Abstract
Exome sequence analysis can be instrumental in identifying the genetic etiology behind atypical disease. We report a patient presenting with microcephaly, dysmorphic features, and intellectual disability with a tentative diagnosis of Dubowitz syndrome. Exome analysis was performed on the patient and both parents. A de novo missense variant was identified in ACTB, c.349G>A, p.E117K. Recent work in Baraitser-Winter syndrome has identified ACTB and ACTG1 mutations in a cohort of individuals and we rediagnosed the patient with atypical Baraitser-Winter syndrome. We performed functional characterization of the variant actin and show that it alters cell adhesion and polymer formation supporting its role in disease. We present the clinical findings in the patient, comparison of this patient to other patients with ACTB/ACTG1 mutations, and results from actin functional studies that demonstrate novel functional attributes of this mutant protein.
Keywords: actin, ACTB, Dubowitz, Baraitser-Winter syndrome
Introduction
Exome sequence analysis has been adopted as the method of choice for analysis of patient samples when a clinical diagnosis is not available to support the ordering of a single gene test. This approach is effective with a success rate reported to be approximately 50% [Gilissen et al., 2011; Need et al., 2012] depending on the types of conditions being studied and the samples available. Here we present a female patient with an initial clinical diagnosis of Dubowitz syndrome (MIM# 223370) at the time of presentation. Her major manifestations included microcephaly, dysmorphic facial features, and intellectual disability. Her diagnosis was changed based on the molecular findings to Baraitser-Winter syndrome (BRWS1, MIM# 243310; BRWS2, MIM# 614583) with absence of some of the characteristic findings of this diagnosis including lissencephaly, seizures, and iris/retinal colobomas.
Materials and Methods
Patients
Information on clinical manifestations was collected on the proband. Blood samples were collected on the proband and her parents for DNA analysis. The NHGRI IRB reviewed and approved this study.
DNA Library preparation and sequencing
DNA library preparation and sequencing for the proband and both parents were performed at the NIH Intramural Sequencing Center (NISC) using their exome analysis pipeline. Whole genome libraries with ~280 base inserts were prepared according to the manufacturer’s instructions (TruSeq DNA Sample Preparation v2 method, Illumina Corp, San Diego, CA). Batches of 24 uniquely barcoded libraries were pooled using equal volumes of input and run on a MiSeq with version 1 chemistry at a loading concentration of 6 pM. The run consisted of 25 cycles followed by an index read. The demultiplexed read counts were used to normalize the DNA input for exome capture where six libraries were pooled together. The exome capture was preformed according to the manufacturer’s instructions (Illumina’s TruSeq Exome Enrichment Kit protocol, Illumina Corp, San Diego, CA).
Each captured exome pool was sequenced in two lanes on a HiSeq2000 using version 3 chemistry. At least 40 million paired-end 100 base reads were obtained for each sample. Data were processed using RTA ver. 1.13.48 and CASAVA 1.8.2 (Illumina Corp, San Diego, CA).
The presence/absence of the de novo candidate variant was verified in the proband and both parents using Sanger sequence analysis according to standard protocols [Johnston et al., 2005]. The variant has been submitted to the ACTB locus specific database (www.lovd.nl/ACTB).
Read mapping, variant calling and annotation
Reads were aligned to a human reference sequence (UCSC assembly hg18, NCBI build 36) using efficient large-scale alignment of nucleotide databases (ELAND). Reads that aligned uniquely were grouped into genomic sequence intervals of about 100 kb, and reads that failed to align were binned with their paired-end mates. Reads in each bin were subjected to a Smith-Waterman-based local alignment algorithm, cross_match using the parameters –minscore 21 and –masklevel 0 to their respective 100 kb genomic sequence (http://www.phrap.org). Genotypes were called at all positions where there were high-quality sequence bases (Phred-like Q20 or greater) using a Bayesian algorithm (Most Probable Genotype – MPG)[Teer et al., 2010].
Lymphocyte Adhesion Assays
Human fibronectin was purified from outdated human platelets as previously described [Ruoslahti et al., 1982]. Dishes were coated with 20 ug/mL fibronectin overnight at 4 °C. 1×105 lymphoblasts from individual W2O.1 (proband), W2O.2 (father), or W2O.3 (mother) were plated and allowed to adhere for 48 hours at 37 °C. For adhesion assays, 15 fields of view (FOV) were counted prior to, and immediately following a gentle wash with PBS. The cells were visualized by staining with a Texas Red conjugated phalloidin (Invitrogen, Grand Island, NY) and manually counted. Percent adhesion was calculated by dividing the number of cells remaining post-wash by the number of cells adhered pre-wash.
Lymphocyte Imaging
Glass coverslips were coated with 20 µg/mL fibronectin overnight at 4 °C. Cells from W20.1, W20.2, or W20.3 were plated and allowed to adhere for 2 days. Cells were then fixed in 3.7% paraformaldehyde in PBS, permeabilized in 0.5% Triton X-100 in universal buffer (UB: 150 mM NaCl, 50 mM Tris pH 7.6, 0.01% NaN3) for 10 minutes and washed in a tris based buffer. For actin staining, the cells were incubated with a Texas-Red conjugated phalloidin, washed, and mounted. Images were captured with a confocal microscope (model LSM 510; Carl Zeiss Microscopy, Thornwood, NY) using a 63X oil objective (Carl Zeiss Microscopy, Thornwood, NY) with an NA of 1.4. Images were obtained using the LSM Image Browser (Carl Zeiss Microscopy, Thornwood, NY) as described [Peng et al., 2010].
Yeast in vivo Studies
A haploid yeast strain expressing mutant p.E117K (E117K) actin as the only actin in the cell was constructed using a site-directed mutagenesis approach followed by selection of the appropriate strain based on the use of nutritional markers as previously described [Bergeron et al., 2011]. Measurement of cell growth in rich liquid YPD medium and on agar plates under different stress or nutrient conditions was performed as described [Bryan et al., 2006]. To visualize the actin cytoskeleton, cells were fixed in 3% formaldehyde and stained with 0.5 µM Alexa 488-phalloidin (Invitrogen, Grand Island, NY) overnight at 4 °C. Stained cells were observed with a Zeiss confocal microscope as described [McKane et al., 2005]. The 3-D images of the cells were stacked using ImageJ v1.47e (rsweb.nih.gov/ij/) into 2-D images, displays were used to assess cell size and cytoskeletal normality and to quantitate these parameters.
In vitro Actin Studies
Actin purification
The wild type (WT) and E117K actins were purified from baker’s yeast cake purchased from a local grocery store (WT) or from mutant cells grown in the laboratory using a combination of DNase I-agarose affinity chromatography, DE52 anion-exchange chromatography, and polymerization/depolymerization cycling as described [Malloy et al., 2012] with one modification. Extensive work (data not shown) has demonstrated that there is no difference in the behavior of WT actin purified from either commercially purchased yeast cakes or cells grown under the same culture conditions as those expressing the mutant actins. As a final step in an attempt to prepare G-actin (globular actin as opposed to filamentous or F-actin), we centrifuged the actin preparation to remove actin oligomers. Yet, with E117K actin, light scattering showed extensive evidence for residual actin oligomers/aggregates. We then filtered the “G-actin” preparation with a Whatman Anotop 10 0.1 µm pore size filter immediately before polymerization experiments to try to remove these bodies. The concentration of G-actin was determined from the absorbance at 290 nm (ε290nm=0.63M−1cm−1). Actin was stored in G buffer (10 mM Tris-HCl pH 7.5, 0. 2 mM CaCl2, 0.2 mM ATP, and 0.1 mM DTT) at 4 °C and used within two days.
Actin polymerization and filament visualization by electron microscopy
Polymerization of G-actin in a total volume of 120 µl was induced by the addition of MgCl2 and KCl to final concentrations of 2 mM and 50 mM, respectively. Polymerization was performed at 25 °C. The change in light scattering intensity as an indication of polymerization was recorded using a FluoroMax-3 fluorescence spectrometer with a four-sample exchanger and thermostated control (HORIBA Jobin Yvon, Inc., Edison, NJ). The excitation and emission wavelength were both set at 360 nm.
To visualize actin filaments, a 2 µl sample from the polymerization reaction, following attainment of steady state, was applied to a carbon-coated Formvar grid followed by negative staining with 1% uranyl acetate. The images were captured with a JEM-1230 transmission electron microscope (Jeol Inc, Tokyo, Japan) equipped with an UltraScan camera (Gatan, Pleasanton, CA) in the University of Iowa Central Microscopy Facility. All experiments were repeated at least twice with similar results.
Dynamic light scattering
A 20 µl sample of G-actin in G buffer at 1 µM concentration, filtered as described above, was placed in a cuvette, and the dynamic light scattering was monitored over time with a DynaPro Nanostar (Wyatt Technology, Santa Barbara, CA) temperature controlled instrument at 25 °C. The data were recorded and further analyzed with a Dynamics 7.1.7 software package (Wyatt Technology, Santa Barbara, CA). The experiments with individual actin preparations were repeated twice with similar outcomes.
Molecular Modeling
Assessment of the perturbation of local structure based on charge and mass differences was performed with PyMOL software (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC, Portland, OR).
Results
Clinical Report
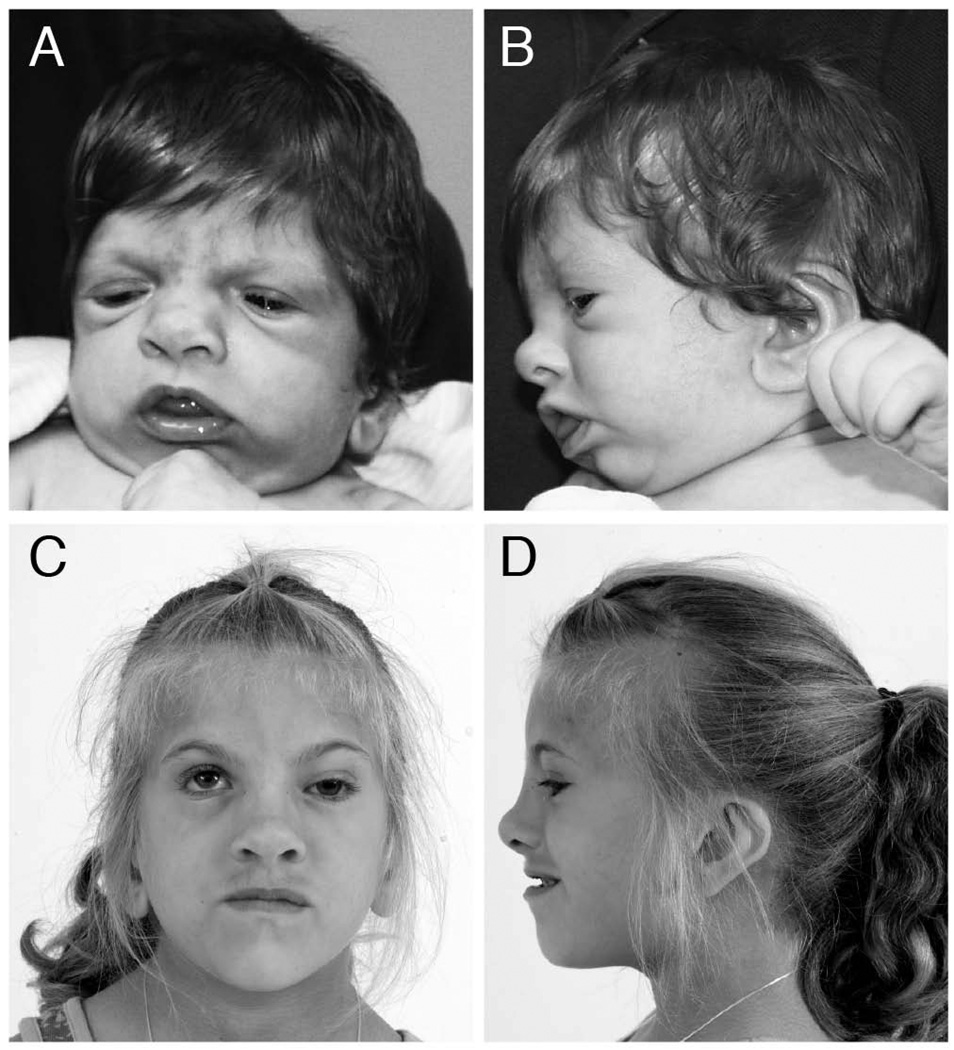
The patient was born at 40 weeks’ estimated gestational age by repeat Cesarean to a healthy 28-year-old with two prior pregnancies and no miscarriages (G3P2A0). The father was 29 years old and the family history was negative for similarly affected individuals. At birth, she weighed 2.4 kg (small for gestational age), and she was noted to have low placed ears, unilateral ptosis, low anterior hairline and mild hypertrichosis. All her growth parameters plotted at <3rd centile up until 2 years of age after which her height and weight normalized but her occipital frontal circumference (OFC) remained at the 3rd – 10th centile (OFC 49 cm at 7 years). On examination at 4 years 5 months, she had metopic ridging with apparent microcephaly and a hoarse voice (Figure 1). Notable craniofacial findings included: asymmetric positioning of the globes with left ptosis, short palpebral fissures and apparent widely spaced eyes; posteriorly rotated ears with abnormally shaped pinnae; broad nasal root, long columella, alae nasae flaring; prominent tongue, wide mouth, and bifid uvula. Limb findings included distally placed thumbs, small thenar eminence, camptodactyly of digits 3, 4, and 5, and prominent fingertip pads. She had small fifth toenails with fifth digit clinodactyly.
Figure 1.
Craniofacial findings of patient. A: Frontal and B: Lateral views at 2 months of age show metopic ridging, low anterior hair line, broad nasal root, short palpebral fissures, low placed ears with prominent lobes, micrognathia. C: Frontal and D: Lateral views at 7 years of age show left ptosis, broad nasal root, prominent tip and long columella, and low placed ears with prominent lobes.
Her cognitive evaluation at 6 years showed functioning at the level average for a 3 year 6 month old. At 7 years, she has obsessive-compulsive behaviors and hyperactivity, treated successfully with guanfacine.
Medical issues included severe myopia in her right eye treated with a contact lens, and optic nerve asymmetry. Otolaryngology evaluation showed recurrent middle ear effusion with small external auditory canals. Hearing evaluation showed mild conductive hearing loss (500–2000 Hz) of 25 db. She had treated left vertical talus.
Normal laboratory test results included: chromosome analysis, FISH for sub-telomere analysis, chromosome microarray analysis, 7-dehydrocholesterol, and immunoglobulins A and M, IGFI, IGFII and IGFBPIII levels. Radiological studies included: bone age at chronologic age 2 years (delayed); skeletal survey (normal, except shallow acetabulae), MRI scan of the brain at 34 months (normal), renal ultrasound (normal). Echocardiogram showed a bicuspid aortic valve.
Exome Sequence Analysis
Exome sequence data were generated for the proband and both parents. A total of 164,694 quality variants, MPG ≥10 and MPG/coverage ≥0.5, were identified in the proband. Filtering for quality across all three individuals in the trio reduced the variant count to 131,656. The analysis was restricted to variants predicted to result in non-synonymous, nonsense, frame shifting, or splice site alterations. The initial filtering strategy was based on the hypothesis that the likely causative variant would be a de novo mutation in the proband. A total of three variants were identified that were apparently de novo in the patient and not present in a control cohort of 951 individuals or the NHLBI exome server dataset (evs.gs.washington.edu; 12/20/2012). Two variants were in highly repetitive regions and were removed from the analysis. The remaining variant was a novel variant in ACTB (MIM# 102630, NM_001101.3) c.349G>A, which predicts p.E117K. This variant was confirmed to be de novo in the proband. Parentage was confirmed by the thousands of variants in common between the child and her parents on the exome trio analysis.
Functional Studies of ACTB p.E117K
Mutations in ACTB and other actins have been documented to cause disease and the discovery of a mutation in ACTB in this patient led us to determine if we could document functional defects in the patient’s cells that could be attributed to defective actin. Using lymphocytes obtained from the patient and both parents, we assessed the ability of the cells to adhere to a fibronectin-coated surface. The patient’s cells demonstrated a significantly decreased ability to adhere compared with that of the parents (Supp. Figure S1). Additionally, while the parent’s cells formed actin rich protrusions the patient’s cells demonstrated few protrusive structures (Supp. Figure S2). It is likely that the reduction in protrusive structures, with the concomitant reduction in contact with the fibronectin coated surface, reduced the adhesive properties of the cells.
To study the biochemical consequences of the p.E117K mutation we cloned the mutation into the actin from the budding yeast S. cerevisiae and created a strain expressing only the mutant actin. Yeast actin is 90% identical in sequence to human β-actin and it is an essential protein. We first assessed the effect of the mutation on growth of yeast in liquid rich medium and under various stress conditions, which are often sensitive to actin cytoskeletal defects. Growth of the mutant cells in this medium levels off at a much lower cell density than WT cells (Supp. Figure S3). In yeast, proper cytoskeletal function is required for growth on glycerol as a sole carbon source (a measure of mitochondrial function) or growth in hyperosmolar medium. In both of these environments, growth of the mutant cells was severely affected relative to WT cells.
Altered actin cytoskeletal behavior in yeast can result in the generation of larger and rounder cells than WT yeast due to loss of a polarized cytoskeleton and resulting isotropic growth. The average sizes of the E117K and WT cells were 18.8 ± 6.4 and 11.4 ± 3.1 µm2, respectively (Supp. Figure S4). The extent to which cytoskeletal polarity had been compromised was determined by examining fixed cells after visualization of the cytoskeleton with fluorescent phalloidin. The normal yeast actin cytoskeleton early in the cell cycle displays endocytic actin patches in the bud membrane and polarized actin cables running along the longitudinal axis of the budding cell. The mutant cells showed virtually complete loss of this polarization (Supp. Figure S5). About half the cells had short randomly organized actin aggregates or short filament fragments, and the other half had, in addition, a few longer polarized actin cables but to a much lesser extent than observed with WT cells. This disruption is consistent with the larger, rounder cells described above.
The disorganized short actin cables suggested the mutation might have interfered with filamentous-actin (F-actin) dynamics. To further explore this possibility, we compared the ability of WT and mutant cells to grow in the presence of latrunculin A. This drug binds to G-actin ultimately resulting in the depolymerization of actin in the cell. The mutant cells were almost completely resistant to the concentrations of drug tested (Supp. Figure S6), suggesting that the p.E117K mutation might result in strengthened actin monomer-monomer interactions and increased filament stability.
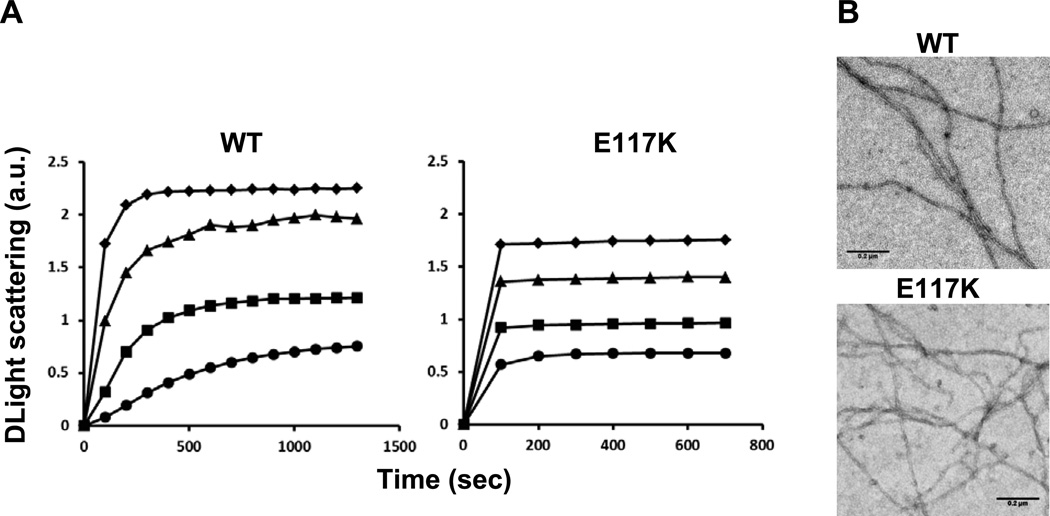
The in vivo studies suggested that the p.E117K mutation adversely affected normal actin polymerization and its regulation. To assess the extent to which this was true, both WT and mutant actin were purified. Yields of mutant actin were substantially decreased, often only about 10% of what we normally achieve. Furthermore, initial attempts to follow actin polymerization using light scattering increase as an assay indicated that the actin could not be initially converted to its monomeric G-actin form by dialysis against G-buffer (0.2 mg/ml E117K, 1.4 absorbance units (au); 0.2 mg/ml WT actin, 0.1 au). To circumvent this problem, we subjected the mutant actin solution to filtration through a 0.1 µm pore size filter to remove residual aggregates and filaments. Polymerization was then initiated by the addition of KCl and MgCl2. Figure 2 shows normal polymerization kinetics for increasing amounts of WT G-actin. In comparison, polymerization of mutant actin was almost complete in the time it took to add salt and restart the fluorimeter. This behavior suggested the light scattering increase resulted from annealing of actin oligomers or aggregates into filaments, thereby bypassing the need for the normally rate-limiting filament nucleation step. To determine if this hypothesis was true and the result did not represent some amorphous aggregation process, we examined negatively stained samples of the mixtures under the electron microscope at the end of the polymerization reaction. In both WT and mutant actin samples, normal appearing filaments were present.
Figure 2.
Effect of the p.E117K mutation on polymerization of purified actin. Purified yeast G-actin was subject to centrifugation followed by filtration as described in the methods. Polymerization was initiated by the addition of MgCl2 and KCl as described, and the increase in light scattering was followed over time as an indication of polymerization. Shown is the net change in light scattering for each sample. Key: ● 3.5 µM; ■ 4.6 µM; ▲ 5.8 µM; ◆ 6.9 µM. B. Electron micrographs of samples taken from the 3.5 µM reaction following attainment of steady state.
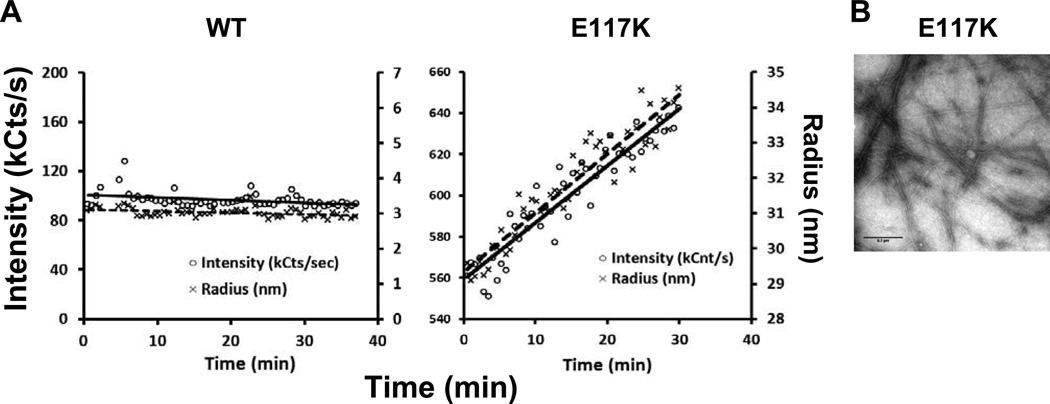
The presumptive G-actin self-assembling behavior described above led to us to use dynamic light scattering to further examine the nature and behavior of the aggregates that might be responsible. Figure 3 shows that over time, WT G-actin behaves as a stable population of 43 kD monomers. The behavior of the mutant “monomer” solution, however, was much different. Immediately following polymerization, the residual scattering began at a higher value, indicative of larger bodies, and increased linearly over time, even though the actin was in G-buffer. Examination of this material showed the presence of short, well-formed filaments as if the smaller aggregates were still capable of annealing into bona fide filament structures. Thus, the mutation seemed to impart unusual stability onto actin nuclei or oligomers that facilitated their assembly into longer filaments.
Figure 3.
Dynamic light scattering analyses of filtered WT and mutant G-actin samples. A: One micromolar G-actin samples were placed in a dynamic light scattering apparatus and the change in scattering was followed over time at room temperature as described in the methods. For each panel, the left vertical axis depicts the signal intensity, and the right vertical axis depicts the average radius of bodies in the sample. B: Short filaments were present in the 6.9 mM mutant G-actin sample in G-buffer after 1 hr. No filaments were present in the WT sample.
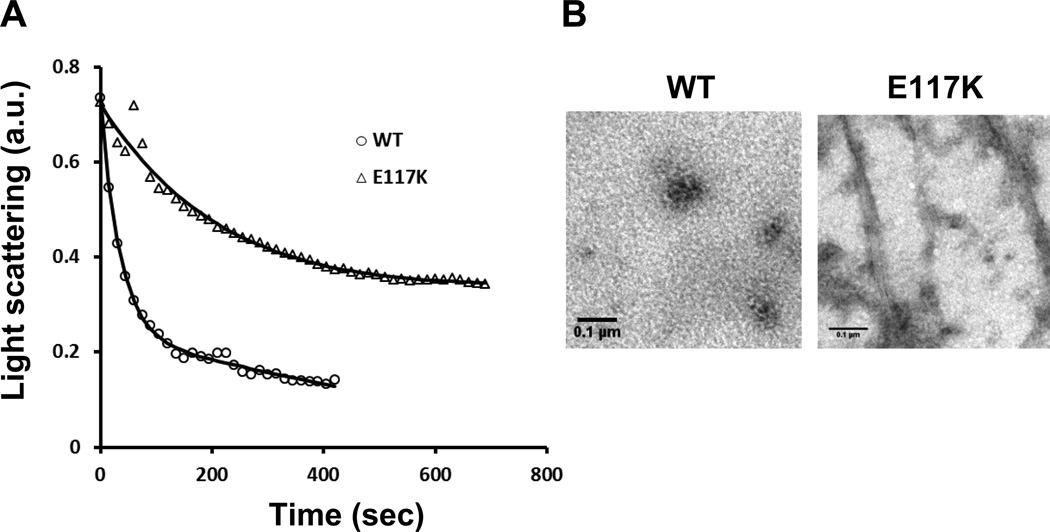
Finally, in light of the aberrant G-actin behavior and the resistance of the mutant cells to latrunculin A, we determined the extent to which latrunculin A, which sequesters actin monomers and leads to F-actin depolymerization, could drive mutant F-actin into a monomeric state in vitro. Figure 4 demonstrates that exposure of WT F-actin to latrunculin A in filament stabilizing buffer resulted in complete filament disassembly. Conversely, substantial residual light scattering remained with the mutant actin sample, and EM visualization showed the presence of distinct filaments and filament bundles, in agreement with the hyperstability phenotype described above.
Figure 4.
Latrunculin A-dependent depolymerization of 3.5 mM WT and mutant F-actin samples. A: Time course of WT and mutant F-actin depolymerization. 20 mM latrunculin A was added to each sample, and the decrease in light scattering at room temperature was followed over time. B: Electron micrographs of the WT and mutant actin samples at the end of the depolymerization reaction showing residual filament bundles in the mutant, but not WT, sample.
Discussion
The patient reported here presented with a diagnosis of Dubowitz Syndrome based on the combination of microcephaly, ptosis, low set ears, and intellectual delay [Kuster and Majewski, 1986]. As the genetic etiology of Dubowitz syndrome is not understood, the patient and her parents were subjected to exome sequence analysis and a de novo alteration in ACTB was identified in the patient, c.349G>A, p.E117K. Although this mutation has not previously been reported, six other mutations in ACTB have been reported to cause disease. An individual was reported with p.E364K associated with neutrophil dysfunction and recurrent infection, but no reported dysmorphic or cognitive abnormalities [Nunoi et al., 1999]. Interestingly, p.E364K does not affect in vitro actin polymerization and is in a region of the protein important for profilin binding, suggesting this alteration may have a very different effect on the cellular function of actin resulting in the distinctly different phenotype. A set of twins was described with p.R183W [Procaccio et al., 2006]. Their phenotypes included cleft palate and lip, widely spaced eyes, esophageal motility issues, severe bilateral sensory hearing loss, and progressive delayed onset generalized dystonia. The p.R183W variant has also been shown to affect actin polymerization. In addition, four alterations in ACTB have been reported in patients with BRWS1 and some of these alterations have been shown to result in increased filamentous actin in lymphoblasts and resistance to latrunculin A suggesting an effect on actin polymerization [Riviere et al., 2012]. The phenotype of the patient presented here overlaps the phenotypic features reported for individuals with mutations in ACTB (MIM# 102630) and ACTG1 (MIM# 102560) Table 1 [Riviere et al., 2012]. However, the patient reported here does not exhibit some of the common features of BRWS1 or 2 including lissencephaly, seizures, sensorineural hearing loss and iris/retinal colobomas.
Table 1.
Phenotypic overlap of patient W20.1 presented here and individuals reported to have mutations in ACTB and ACTG1 in the literature [Riviere et al., 2012]
| BRWS1 ACTB |
BRWS2 ACTG1 |
W20.1 | |
|---|---|---|---|
| Growth | |||
| Short Stature | 6/10 | 3/7 | + |
| Microcephaly postnatal | 6/9 | 4/7 | + |
| Neurological | |||
| Intellectual Disability | 9/9 | 5/5 | + |
| Hearing loss | 4/8 | 5/6 | +, conductive |
| Seizures | 9/9 | 7/8 | − |
| Facial dysmorphism | |||
| Metopic ridging | 8/10 | 7/7 | + |
| Widely spaced eyes | 10/10 | 7/8 | + |
| High-arched eyebrows | 10/10 | 7/7 | − |
| Ptosis, congenital | 10/10 | 8/8 | +, unilateral |
| Eyes | |||
| Coloboma | 6/10 | 5/7 | − |
| Lissencephaly-type pachygria | 8/8 | 7/7 | − |
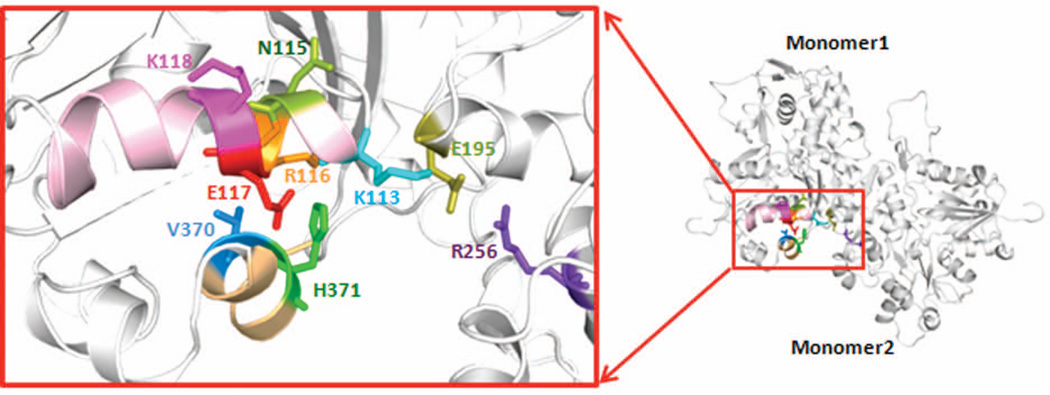
Figure 5 shows that the p.E117K mutation in β-actin falls in a system of residues consisting of a helix (residues 113–118), which appears to interact across the strand-strand interface of the actin filament with residues 256 and 195 in another monomer. Studies have shown that the cross-strand interaction may be important in regulating actin filament stability, and the helix may provide an allosteric conduit for transmitting conformation changes between the filament interior and binding sites for F-actin regulatory proteins such as Arp2/3 complex and formin in the area of residue 118 [Kruth and Rubenstein, 2012; Malloy et al., 2012]. While no alterations have previously been reported in residue 117, a number of pathogenic mutations originating in five of the six human actin paralogs cluster in this structural element. These mutations have been shown to give rise to a number of diseases including thoracic aortic aneurysm and dissection (MIM# 611788; ACTA2, MIM# 102620, [Guo et al., 2007]), nemaline myopathy (MIM# 161800; ACTA1, MIM# 102610, [Laing et al., 2009; Nowak et al., 1999; Sparrow et al., 2003]), early onset nonsyndromic deafness, DFNA 20/26 (MIM# 604717; ACTG1, MIM# 102560, [Morin et al., 2009; Zhu et al., 2003]) and BRWS1 and 2 (ACTB, ACTG1, [Riviere et al., 2012]). The adhesion defects we documented in cells from the patient with the p.E117K mutation coupled with the effects of this mutation on the behavior of the mutant yeast in vivo and on the behavior of purified mutant actin in vitro are all consistent with the ACTB p.E117K mutation being the cause of the disorder in this patient.
Figure 5.
Structural model of the pathogenic helix containing residue 117 and the residues with which the helix interacts. The structure was generated based on the Oda F-actin trimer model [Oda et al., 2009] using PyMol as described.
A particularly striking result from our yeast in vivo and in vitro studies is the severity of the effects of this mutation on actin polymerization behavior compared with other mutations in this actin structural element referred to above. A clue to the reason for this added severity may be found from inspection of the actin structure in the area of the mutation. Figure 5 shows that residue 117, of all the residues in the 113–118 helix, interacts with residue 371 of the same monomer, very near the actin C-terminus. The substitution of glutamate with lysine should cause substantial perturbation of local structure because of both charge and mass differences based on modeling studies using PyMOL. The C-terminal peptide has been previously shown to be important in the allosteric behavior governing actin polymerization and is an important factor in establishing a proper monomer-monomer interface longitudinally in one strand of the actin filament [Crosbie et al., 1994; Hegyi et al., 1998; Johannes and Gallwitz, 1991; Kim et al., 1998; Orlova and Egelman, 1995; Phan et al., 1997].
Proper actin cytoskeletal behavior requires careful regulation of actin filament assembly both temporally and spatially. In the cell, this regulation is generally controlled by a series of actin binding proteins. Most mutations we have studied in the 113–118 helix seem to affect the interaction between actin and these regulatory proteins with lesser effects on the actin’s inherent polymerization properties. In the case of the K118 mutants (ACTG1, deafness), for example, K118M and WT actins polymerized with essentially the same kinetics, while K118N actin polymerized about twice as fast [Bryan and Rubenstein, 2009; Kruth and Rubenstein, 2012; Morin et al., 2009]. The N115T actin (ACTA2, p.N117T, aortic aneurysm) polymerized slightly more slowly than WT but to the same extent [Bergeron et al., 2011]. Conversely, R116Q actin (ACTA2, p.R118Q, aortic aneurysm), adjacent to 117, exhibited a prolonged nucleation phase and slower polymerization. All of these actins transformed to G-actin in low salt buffer, contrary to the case with the E117K mutant, which seemed to be sequestered in small aggregates or oligomers. The more profound effect of the p.E117K mutation on actin polymerization and filament stability may have sufficiently disrupted actin filament dynamics to impart the severity of the effects we have outlined. In agreement with this notion, the ACTG1 p.V370A mutation, which causes deafness [Rendtorff et al., 2006], produced polymerization defects in purified yeast actin very similar to what we observed with the E117K mutation [Bryan et al., 2006]. Modeling showed that V370 is part of a system that can interact with mutant K117, in line with this theory. Furthermore, the ACTA1 p.V372F nemaline myopathy mutation, when cloned into yeast actin (yeast V370F) was incompatible with yeast cell viability [Nowak et al., 1999].
We hypothesize that the distinct disease phenotypes associated with the various β-actin mutations reflect the sensitive allosteric regulation associated with normal actin cytoskeletal dynamics and the sensitivity that developmental processes have to alterations in cytoskeletal behavior. The phenotype associated with a particular mutation will be affected by the extent to which a particular mutation affects the allosteric behavior of this peptide, resulting in either differential effects on filament dynamics or the ability of actin to be controlled by regulatory proteins that interact with this allosteric system. The situation is further complicated by the fact that there are two different nonmuscle actin isoforms in nonmuscle cells, β- and γ- (ACTB and ACTG1), and they are present at different ratios in different cell types [Furness et al., 2005; Hofer et al., 1997; Otey et al., 1987; Vandekerckhove and Weber, 1981]. The absolute amount of the normal vs. mutant β-actin in a particular cell in addition to the β/γ ratio may also be important factors in dictating the extent to which a particular actin mutation affects normal development. The complexity arising from the presence of multiple actin isoforms in the same cell requires the use of cells expressing only a single mutant actin to gain insight into the biochemical and biophysical effects of disease-causing actin mutations [Rubenstein and Mayer, 2012]. The studies described here demonstrate the utility of this approach.
We have identified a novel mutation in ACTB in an individual with phenotypic features that overlap BRWS1. Whether this phenotype should be considered to be an atypical form of BRWS1 or a distinct phenotype awaits the description of additional patients with mutations in this gene, most of whom we predict will be identified by clinical and research exome sequencing. These data also illustrate the challenge with studying Dubowitz syndrome. Patients with this diagnosis are clinically and molecular heterogeneous (Biesecker and Stewart, unpublished data). As was the case for the current patient, we expect that many patients currently diagnosed with Dubowitz syndrome will be reclassified based on molecular data. Like many genes, alterations in ACTB cause a spectrum of phenotypes, both overlapping and distinct. Further functional analysis of the actin alterations identified in patients with BRWS will be required to fully understand the correlation between phenotype and actin function.
Supplementary Material
Acknowledgments
We thank Julia Fekecs for graphic design services. PAR acknowledges support from NIH NIDCD grant DC008803. KAD acknowledges support from the National Science Foundation Grant 1120478. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
LGB is an uncompensated advisor to the Illumina Corp. and his group receives in-kind support for collaborative work distinct from that described here.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
Conflict of interest
All other authors declare no conflict of interest.
References
- Bergeron SE, Wedemeyer EW, Lee R, Wen KK, McKane M, Pierick AR, Berger AP, Rubenstein PA, Bartlett HL. Allele-specific effects of thoracic aortic aneurysm and dissection alpha-smooth muscle actin mutations on actin function. J Biol Chem. 2011;286:11356–11369. doi: 10.1074/jbc.M110.203174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan KE, Rubenstein PA. Allele-specific effects of human deafness gamma-actin mutations (DFNA20/26) on the actin/cofilin interaction. J Biol Chem. 2009;284:18260–18269. doi: 10.1074/jbc.M109.015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan KE, Wen KK, Zhu M, Rendtorff ND, Feldkamp M, Tranebjaerg L, Friderici KH, Rubenstein PA. Effects of human deafness gamma-actin mutations (DFNA20/26) on actin function. J Biol Chem. 2006;281:20129–20139. doi: 10.1074/jbc.M601514200. [DOI] [PubMed] [Google Scholar]
- Crosbie RH, Miller C, Cheung P, Goodnight T, Muhlrad A, Reisler E. Structural connectivity in actin: effect of C-terminal modifications on the properties of actin. Biophys J. 1994;67:1957–1964. doi: 10.1016/S0006-3495(94)80678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness DN, Katori Y, Mahendrasingam S, Hackney CM. Differential distribution of beta- and gamma-actin in guinea-pig cochlear sensory and supporting cells. Hear Res. 2005;207:22–34. doi: 10.1016/j.heares.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo DC, Pannu H, Tran-Fadulu V, Papke CL, Yu RK, Avidan N, Bourgeois S, Estrera AL, Safi HJ, Sparks E, Amor D, Ades L, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- Hegyi G, Mak M, Kim E, Elzinga M, Muhlrad A, Reisler E. Intrastrand cross-linked actin between Gln-41 and Cys-374. I. Mapping of sites cross-linked in F-actin by N-(4-azido-2-nitrophenyl) putrescine. Biochemistry. 1998;37:17784–17792. doi: 10.1021/bi981285j. [DOI] [PubMed] [Google Scholar]
- Hofer D, Ness W, Drenckhahn D. Sorting of actin isoforms in chicken auditory hair cells. J Cell Sci. 1997;110(Pt 6):765–770. doi: 10.1242/jcs.110.6.765. [DOI] [PubMed] [Google Scholar]
- Johannes FJ, Gallwitz D. Site-directed mutagenesis of the yeast actin gene: a test for actin function in vivo. Embo J. 1991;10:3951–3958. doi: 10.1002/j.1460-2075.1991.tb04965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Olivos-Glander I, Killoran C, Elson E, Turner JT, Peters KF, Abbott MH, Aughton DJ, Aylsworth AS, Bamshad MJ, Booth C, Curry CJ, et al. Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. Am J Hum Genet. 2005;76:609–622. doi: 10.1086/429346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Phillips M, Hegyi G, Muhlrad A, Reisler E. Intrastrand cross-linked actin between Gln-41 and Cys-374. II. Properties of cross-linked oligomers. Biochemistry. 1998;37:17793–17800. doi: 10.1021/bi9812874. [DOI] [PubMed] [Google Scholar]
- Kruth KA, Rubenstein PA. Two deafness-causing (DFNA20/26) actin mutations affect Arp2/3-dependent actin regulation. J Biol Chem. 2012;287:27217–27226. doi: 10.1074/jbc.M112.377283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster W, Majewski F. The Dubowitz syndrome. Eur J Pediatr. 1986;144:574–578. doi: 10.1007/BF00496039. [DOI] [PubMed] [Google Scholar]
- Laing NG, Dye DE, Wallgren-Pettersson C, Richard G, Monnier N, Lillis S, Winder TL, Lochmuller H, Graziano C, Mitrani-Rosenbaum S, Twomey D, Sparrow JC, et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1) Hum Mutat. 2009;30:1267–1277. doi: 10.1002/humu.21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloy LE, Wen KK, Pierick AR, Wedemeyer EW, Bergeron SE, Vanderpool ND, McKane M, Rubenstein PA, Bartlett HL. Thoracic aortic aneurysm (TAAD)-causing mutation in actin affects formin regulation of polymerization. J Biol Chem. 2012;287:28398–28408. doi: 10.1074/jbc.M112.371914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKane M, Wen KK, Boldogh IR, Ramcharan S, Pon LA, Rubenstein PA. A mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. J Biol Chem. 2005;280:36494–36501. doi: 10.1074/jbc.M506970200. [DOI] [PubMed] [Google Scholar]
- Morin M, Bryan KE, Mayo-Merino F, Goodyear R, Mencia A, Modamio-Hoybjor S, del Castillo I, Cabalka JM, Richardson G, Moreno F, Rubenstein PA, Moreno-Pelayo MA. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum Mol Genet. 2009;18:3075–3089. doi: 10.1093/hmg/ddp249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, Meisler MH, Goldstein DB. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak KJ, Wattanasirichaigoon D, Goebel HH, Wilce M, Pelin K, Donner K, Jacob RL, Hubner C, Oexle K, Anderson JR, Verity CM, North KN, et al. Mutations in the skeletal muscle alpha-actin gene in patients with actin myopathy and nemaline myopathy. Nat Genet. 1999;23:208–212. doi: 10.1038/13837. [DOI] [PubMed] [Google Scholar]
- Nunoi H, Yamazaki T, Tsuchiya H, Kato S, Malech HL, Matsuda I, Kanegasaki S. A heterozygous mutation of beta-actin associated with neutrophil dysfunction and recurrent infection. Proc Natl Acad Sci U S A. 1999;96:8693–8698. doi: 10.1073/pnas.96.15.8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Orlova A, Egelman EH. Structural dynamics of F-actin: I. Changes in the C terminus. J Mol Biol. 1995;245:582–597. doi: 10.1006/jmbi.1994.0048. [DOI] [PubMed] [Google Scholar]
- Otey CA, Kalnoski MH, Bulinski JC. Identification and quantification of actin isoforms in vertebrate cells and tissues. J Cell Biochem. 1987;34:113–124. doi: 10.1002/jcb.240340205. [DOI] [PubMed] [Google Scholar]
- Peng X, Cuff LE, Lawton CD, DeMali KA. Vinculin regulates cell-surface E-cadherin expression by binding to beta-catenin. J Cell Sci. 2010;123:567–577. doi: 10.1242/jcs.056432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan BC, Peyser YM, Reisler E, Muhlrad A. Effect of complexes of ADP and phosphate analogs on the conformation of the Cys707-Cys697 region of myosin subfragment 1. Eur J Biochem. 1997;243:636–642. doi: 10.1111/j.1432-1033.1997.00636.x. [DOI] [PubMed] [Google Scholar]
- Procaccio V, Salazar G, Ono S, Styers ML, Gearing M, Davila A, Jimenez R, Juncos J, Gutekunst CA, Meroni G, Fontanella B, Sontag E, et al. A mutation of beta - actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am J Hum Genet. 2006;78:947–960. doi: 10.1086/504271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendtorff ND, Zhu M, Fagerheim T, Antal TL, Jones M, Teslovich TM, Gillanders EM, Barmada M, Teig E, Trent JM, Friderici KH, Stephan DA, et al. A novel missense mutation in ACTG1 causes dominant deafness in a Norwegian DFNA20/26 family, but ACTG1 mutations are not frequent among families with hereditary hearing impairment. Eur J Hum Genet. 2006;14:1097–1105. doi: 10.1038/sj.ejhg.5201670. [DOI] [PubMed] [Google Scholar]
- Riviere JB, van Bon BW, Hoischen A, Kholmanskikh SS, O'Roak BJ, Gilissen C, Gijsen S, Sullivan CT, Christian SL, Abdul-Rahman OA, Atkin JF, Chassaing N, et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat Genet. 2012;44:440–444. doi: 10.1038/ng.1091. S441–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein PA, Mayer EA. Familial visceral myopathies: from symptom-based syndromes to actin-related diseases. Gastroenterology. 2012;143:1420–1423. doi: 10.1053/j.gastro.2012.10.031. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E, Hayman EG, Pierschbacher M, Engvall E. Fibronectin: purification, immunochemical properties, and biological activities. Methods Enzymol. 1982;82(Pt A):803–831. doi: 10.1016/0076-6879(82)82103-4. [DOI] [PubMed] [Google Scholar]
- Sparrow JC, Nowak KJ, Durling HJ, Beggs AH, Wallgren-Pettersson C, Romero N, Nonaka I, Laing NG. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1) Neuromuscul Disord. 2003;13:519–531. doi: 10.1016/s0960-8966(03)00101-9. [DOI] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, Margulies EH, Green ED, Collins FS, Mullikin JC, et al. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekerckhove J, Weber K. Actin typing on total cellular extracts: a highly sensitive protein-chemical procedure able to distinguish different actins. Eur J Biochem. 1981;113:595–603. doi: 10.1111/j.1432-1033.1981.tb05104.x. [DOI] [PubMed] [Google Scholar]
- Zhu M, Yang T, Wei S, DeWan AT, Morell RJ, Elfenbein JL, Fisher RA, Leal SM, Smith RJ, Friderici KH. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26) Am J Hum Genet. 2003;73:1082–1091. doi: 10.1086/379286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.