. However, biochemical studies are now being initiated that will shed new light on their functions. As a first step, the RAD51D–XRCC2 complex was purified and shown to bind preferentially to single-stranded DNA (ssDNA) and to exhibit a DNA-stimulated ATPase activity (42). It has also been reported that RAD51B exhibits protein kinase activity that can phosphorylate p53, cyclin E, and cdk2 (52). Because RAD51B is induced by both ionizing and UV irradiation and its over-expression causes a delay at G1, a role for RAD51B in cell cycle progression was postulated.
In this paper, we describe the purification of a complex containing human RAD51C and XRCC3 proteins and show that this complex binds preferentially to ssDNA.
Materials and Methods
Purification of RAD51C and XRCC3.
Recombinant RAD51C-His10 and XRCC3-His6 were copurified from five 1.25-liter spinner flasks of Sf9 cells (1 × 106 per ml) infected with the RAD51C-His10 and XRCC3-His6 baculoviruses (moi 10) for 4 days at 27°C. Cells were harvested, frozen in dry ice/ethanol and stored at −20°C. The cell paste was resuspended in 150 ml of T buffer (20 mM Tris⋅HCl, pH 8.0/0.5 M NaCl/10% glycerol/0.02% Triton X-100) containing 5 mM imidazole and protease inhibitors. The suspension was lysed by using a Dounce homogenizer (20 strokes), sonicated, and then homogenized a second time. Insoluble material was removed by centrifugation (35,000 rpm for 1 h in a Beckman 45 Ti rotor). The supernatant was loaded on a 20-ml Talon column and washed with 200 ml of T buffer containing 50 mM imidazole. RAD51C–XRCC3 was eluted with a 200-ml linear gradient of 0.05–1.0 M imidazole in T buffer. Fractions containing the RAD51C and XRCC3 proteins were identified by SDS/PAGE, pooled, concentrated, and dialyzed against storage buffer (20 mM Tris-acetate, pH 7.5/10% glycerol/1 mM DTT) containing 400 mM KOAc. The proteins were then further purified by using a 24 ml Superdex 200 gel filtration column (Amersham Pharmacia), which was eluted with the same buffer, and stored at −80°C.
Protein for Animal Immunization.
Denatured recombinant RAD51C-His10 and XRCC3-His6 proteins were used for antibody production. They were purified from 2.4 liters of E. coli FB810 recA− pLysS (53) carrying either pRAD51C-16b or pXRCC3-28c in Luria broth supplemented with 100 μg/ml carbenicillin and 25 μg/ml chloramphenicol. At OD650 = 0.5, RAD51C-His10 or XRCC3-His6 synthesis was induced by the addition of 1 mM isopropyl β-d-thiogalactoside (IPTG). After 4 h, the cells were harvested by centrifugation, frozen in dry ice/ethanol, and stored at −80°C. The cell paste was resuspended in 100 ml of T buffer containing 5 mM imidazole and protease inhibitors, and aliquots were lysed by sonication. Insoluble material was recovered by centrifugation, resuspended in T buffer containing 8 M urea and 5 mM imidazole (12 h at room temperature), and centrifuged at 35,000 rpm for 30 min in a Beckman 45 Ti rotor. The supernatant was loaded on a 20-ml Talon column equilibrated in the same buffer and washed with 150 ml of T buffer containing 30 mM imidazole; RAD51C or XRCC3 were eluted with a 200-ml linear gradient of 0.03–1.0 M imidazole in T buffer. The purified RAD51C or XRCC3 (≈2.5 mg) were then electrophoresed through SDS/PAGE, and the protein band was excised and electroeluted in 25 mM Tris, 190 mM glycine, and 0.05% SDS for 1 h at 150 V. The denatured protein was then used to immunize rabbit and mice.
Immunoprecipitations.
Baculovirus expression systems.
Sf9 cells were coinfected with RAD51C-His10 and/or XRCC3-His6 baculovirus and harvested after 4 days. The cells were resuspended in lysis buffer (50 mM Tris⋅HCl, pH 7.5/0.5 M NaCl/0.5% Nonidet P-40) containing protease inhibitors, incubated for 30 min on ice, and then lysed by sonication. Insoluble material was removed by high-speed centrifugation. Protein complexes in the supernatant (equivalent to ≈3 × 106 cells) were pulled down for 1.5 h at 4°C by using preimmune serum or polyclonal Abs (pAbs) raised against RAD51C or XRCC3 cross-linked to aminolink beads (Pierce). Complexes were washed 4× in lysis buffer containing NaCl, and visualized by Western blotting by using an anti-histidine mAb (Clontech).
HeLa extracts.
Extracts were prepared from 2 × 107 HeLa cells, and protein complexes were immunoprecipitated essentially as described above. The complexes were washed and visualized by Western blotting by using anti-RAD51C or anti-XRCC3 mAbs followed by enhanced chemiluminescence (ECL; NEN).
DNA Binding Assays.
Reactions (10 μl) contained 10 nM DNA in binding buffer (20 mM triethanolamine-HCl, pH 7.5/2 mM ATP/1 mM Mg(OAc)2/1 mM DTT/100 μg/ml BSA). After 5 min at 37°C, the indicated amount of RAD51C–XRCC3 was added (2 μl), and incubation was continued for a further 10 min. Protein–DNA complexes were analyzed by 6% PAGE using TBE buffer (90 mM Tris/90 mM boric acid/2 mM EDTA, pH 8.3) followed by autoradiography. DNA substrates were prepared by annealing a 32P-labeled oligonucleotide (100 nt in length) with appropriate complementary sequences. The sequence of the 100-mer is 5′-GGGCGAATTGGGCCCGACGTCGCATGCTCCTCTAGACTCGAGGAATTC GGTACCCCGGGTTCGAAATCGATAAGCTTACAGTCTCCATTTAAAGGACAAG-3′. DNA concentrations are expressed in terms of moles of DNA molecules.
Electron Microscopy.
For visualization of protein-DNA complexes, reactions (10 μl) contained 5 μM (expressed in nucleotides) DNA in 20 mM triethanolamine-HCl, pH 7.5/2 mM ATP/1 mM Mg(OAc)2/1 mM DTT. After 5 min at 37°C, RAD51C–XRCC3 (0.5 μM) was added, and incubation was continued for a further 10 min. Protein–DNA complexes were fixed by addition of glutaraldehyde to 0.2% followed by 15 min incubation at 37°C. Samples were diluted and washed in 5 mM Mg(OAc)2 before uranyl acetate staining (54) and visualized at a magnification of ×20,500 by using a Philips C100 electron microscope.
Results
Purification of RAD51C–XRCC3.
The RAD51C and XRCC3 genes were PCR amplified from a human testis cDNA library and cloned into pET16b and pET28c. In these constructs, the RAD51C and XRCC3 protein sequences were linked to decahistidine and hexahistidine tags at their amino-terminal ends, respectively. The His-tagged RAD51C and XRCC3 genes from the resultant clones (pRAD51C-16b and pXRCC3-28c) were then subcloned into pFASTBAC1 to produce the recombinant baculoviruses RAD51C-His10 and XRCC3-His6. The sequences of RAD51C and XRCC3 were confirmed to be identical to those published previously (25, 31).
Initial attempts to purify RAD51C-His10 in E. coli or baculovirus-infected insect cells were unsuccessful because the protein was found to be mostly insoluble. However, because RAD51C interacts with XRCC3 (31), we coinfected Sf9 cells with the RAD51C-His10 and XRCC3-His6 baculoviruses and found that RAD51C-His10 was considerably more soluble. This strategy enabled us to copurify RAD51C-His10 and XRCC3-His6 by using Talon affinity chromatography. Because of an extensive imidazole wash before elution from Talon, RAD51C and XRCC3 were highly pure after this first step (Fig. 1A, lanes c–i). However, further purification by gel filtration was necessary to remove minor contaminants. When analyzed by gel filtration through superdex 200, RAD51C–XRCC3 exhibited a broad elution profile (data not shown). During all chromatographic steps, RAD51C and XRCC3 coeluted with an apparent 1:1 stoichiometry. Because RAD51C-His10 (45 kDa) and XRCC3-His6 (40 kDa) are similar in size, the presence of the two proteins was confirmed by using mAbs raised against XRCC3 and RAD51C, respectively (Fig. 1B, lanes a and b). The protein preparation was free of nuclease activities, as monitored by the release of acid-soluble counts from 5′-32P-end-labeled single- or double-stranded oligonucleotides.
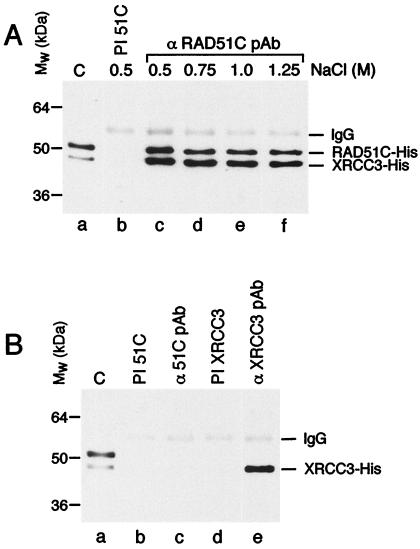
Figure 1.
Copurification of RAD51C and XRCC3 from baculovirus-infected insect cells. (A) Purification of RAD51C-His10 and XRCC3-His6. Lane a, molecular weight markers. Lanes b–j, elution profile from the Talon column. Proteins were visualized by SDS/PAGE followed by Coomassie blue staining. (B) Western blot of purified RAD51C-His10 and XRCC3-His6 using mAbs raised against RAD51C or XRCC3 (mAbs 2H11 and 10F1, respectively).
Interactions Between RAD51C and XRCC3.
Because XRCC3 and RAD51C coelute during chromatography, we reasoned that the two proteins may have interacted during expression in the Sf9 cells. To confirm this interaction, extracts from coinfected Sf9 cells were analyzed by immunoprecipitation by using a pAb raised against RAD51C. Pull-down complexes were washed extensively with buffer containing 0.5–1.25 M NaCl and analyzed by Western blotting with an anti-histidine mAb. XRCC3 was found to interact with RAD51C under all conditions tested, suggesting strong interactions between the two proteins (Fig. 2A, lanes c–f). Control reactions showed that the anti-RAD51C pAb was specific for RAD51C because it failed to pull down XRCC3 from Sf9 cells infected only with the XRCC3-His6 virus (Fig. 2B, lane c). The XRCC3 could, however, be immunoprecipitated by using an anti-XRCC3 pAb (lane e).
Figure 2.
RAD51C forms a stable complex with XRCC3 in insect cells. (A) Coimmunoprecipitation of RAD51C with XRCC3. Sf9 cells were coinfected with RAD51C-His10 and XRCC3-His6 baculovirus. Immunoprecipitations were carried out as described in Materials and Methods by using preimmune serum (lane b) or a pAb raised against RAD51C (lanes c–f). Complexes were washed in lysis buffer with the indicated concentration of NaCl, and visualized by Western blotting using an anti-histidine mAb. Lane a, purified RAD51C-His10–XRCC3-His6 complex (50 ng). (B) Specificity of the anti-RAD51C pAb. Sf9 cells were infected with an XRCC3-His6 baculovirus, and cell-free extracts were prepared in lysis buffer. Lane a, control showing purified RAD51C-His10–XRCC3-His6 complex (50 ng). Lanes b–e, pull downs by using preimmune RAD51C serum (lane b), anti-RAD51C pAb (lane c), preimmune XRCC3 serum (lane d), or anti-XRCC3 pAb (lane e). The immunoprecipitates were analyzed by Western blotting using an anti-histidine mAb and ECL.
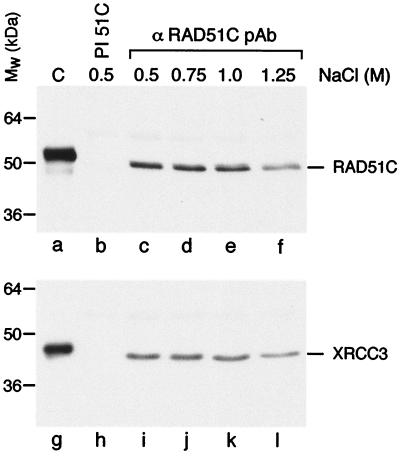
We next investigated whether stable RAD51C–XRCC3 complexes were also present in mammalian cells. Immunoprecipitation analyses were again carried out using the RAD51C pAb, and pull-down complexes were probed by Western blotting by using RAD51C (2H11) and XRCC3 (10F1) mAbs. These mAbs were specific for RAD51C and XRCC3, respectively, and did not cross-react with any of the other RAD51 paralogs. Endogenous XRCC3 and RAD51C coimmunoprecipitated from HeLa extracts (Fig. 3, lanes c–f and i–l). Again the RAD51C–XRCC3 interaction was found to be stable to 1.25 M NaCl, indicating that the interactions were not because of the presence of contaminating DNA. Control experiments showed that preimmune antisera were unable to pull down RAD51C or XRCC3 (lanes b and h). In related experiments, we found that a pAb raised against XRCC3 also pulled down RAD51C from the HeLa extract (data not shown). These results show that the human RAD51C and XRCC3 proteins form a stable complex.
Figure 3.
Coimmunoprecipitation of endogenous RAD51C and XRCC3 from HeLa cell-free extracts. Protein complexes were precipitated from HeLa cell-free extracts by using preimmune serum (lanes b and h) or pAbs raised against RAD51C (lanes c–f and i–l). The complexes were washed in buffer containing NaCl, as indicated, and visualized by Western blotting using anti-RAD51C (lanes a–f) or anti-XRCC3 mAbs (lanes g–l). Lanes a and g, purified RAD51C-His10–XRCC3-His6 complex (50 ng). The His-tagged controls migrate more slowly than the endogenous RAD51C or XRCC3 from HeLa.
Electron Microscopic Visualization of RAD51C–XRCC3.
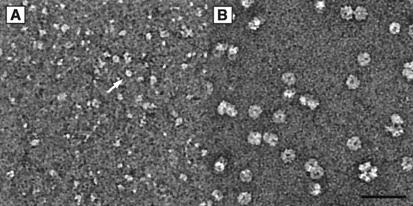
Purified RAD51C–XRCC3 complex was visualized by electron microscopy after negative staining with uranyl acetate; we observed that the protein mainly formed disperse and irregular structures. In some instances, however, ring-like structures could be seen that contained a central cavity (Fig. 4A, white arrow). Because the protein preparation is highly purified and essentially homogeneous, we believe that it is unlikely that these ring-like structures are due to contaminant proteins. For size comparison, we spread the human meiosis-specific RecA homolog DMC1, which is known to form octameric rings with a diameter of about 140 Å and a central hole of 25Å (55). As expected, the recombinant DMC1 rings (molecular mass of 324 kDa), as shown in Fig. 4B, were considerably larger than the observed RAD51C–XRCC3 complexes. The complexes observed by electron microscopy may represent oligomeric forms of the individual subunits (45 kDa and 40 kDa), or heterodimeric (85 kDa) and oligomeric forms of RAD51C–XRCC3.
Figure 4.
Visualization of RAD51C–XRCC3 by electron microscopy. (A) RAD51C–XRCC3 complex. (B) Human DMC1 protein. The white arrow indicates a RAD51C–XRCC3 structure containing a cavity. The bar represents 50 nm.
DNA Binding by RAD51C–XRCC3.
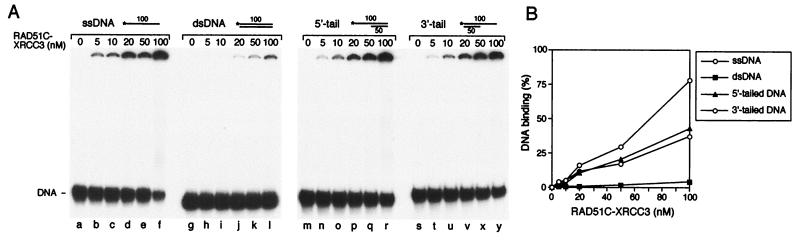
When purified RAD51C–XRCC3 complex was incubated with ssDNA (100 nucleotides in length), we observed the formation of protein–DNA networks that failed to enter polyacrylamide gels (Fig. 5A, lanes b–f). The protein complex bound preferentially to ssDNA; duplex DNA was bound poorly under the same conditions (lanes h–l). Indeed, quantification of the DNA–protein complexes formed with each substrate at 100 nM RAD51C–XRCC3 indicated that only 4% of the input duplex DNA was stably bound, compared with 75% for the ssDNA (Fig. 5B). RAD51C–XRCC3 bound 5′- or 3′-tailed DNA substrates at a level that was intermediate of that observed with the ssDNA and double-stranded DNA substrates (Fig. 5A, lanes n–r and t–y, respectively, and Fig. 5B).
Figure 5.
DNA binding by RAD51C–XRCC3. (A) Reactions (10 μl) contained single-stranded (lanes a–f), double-stranded (lanes g–l), 5′-tailed (lanes m–r), or 3′-tailed (lanes s–y) DNA in binding buffer, and the indicated amounts of RAD51C–XRCC3. Protein–DNA complexes were analyzed by PAGE. 5′-32P-end labels are indicated with asterisks. (B) The gels shown in A were quantified by phosphorimaging.
In an attempt to resolve the protein–DNA complexes that failed to enter polyacrylamide gels, complexes were also analyzed by electrophoresis through 0.8% agarose gels. Again, however, the complexes were found to resolve poorly and tended to smear up the gel (data not shown), a result that is characteristic of the formation of networks containing a variable number of DNA molecules. Interestingly, the DNA binding properties of RAD51C–XRCC3 are similar to those exhibited by the human RAD52 protein, which has also been shown to aggregate ssDNA (4, 56).
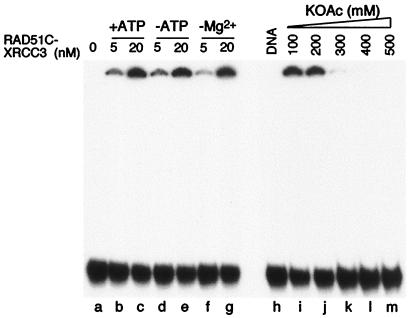
The binding of ssDNA by RAD51C–XRCC3 showed no requirement for ATP (Fig. 6, compare lanes b and c with d and e) or divalent metal ions (lanes f and g). Omission of Mg2+, however, resulted in a decrease in the amount of protein–DNA aggregates that were formed (compare lanes b and f). The formation of protein–DNA aggregates was eliminated by inclusion of salt at concentrations in excess of 200 mM (lanes i–m).
Figure 6.
Requirements for RAD51C–XRCC3-mediated DNA aggregation. DNA binding reactions were carried out as described in Fig. 5 by using binding buffer modified as indicated. In lanes i–m, the concentration of RAD51C–XRCC3 was 20 nM.
When complexes formed between RAD51C–XRCC3 and DNA were visualized by electron microscopy, we observed that DNA aggregation was mediated by the formation of large protein complexes. As shown in Fig. 7 A and B, with gapped circular and tailed linear plasmid DNA, respectively, RAD51C–XRCC3 promoted the formation of large DNA networks. Such networks would fail to enter polyacrylamide gels, as shown in Fig. 5. Similarly, networks were formed by the actions of RAD51C–XRCC3 on form I plasmid DNA (Fig. 7D), which are known to exhibit transient regions of ssDNA. We did not observe the binding of RAD51C–XRCC3 to linear duplex DNA (Fig. 7C). These results indicate that the interactions of RAD51C–XRCC3 with various DNA substrates are mediated by binding to ssDNA.
Figure 7.

Electron microscopic visualization of complexes formed between RAD51C–XRCC3 and DNA. Reactions contained RAD51C–XRCC3, with pPB4.3-gapped circular DNA (A), pPB4.3-tailed linear DNA (B), pDEA-7Z linear duplex DNA (C), or pPB4.3-supercoiled DNA (D). The bar represents 100 nm.
Discussion
In this work, we have described the purification and preliminary characterization of the RAD51C–XRCC3 complex. Together with the purification of RAD51D–XRCC2 (42) and RAD51B (52), these studies will facilitate further analyses of the roles of the RAD51 paralogs in recombination and DNA repair. During initial purification trials, we observed that RAD51C was mostly insoluble unless coexpressed with XRCC3. Their coexpression, by using baculovirus vectors, however, allowed purification of a homogeneous protein complex containing stoichiometric amounts of each subunit. The stable association of RAD51C and XRCC3 was also observed with endogenous human proteins in HeLa cell-free extracts.
In response to DNA damaging agents, many of the known recombination proteins have been shown to colocalize to nuclear foci. These foci presumably identify sites where double-strand break repair is taking, or has recently taken, place. RAD51 has been shown to colocalize with histone H2AX (57), RP-A (58), RAD52 (59), RAD54 (60), BRCA1 (61), and BRCA2 (62). Interestingly, the formation of RAD51 foci after DNA damage occurs at reduced levels in chicken DT40 cells defective in RAD51B, RAD51C, RAD51D, XRCC2, or XRCC3 (49, 50). Moreover, RAD51 foci are absent in irs1SF cells lacking XRCC3 (63). It is thought that the presence of these proteins may facilitate the assembly of RAD51 at sites of DNA damage. Future studies will determine whether the RAD51 paralogs themselves are components of the damage-induced foci.
We have shown that the RAD51C–XRCC3 complex binds ssDNA, and similar observations have been made with RAD51D-XRCC2 (42). The specificity of these interactions may be important for the initiation of recombinational repair as DSBs are resected to expose single-stranded tails. Indeed, the RAD51 paralogs may act as accessory factors that facilitate DSB recognition, or may be involved in the assembly of RAD51 filaments. Similar proposals have been made for RAD52, RAD54, and RP-A. Alternatively, these proteins may be involved in stabilizing or extending heteroduplex joints formed by RAD51. However, one striking feature of the RAD51C–XRCC3 complex is its tendency to aggregate DNA, a property that it shares with both RAD51 and RAD52. Early studies with RecA protein showed that the formation of DNA networks is a prerequisite for homologous pairing, and it is possible that DNA aggregation promoted by RAD51C–XRCC3 may facilitate DNA–DNA contacts during the search for homology between sister chromatids or homologous chromosomes. In this regard, it is noteworthy that the efficiency of gene targeting is reduced in chicken DT40 cells carrying defects in RAD51C or XRCC3 (50).
Although both RAD51C and XRCC3 share limited sequence homology with RAD51, we have not observed the formation of the regular nucleoprotein filaments that are the characteristic signature of a recombinase. Additionally, attempts to show D-loop formation (using single-stranded oligonucleotides and superhelical duplex DNA) by RAD51C–XRCC3 complex have proven negative (M. McIlwraith, J.-Y.M., and S.C.W., unpublished data). These results lead us to conclude that the RAD51C–XRCC3 complex does not facilitate homologous pairing in vitro, although we cannot at this time rule out the possibility that it might promote such interactions in the presence of RAD51 and possibly other RAD51 paralogs. More likely, we suggest that RAD51C–XRCC3 are important constituents of the recombination-promoting complex and, as such, are indirectly involved in both nucleoprotein assembly and the subsequent DNA–DNA interactions required for the establishment of a heteroduplex joint.
In recent studies, conditional RAD52−/−XRCC3−/− DT40 cell lines were generated in which an XRCC3 transgene could be inactivated by Cre-Lox recombination (64). Whereas clones of either single mutant could proliferate, although some die because of spontaneous chromosomal breaks, all cells deficient for both RAD52 and XRCC3 function died a few days after XRCC3 inactivation. Remarkably, the lethal effect of XRCC3 inactivation was overcome by overexpression of RAD51. These results can be interpreted in terms of the way that both RAD52 and XRCC3 may be required as accessory factors that play important, but not essential, roles in RAD51 filament assembly within the context of a DNA damage-induced recombination complex. Whereas the loss of either RAD52 or XRCC3 function can be tolerated, the absence of both functions will result in severe chromosome instability and cell death. Our understanding of the way in which proliferating cells maintain genome stability will therefore be advanced by further studies of the RAD51 paralogs and other protein constituents of the recombination machinery.
Acknowledgments
We thank members of the West laboratory for suggestions. We are grateful to Shunichi Takeda for communication of unpublished results, Ruth Peat and the Imperial Cancer Research Fund (ICRF) cell production unit for tissue culture, Jane Steel for antibody production, and Isabelle Brodeur for advice on immunoprecipitation analyses. We also thank Jacques Dubochet for support and interest. This work was supported by the Imperial Cancer Research Fund, the Swiss National Science Foundation, the Human Frontiers Science Program, and the Swiss-British Council Joint Research Program. J.-Y.M. was supported by postdoctoral fellowships from the National Cancer Institute of Canada and the Imperial Cancer Research Fund.
Abbreviations
- ssDNA
single-stranded DNA
- DSB
double-strand break
- mAb
monoclonal antibody
- pAb
polyclonal antibody
Footnotes
This paper results from the National Academy of Sciences colloquium, “Links Between Recombination and Replication: Vital Roles of Recombination,” held November 10–12, 2000, in Irvine, CA.
References
- 1.Mazin A V, Zaitseva E, Sung P, Kowalczykowski S C. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McIlwraith M J, Van Dyck E, Masson J-Y, Stasiak A Z, Stasiak A, West S C. J Mol Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- 3.Sung P. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 4.Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 5.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Nature (London) 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 6.Petukhova G, Stratton S, Sung P. Nature (London) 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 7.Shinohara A, Ogawa T. Nature (London) 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 8.Baumann P, West S C. J Mol Biol. 1999;291:363–374. doi: 10.1006/jmbi.1999.2954. [DOI] [PubMed] [Google Scholar]
- 9.Mazin A V, Bornarth C J, Solinger J A, Heyer W-D, Kowalczykowski S C. Mol Cell. 2000;6:583–592. doi: 10.1016/s1097-2765(00)00057-5. [DOI] [PubMed] [Google Scholar]
- 10.Song B W, Sung P. J Biol Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- 11.Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P. Mol Cell. 2000;6:563–572. doi: 10.1016/s1097-2765(00)00055-1. [DOI] [PubMed] [Google Scholar]
- 12.Lim D S, Hasty P. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuzuki T, Fujii Y, Sakuma K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonoda E, Sasaki M S, Buerstedde J M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rijkers T, van den Ouweland J, Morolli B, Rolink A G, Baarends W M, van Sloun P P H, Lohman P H M, Pastink A. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi-Iwai Y, Sonoda E, Buerstedde J-M, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J M. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 18.Essers J, Hendriks R W, Swagemakers S M A, Troelstra C, Dewit J, Bootsma D, Hoeijmakers J H J, Kanaar R. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 19.Hays S L, Firmenich A A, Berg P. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung P. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 21.Johnson R D, Symington L S. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovett S T, Mortimer R K. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thacker J. Trends Genet. 1999;15:166–168. doi: 10.1016/s0168-9525(99)01733-3. [DOI] [PubMed] [Google Scholar]
- 24.Cartwright R, Tambini C E, Simpson P J, Thacker J. Nucleic Acids Res. 1998;26:3084–3089. doi: 10.1093/nar/26.13.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W F, et al. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 26.Jones N J, Cox R, Thacker J. Mutat Res. 1987;183:279–286. doi: 10.1016/0167-8817(87)90011-3. [DOI] [PubMed] [Google Scholar]
- 27.Fuller L F, Painter R B. Mutat Res. 1988;193:109–121. doi: 10.1016/0167-8817(88)90041-7. [DOI] [PubMed] [Google Scholar]
- 28.Albala J S, Thelan M P, Prange C, Fan W, Christensen M, Thompson L H, Lennon G G. Genomics. 1997;46:476–479. doi: 10.1006/geno.1997.5062. [DOI] [PubMed] [Google Scholar]
- 29.Rice M C, Smith S T, Bullrich F, Havre P, Kmiec E B. Proc Natl Acad Sci USA. 1997;94:7417–7422. doi: 10.1073/pnas.94.14.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartwright R, Dunn A M, Simpson P J, Tambini C E, Thacker J. Nucleic Acids Res. 1998;26:1653–1659. doi: 10.1093/nar/26.7.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dosanjh M K, Collins D W, Fan W F, Lennon G G, Albala J S, Shen Z Y, Schild D. Nucleic Acids Res. 1998;26:1179–1184. doi: 10.1093/nar/26.5.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pittman D L, Weinberg L R, Schimenti J C. Genomics. 1998;49:103–111. doi: 10.1006/geno.1998.5226. [DOI] [PubMed] [Google Scholar]
- 33.Thompson L H, Schild D. Biochimie. 1999;81:87–105. doi: 10.1016/s0300-9084(99)80042-x. [DOI] [PubMed] [Google Scholar]
- 34.Tsutsui Y, Morishita T, Iwasaki H, Toh H, Shinagawa H. Genetics. 2000;154:1451–1461. doi: 10.1093/genetics/154.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tebbs R S, Zhao Y, Tucker J D, Scheerer J B, Siciliano M J, Hwang M, Liu N, Legerski R J, Thompson L H. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui X, Brenneman M, Meyne J, Oshimura M, Goodwin E H, Chen D J. Mutat Res DNA Repair. 1999;434:75–88. doi: 10.1016/s0921-8777(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 37.Griffin C S, Simpson P J, Wilson C R, Thacker J. Nat Cell Biol. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 38.Johnson R D, Liu N, Jasin M. Nature (London) 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 39.Pierce A J, Johnson R D, Thompson L H, Jasin M. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deans B, Griffin C S, Maconochie M, Thacker J. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pittman D L, Schimenti J C. Genesis. 2000;26:167–173. doi: 10.1002/(sici)1526-968x(200003)26:3<167::aid-gene1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 42.Braybrooke J P, Spink K G, Thacker J, Hickson I D. J Biol Chem. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- 43.Schild D, Lio Y C, Collins D W, Tsomondo T, Chen D J. J Biol Chem. 2000;275:16443–16449. doi: 10.1074/jbc.M001473200. [DOI] [PubMed] [Google Scholar]
- 44.Shu Z G, Smith S, Wang L J, Rice M C, Kmiec E B. Mol Cell Biol. 1999;19:8686–8693. doi: 10.1128/mcb.19.12.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingraham S E, Lynch R A, Kathiresan S, Buckler A J, Menon A G. Cancer Genet Cytogenet. 1999;115:56–61. doi: 10.1016/s0165-4608(99)00070-9. [DOI] [PubMed] [Google Scholar]
- 46.Schoenmakers E F P M, Huysmans C, Van de Ven W J M. Cancer Res. 1999;59:19–23. [PubMed] [Google Scholar]
- 47.Barlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi O P, Kallioniemi A. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 48.Wu G J, Sinclair C S, Paape J, Ingle J N, Roche P C, James C D, Couch F J. Cancer Res. 2000;60:5371–5375. [PubMed] [Google Scholar]
- 49.Takata M, Sasaki M S, Sonoda E, Fukushima T, Morrison C, Albala J S, Swagemakers S M A, Kanaar R, Thompson L H, Takeda S. Mol Cell Biol. 2000;20:6476–6482. doi: 10.1128/mcb.20.17.6476-6482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takata M, Sasaki M S, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson L H, Takeda S. Mol Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies A A, Masson J-Y, McIlwraith M J, Stasiak A Z, Stasiak A, Venkitaraman A R, West S C. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 52.Havre P A, Rice M, Ramos R, Kmiec E B. Exp Cell Res. 2000;254:33–44. doi: 10.1006/excr.1999.4725. [DOI] [PubMed] [Google Scholar]
- 53.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sogo J, Stasiak A, De Bernadin W, Losa R, Koller T. In: Electron Microscopy in Molecular Biology. Sommerville J, Scheer U, editors. Oxford: IRL; 1987. pp. 61–79. [Google Scholar]
- 55.Passy S I, Yu X, Li Z, Radding C M, Masson J-Y, West S C, Egelman E H. Proc Natl Acad Sci USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Dyck E, Hajibagheri N M A, Stasiak A, West S C. J Mol Biol. 1998;284:1027–1038. doi: 10.1006/jmbi.1998.2203. [DOI] [PubMed] [Google Scholar]
- 57.Paull T T, Rogakou E P, Yamazaki V, Kirchgessner C U, Gellert M, Bonner W M. Curr Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 58.Golub E I, Gupta R C, Haaf T, Wold M S, Radding C M. Nucleic Acids Res. 1998;26:5388–5393. doi: 10.1093/nar/26.23.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y L, Li M J, Lee E Y H P, Maizels N. Curr Biol. 1999;9:975–978. doi: 10.1016/s0960-9822(99)80427-8. [DOI] [PubMed] [Google Scholar]
- 60.Tan T L R, Essers J, Cittero E, Swagemakers S M A, de Wit J, Benson F E, Hoeijmakers J H J, Kanaar R. Curr Biol. 1999;9:325–328. doi: 10.1016/s0960-9822(99)80142-0. [DOI] [PubMed] [Google Scholar]
- 61.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 62.Chen J J, Silver D P, Walpita D, Cantor S B, Gazdar A F, Tomlinson G, Couch F J, Weber B L, Ashley T, Livingston D M, Scully R. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 63.Bishop D K, Ear U, Bhattacharyya A, Calderone C, Beckett M, Weichselbaum R R, Shinohara A. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 64.Sonoda E, Takata M, Yamashita Y M, Morrison C, Takeda S. Proc Natl Acad Sci USA. 2001;98:8388–8394. doi: 10.1073/pnas.111006398. [DOI] [PMC free article] [PubMed] [Google Scholar]