Abstract
Background
Gene fusions between the ERG transcription factor and the androgen-regulated gene TMPRSS2 occur in a subset of prostate cancers and contribute to transformation of prostatic epithelial cells. Prior reports have used FISH or quantitative PCR (QPCR) to determine the presence of TMPRSS2-ERG fusions or ERG expression, respectively. Recently, several groups have reported on immunohistochemistry (IHC) to measure ERG expression, which is much more readily performed in clinical practice. However, the prior studies examining ERG expression by IHC had small samples sizes or they failed to clarify the association of ERG protein expression with important clinico-pathological features or prostate cancer-specific mortality. Methods: To address these deficits, we evaluated ERG expression by IHC in 208 radical prostatectomy samples from the Kaiser Permanente Molecular Epidemiology of Fatal Prostate Cancer (MEFPC) study, a case-control study of prostate-cancer specific mortality.
Results
Nuclear ERG expression was seen in neoplastic prostate epithelia in 49 of the samples (23.7%). ERG expression in tumor cells was associated with higher tumor stage (OR=2.0, 95% confidence interval 1.0–4.0, p value= 0.04). ERG immunoreactivity was positively associated with prostate cancer-specific mortality, although the confidence interval was wide (OR=1.9, 95% confidence interval 0.88–4.0, p value =0.10).
Conclusions
Our results demonstrate that ERG protein expression is readily quantifiable with an existing commercial antibody. Evaluating ERG protein expression may improve our ability to identify the subset of more aggressive, invasive prostate cancers.
Introduction
Prostate cancer remains a significant medical problem, and over 32,000 U.S. men are expected to die from the disease this year.1 There is a paucity of data to distinguish between aggressive and indolent prostate cancer, although a number of molecular markers have been studied. v-ets erythroblastosis virus E26 oncogene homolog (ERG) is a member of the ETS family of transcription factors.2,3 In 2005, Tomlins, et al reported that fusions between ERG and TMPRSS2, an androgen receptor (AR) target gene, are common in prostate cancer.4 Recent reports indicate that ERG may contribute to transformation of prostatic epithelial cells and that ERG-expressing prostate cancers may be particularly susceptible to therapeutic strategies involving PARP inhibition.5–8 For all these reasons, understanding ERG expression in prostate tumors may be important in clinical care.
Prior reports have used fluorescence in situ hybridization (FISH) or quantitative PCR (QPCR) to measure the TMPRSS2-ERG gene fusion.9–11 However, these techniques are not readily performed in many clinical laboratories. For QPCR, high tumor content in a specimen or frozen samples is also often necessary. For these reasons, several groups have used immunohistochemistry (IHC) to quantify ERG protein expression.12–16 However, the association between ERG expression and important clinico-pathological features of this disease remains unclear. Additionally, none of the prior reports included sufficient numbers of patients with prostate cancer-specific mortality to determine this association. In this study, we sought to examine how ERG protein expression by IHC is associated with clinico-pathological and mortality outcomes.
Materials and Methods
Case-Control Study Description
The Molecular Epidemiology of Fatal Prostate Cancer (MEFPC) study is a population-based case-control study conducted in three Kaiser Permanente regions. This analysis focused on the subjects from the Kaiser Permanente Northwest (KPNW) and Kaiser Permanente Northern California (KPNC) regions. Men who had prostatectomy as part of prostate cancer treatment and who died from 1971–2001 at KPNW (546) and from 1980–2001 at KPNC (1,026) were selected from the KPNW electronic cancer registry files and National Cancer Institute (NCI) Surveillance, Epidemiology, and End Results (SEER) program files (for KPNC). We then restricted the group to men coded as dying from prostate cancer or from a list of immediate causes for which prostate cancer might be the underlying cause (eg. unknown cause, pneumonia, renal failure) (1,006). From the subset with formalin fixed paraffin-embedded tumor tissue in health plan archives, the medical records of men who were diagnosed with prostate cancer before age 81 years; were Caucasian, African-American, or Hispanic race; and were members of the health plan when diagnosed and for at least 12 months following their diagnoses or until death, if death occurred within 12 months, were evaluated using a cause-of-death algorithm developed for this study to select men whose deaths were due to prostate cancer (192). Of these 192 cases, tumor tissue for 99 cases was available for this secondary analysis.
Controls (n=109 for this analysis) were originally matched to study cases on health plan, race, tumor SEER stage at diagnosis as recorded in the health plan tumor registries, age at diagnosis, year of diagnosis, and duration of health plan membership and had to be alive at the time of their matched cases’ death. We abstracted medical records for prostate cancer tumor characteristics, treatment outcome, co-morbidities, clinical characteristics, and demographics. We collected other pertinent information from automated laboratory, cancer registry, and health plan membership files. Not all subjects in this secondary analysis were in matched case-control sets.
ERG IHC and Staining Interpretation
The H&E slides from prostatectomy tissue for all subjects were uniformly reviewed by a single genitourinary pathologist (BK) to confirm the diagnosis and provide a uniform Gleason grade, pathologic stage, surgical margin status, extent of invasion (capsular, extracapsular, perineural, seminal vesicle) and to select the tumor block containing the highest Gleason grade tumor for each subject. Immunohistochemistry analyses were performed in a single laboratory blinded to outcome and clinico-pathological features. A subject was excluded if the tumor tissue specimen was insufficient for prostate cancer diagnosis or IHC analysis.
PC3 or VCaP cell lines were embedded in Histogel. IHC staining of cell lines and prostate tissue samples from patients was performed for ERG. Five micron FFPE tissue sections were de-paraffinized with xylene and rehydrated through a graded alcohol series. Heat induced epitope retrieval (HIER) was performed by immersing the tissue sections at 98°C for 20 minutes in 10 mM citrate buffer (pH 6.0) with 0.05% Tween. IHC staining was performed using the Histostain Plus Kit from Zymed/Invitrogen according to manufacturer’s instructions. Briefly, slides were treated with 3% hydrogen peroxide and Zymed solution A for 10 minutes each, and exposed to primary antibodies for ERG (1:200 dilution, sc-353 from Santa Cruz) for 1 hour at room temperature. Slides were exposed to Zymed solutions B and C for 10 minutes each and DAB chromagen (Dako) for 5 minutes. Slides were counterstained with Hematoxylin (Fisher, Harris Modified Hematoxylin) at a 1:17 dilution for 2 minutes at RT, blued in 1% ammonium hydroxide for 1 minute at room temperature, dehydrated, and mounted with Acrymount. Consecutive sections with the omitted primary antibody were used as negative controls. Images were captured using an Olympus DP70 microscope at 40x magnification. Only subjects with nuclear ERG immunoreactivity were classified as ERG positive.
Data Analysis
For the main analysis of ERG expression in relation to prostate cancer mortality, we used a dichotomous marker cutpoint (positive or negative for nuclear ERG immunoreactivity). We performed standard conditional and unconditional logistic regression analyses; results were similar and therefore, unconditional results are presented here. We initially performed univariate analyses in order to determine unadjusted odds ratios. Analyses were repeated after adjusting for matching variables and Gleason grade. We also evaluated factors known or suspected to be confounders or modifiers of the association of interest. The final model contained variables for participating site, age, diagnosis year, race, AJCC tumor stage, Gleason grade and seminal vesicle invasion status.
Results
ERG Protein Expression is Only Apparent in TMPRSS2-ERG Expressing Cells
We first sought to determine the specificity of the anti-ERG antibody. Therefore, we performed ERG IHC staining using paraffin-embedded VCaP prostate cancer cells that harbor the TMPRSS2-ERG fusion and PC3 prostate cancer cells that do not express TMPRSS2-ERG or full-length ERG.17 ERG expression was only detected in VCaP cells and was limited to the nucleus in these cells (Fig 1).
Figure 1. Immunohistochemical expression of ERG in prostate cancer cell lines.
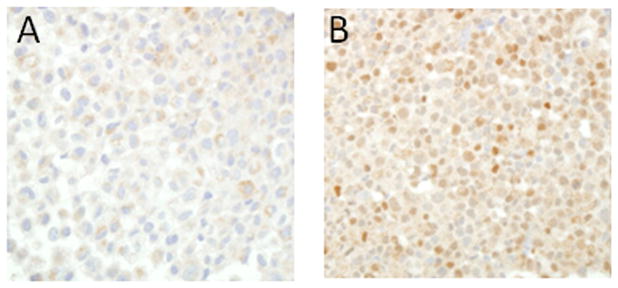
Representative images from A) PC3 cells that do not express ERG and B) VCaP cells that harbor the TMPRSS2-ERG fusion and express ERG are shown.
ERG Expression in Prostate Cancer Patient Specimens
Subjects were diagnosed through prostate-specific antigen (PSA) screening, digital rectal examination (DRE), or clinical evaluation for prostate cancer symptoms. As expected, cases and the controls had similar distributions of the matching variables race and age at diagnosis (Table 1). Although our original study design included matching controls to cases on tumor registry SEER stage, the more precise AJCC pathologic staging performed by the study pathologist supplemented by medical record review resulted in larger proportions of AJCC stages 3 and 4 in cases than in controls. A higher proportion of cases than controls had high Gleason grade.
Table 1.
Demographic and clinical information on study subjects
| Characteristic | Cases N= 99 | Controls N=109 | P-value** |
|---|---|---|---|
| N (%) | N (%) | ||
| Race | 0.72 | ||
| Caucasian | 82 (83) | 86 (79) | |
| African-American | 8 (8) | 14 (13) | |
| Hispanic | 9 (9) | 9 (8) | |
| Age at prostate cancer diagnosis (years) | 0.56 | ||
| 40–59 | 30 (30) | 26 (24) | |
| 60–64 | 16 (16) | 18 (17) | |
| 65–69 | 32 (32) | 46 (42) | |
| 70–80 | 21 (21) | 19 (17) | |
| Gleason grade | <0.0001 | ||
| 2 – 6 | 8 (8) | 32 (29) | |
| 7 (3/4) | 38 (39) | 48 (44) | |
| 7 (4/3) | 20 (20) | 14 (13) | |
| 8 – 10 | 32 (33) | 15 (14) | |
| AJCC stage at diagnosis | 0.02 | ||
| 2 | 36 (37) | 54 (50) | |
| 3 | 47 (48) | 47 (43) | |
| 4 | 16 (16) | 8 (7) | |
| Test leading to diagnosis | 0.02 | ||
| PSA only | 1 (1) | 2 (2) | |
| DRE only | 10 (10) | 17 (16) | |
| DRE and PSA | 15 (15) | 33 (30) | |
| Symptomatic | 73 (74) | 57 (52) | |
| Recurrence in controls | |||
| None | 63 (58) | ||
| Biochemical recurrence only* | 30 (28) | ||
| Other recurrence | 16 (15) | ||
| Site | 0.93 | ||
| KPNW | 26 (26) | 28 (26) | |
| KPNC | 73 (74) | 81 (74) | |
| Capsular invasion | 97 (98) | 104 (96) | 0.47 |
| Extracapsular invasion | 54 (55) | 39 (36) | 0.0008 |
| Seminal vesicle invasion | 37 (37) | 23 (21) | 0.002 |
| Positive lymph nodes | 16 (16) | 8 (7) | 0.10 |
| Positive margins | 50(51) | 54(50) | 0.39 |
| Perineural invasion | 85 (86) | 79 (73) | 0.001 |
All p-values are based on the Pearson chi-square test.
PSA > 0.2ng/mL after prostatectomy
KPNW= Kaiser Permanente Northwest
KPNC= Kaiser Permanente Northern California
We evaluated ERG expression in these prostate cancer patient specimens. Only samples with nuclear ERG expression by IHC were considered positive. A representative sample is shown in Fig 2. Overall, 49 (23.6%) of the 208 patient samples had nuclear ERG expression in prostate cancer cells (26 cases (26.3%) and 23 controls (17.3%)). ERG expression was also commonly detected in endothelial cells. However, ERG nuclear expression in benign prostate epithelial cells was a rare event, occurring in only 4.8% of samples.
Figure 2. Immunohistochemical expression of ERG in prostate tissue specimens.
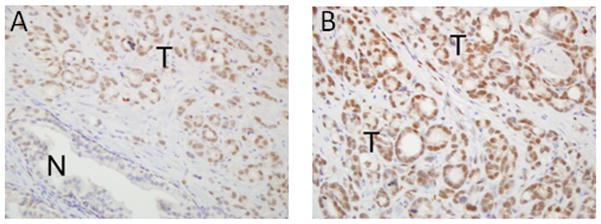
Representative images with A) low-power and B) high-power magnification depicting ERG expression in patient tissue samples are shown. N= normal prostatic epithelium. T= prostate tumor epithelium.
Association Between ERG Expression and Clinico-Pathological Features
Next, we determined the association between ERG expression in the prostate cancer samples and known clinico-pathological features associated with prostate cancer recurrence. ERG expression was not associated with Gleason grade (Gleason grade ≥8 vs. <8, OR= 0.7, 95% confidence interval 0.31–1.57, p value= 0.38) or surgical margin status (OR=1.35, 95% confidence interval 0.70–2.61, p value=0.37). However, ERG expression was associated with a statistically significantly increased risk of higher stage tumors (AJCC stage ≥3 vs. <3, OR=2.0, 95% confidence interval 1.0–4.0, p value= 0.04).
Association Between ERG Expression and Death from Prostate Cancer
Finally, we examined the association between ERG expression in prostate cancer samples and the risk of death from prostate cancer. After adjustment for Kaiser Permanente treatment site, age at prostate cancer diagnosis, diagnosis year, race, AJCC tumor stage, seminal vesicle invasion, and Gleason grade (categorized as <7, 7(3/4), 7(4/3), 8–10), patients whose tumors expressed ERG had a 90% higher likelihood of prostate cancer-specific mortality than those with ERG-negative tumors; however, this association was not statistically significant at α<=0.05 (OR=1.9, 95% confidence interval 0.88–4.0, p value =0.10) (Table 2).
Table 2.
The relationship between ERG expression and fatal prostate cancer
| Marker | Unadjusted | Adjusted for matching variables* | Adjusted for matching variables plus Gleason grade** |
|---|---|---|---|
| ERG nuclear | 1.4 (0.73–2.6) | 1.4 (0.67–2.7) | 1.9 (0.88–4.0) |
| staining (Yes vs. no) | p=0.31 | p-=0.39 | p=0.10 |
Kaiser Permanente treatment site, age, diagnosis year, race, AJCC tumor stage, seminal vesicle invasion
Gleason grade categorized as <7, 7(3/4), 7(4/3), 8–10
Discussion
Although it is estimated that 241,780 men will be diagnosed with prostate cancer in the United States this year, only a small proportion are likely to die from the disease.1 This demonstrates the need to clarify the molecular mechanisms of prostate cancer progression. The ERG transcription factor may be such a driver.7,18 However, the role of ERG in fatal prostate cancer remained unclear in prior studies. In this report, we evaluated ERG protein expression and determined the association of ERG expression with known clinico-pathological features associated with prostate cancer aggressiveness and with prostate cancer-specific mortality. ERG expression was associated with high tumor stage. ERG expression was associated with a higher risk of death from prostate cancer though that association did not reach statistical significance.
Most prior studies examining patient outcomes after radical prostatectomy include patients treated in academic medical centers; such studies may not be generalizable to prostate cancer patients in the general community because in many cases patients are operated on by a single surgeon.19 However, in the United States, most prostatectomies are performed outside of academic medical centers. Our study is unique because we used specimens from patients treated and followed within the Kaiser Permanente health system. Kaiser Permanente is one of the nation’s largest health maintenance organizations, and a broad cross-section of patients is treated in this health system. Indeed, nearly 20 percent of the patients in our series were non-Caucasian. Thus, the patients included in this report may be more representative of typical U.S. patients who undergo radical prostatectomy than prior reports.
We evaluated ERG protein expression by IHC using both cell lines and prostate cancer clinical samples. Our results demonstrate that ERG expression detected by this antibody was specific. Indeed, we only detected ERG expression in VCaP cells that harbor the TMPRSS2-ERG fusion and not in ERG-null PC3 cells. ERG expression in clinical samples was also predominantly found in cancer cells. ERG expression was a rare event in normal prostate epithelial cells, but endothelial cells did express ERG, an observation which has been reported previously.15
The frequency of ERG expression in our patient samples (23.7%) was lower than prior reports that ranged from 44%–65%.13,15,16,20 One possible explanation is a difference in the antibodies used or the age of our tissue specimens. It is also possible that the antibody we used did not detect all possible rearrangements or translocations involving ERG. These factors may have contributed to the lower frequency of ERG expression in our report. However, our ERG staining results in the cell lines and patient samples confirm the specificity of staining with this antibody (Figs 1 and 2).
ERG has been linked to prostate cancer migration and invasion previously.5,6,21 This is consistent with our finding that ERG expression was associated with higher tumor stage, which matches a recent report by Pettersson, et al.16 There were no statistically-significant associations between ERG positivity and other known clinico-pathological features associated with cancer recurrence or death.
The lack of association of ERG expression with higher tumor grade was also seen by Pettersson, et al but contrasts with the results of Furasato, et al.16,20 Our fatal cases had predominantly higher-grade prostate cancers and very few low grade cancers. This lack of variability may have contributed to the lack of association of ERG expression with higher tumor grade. Hoogland, et al did not find an association with ERG expression and either grade or stage.15 However, their findings of the lack of association between ERG expression and stage may be explained by the paucity of higher stage tumors in that series.15
Finally, prostate cancer death outcomes were known for all of the patients in our study. ERG expression was associated with an increased risk of prostate cancer-specific mortality. However, this association did not reach conventional statistical significance (Table 2). Pettersson, et al recently performed a meta-analysis to determine the association between ERG expression and prostate cancer outcomes, including “fatal prostate cancer.”16 This composite endpoint included development of prostate cancer metastases or death from prostate cancer. Eighty-five men in their series experienced one of these outcomes. ERG expression was not associated with an increased risk of fatal prostate cancer in that report. However, it is unclear what proportion of these 85 patients died from prostate cancer. Choice of endpoints may have contributed to the difference in results between our study and that of Pettersson, et al.
While our ERG expression measurements were based on the largest numbers of patients with death outcomes from prostate cancer, it is possible that the low frequency of ERG expression contributed to the lack of association between ERG expression and death from prostate cancer. A larger sample size with more prostate cancer death events may be necessary to definitively answer the question. Nonetheless, higher stage was associated with fatal prostate cancer in our series (Table 1), and ERG was more commonly expressed in higher stage tumors. Therefore, it is possible that ERG may contribute to the molecular basis for these more advanced prostate cancers, many of which progress to the lethal form of this disease.
Conclusions
Development of ERG inhibitors or strategies to personalize the treatment of ERG-expressing prostate cancer is an active area of research.8 However, it will be critical to accurately identify ERG-expressing tumors using assays that may be performed routinely in clinical pathology laboratories. Our results demonstrate that ERG expression may be readily quantified with IHC using a commercially available antibody. The ease of use of ERG IHC makes this approach promising for clinical use. ERG expression was not prognostic in our study. However, in the future, we hope that measuring ERG expression will enable us to match therapy to target in patients with ERG-expressing prostate cancers and that such a strategy will improve patient outcomes.
Acknowledgments
We wish to acknowledge the patients who agreed to participate in the Kaiser Permanente Molecular Epidemiology of Fatal Prostate Cancer (MEFPC) study and Chris Koontz for help with manuscript preparation and submission.
Grant Support: This publication was made possible with support from the Oregon Clinical and Translational Research Institute (OCTRI) grant number UL1 RR024140 (JA, SW) and grant number KL2 RR024141 (JA) from the National Center for Research Resources (NCRR) and the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). This work was also supported by a Wayne D. Kuni & Joan E. Kuni Foundation Kuni Scholar Award (JA), a Prostate Cancer Foundation Young Investigator Award (JA), and NIH R01 CA100743 (SW).
Footnotes
Conflict of Interest Statement: There are no conflicts of interest to disclose.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nature reviews Urology. 2009;6:429–39. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 3.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 5.Klezovitch O, Risk M, Coleman I, et al. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105:2105–10. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carver BS, Tran J, Gopalan A, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41:619–24. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King JC, Xu J, Wongvipat J, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–78. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 10.FitzGerald LM, Agalliu I, Johnson K, et al. Association of TMPRSS2-ERG gene fusion with clinical characteristics and outcomes: results from a population-based study of prostate cancer. BMC Cancer. 2008;8:230. doi: 10.1186/1471-2407-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam RK, Sugar L, Yang W, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. British journal of cancer. 2007;97:1690–5. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furusato B, Tan SH, Young D, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 2010;13:228–37. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12:590–8. doi: 10.1593/neo.10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Leenders GJ, Boormans JL, Vissers CJ, et al. Antibody EPR3864 is specific for ERG genomic fusions in prostate cancer: implications for pathological practice. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:1128–38. doi: 10.1038/modpathol.2011.65. [DOI] [PubMed] [Google Scholar]
- 15.Hoogland AM, Jenster G, van Weerden WM, et al. ERG immunohistochemistry is not predictive for PSA recurrence, local recurrence or overall survival after radical prostatectomy for prostate cancer. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25:471–9. doi: 10.1038/modpathol.2011.176. [DOI] [PubMed] [Google Scholar]
- 16.Pettersson A, Graff RE, Bauer SR, et al. The TMPRSS2:ERG Rearrangement, ERG Expression, and Prostate Cancer Outcomes: A Cohort Study and Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2012;21:1497–509. doi: 10.1158/1055-9965.EPI-12-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartzman J, Mongoue-Tchokote S, Gibbs A, et al. A DNA methylation microarray-based study identifies ERG as a gene commonly methylated in prostate cancer. Epigenetics. 6 doi: 10.4161/epi.6.10.17727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carver BS, Chapinski C, Wongvipat J, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA : the journal of the American Medical Association. 1999;281:1591–7. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 20.Furusato B, Tan SH, Young D, et al. ERG oncoprotein expression in prostate cancer: clonal progression of ERG-positive tumor cells and potential for ERG-based stratification. Prostate Cancer Prostatic Dis. 13:228–37. doi: 10.1038/pcan.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]


