Abstract
Although simple tandem repeats (STRs) comprise ~2% of the human genome and represent an important source of polymorphism, this class of variation remains understudied. We have developed a cost-effective strategy for performing targeted enrichment of STR regions that utilizes capture probes targeting the flanking sequences of STR loci, enabling specific capture of DNA fragments containing STRs for subsequent high-throughput sequencing. Utilizing a capture design targeting 6,243 STR loci <94bp and multiplexing eight individuals in a single Illumina HiSeq2000 sequencing lane we were able to call genotypes in at least one individual for 67.5% of the targeted STRs. We observed a strong relationship between (G+C) content and genotyping rate. STRs with moderate (G+C) content were recovered with >90% success rate, while only 12% of STRs with ≥80% (G+C) were genotyped in our assay. Analysis of a parent-offspring trio, complete hydatidiform mole samples, repeat analyses of the same individual, and Sanger sequencing-based validation indicated genotyping error rates between 7.6–12.4%. The majority of such errors were a single repeat unit at mono- or dinucleotide repeats. Altogether, our STR capture assay represents a cost-effective method that enables multiplexed genotyping of thousands of STR loci suitable for large scale population studies.
Keywords: microsatellite, repeat variation, genome instability, high-throughput sequencing, sequence capture
Introduction
Common repetitive DNA elements comprise approximately half of the entire sequence of the human genome. A subset of these elements are tandemly repeated sequences, defined as two or more contiguous copies of a sequence arranged in a head-to-tail pattern. Often termed “simple tandem repeats” (STRs), they comprise ~2% of the human genome, an amount that exceeds protein-coding sequences [Lander et al. 2001] and almost one third of RefSeq genes have a STR within 1kb of their transcription start site (TSS). STRs are mutational hotspots [Fonville et al. 2011] and therefore represent an abundant source of genetic variation. It has been estimated that length variations of STRs account for approximately 25% of insertion/deletion polymorphisms in the human genome [Pang et al. 2013]. This includes microsatellite repeats, defined as particularly long and pure STRs with a period of 2–6bp, which because of their tendency to be highly polymorphic have found common usage in linkage studies and forensics. In addition several human disorders have been shown to be caused by unstable repeats in the promoter or 5′UTR regions of genes, including progressive myoclonus epilepsy involving CCCCGCCCCGCG expansions upstream CSTB [OMIM# 601145], SCA12 due to a CAG expansion upstream PPP2R2B [OMIM# 604326], or Fragile X syndrome caused by a CGG expansion in the 5′UTR of FMR1 [OMIM# 300624], [Mirkin 2007; Siwach and Ganesh 2008]. Repeat contractions have also been observed in non-human phenotypes such as pituitary dwarfism in dogs due to a deletion of an intronic 7bp repeat in LHX3 [OMIM# 600577] [Voorbij et al. 2011]. In addition to causing overt disease, length variation in tandem repeats in regulatory regions has also been shown to play a role in phenotypic adaptation [Gemayel et al. 2010; Vinces et al. 2009], and can also regulate nearby gene expression levels [Borel et al. 2012].
Despite this evidence for functional effects, STRs remain very poorly studied compared to other classes of genetic variation, and as a result their influence on human phenotypes is almost certainly under-estimated. This is largely due to their repetitive and highly variable nature which, in contrast to single nucleotide polymorphisms (SNPs) and copy number variations (CNVs), has made them inaccessible to current high throughput genotyping methods. SNP genotyping methods used in genome-wide association studies (GWAS) are typically not informative for STR length due to the high mutation rate of STRs compared to SNPs. While estimates for the SNP mutation rate put this at between 1.2×10−8 and 2.3×10−8 per site per meiosis [Kondrashov 2003; Sun et al. 2012; Campbell et al. 2012], estimates derived from studies of >1 million transmissions in large Icelandic pedigrees have shown the mutation rate for STRs to vary between 2.5×10−4 and 3.5×10−4 for dinucleotide repeats, and 8.5×10−4 to 15×10−4 for tetranucleotide repeats [Sun et al. 2012], in agreement with previous more limited studies [Weber and Wong 1993; Xu et al. 2000; Ellegren 2004]. This evidence documenting the elevated mutability and multi-allelic nature of STRs suggests that many of these genetic variations are unlikely to be effectively tagged by SNPs [Burgner et al. 2003]. Copy number variations are commonly assessed using array-CGH. However, this technique tends to saturate at high copy number, and the non-unique nature of STRs and large size of probes typically used on arrays means that this technology is poorly suited for genotyping the vast majority of STRs. The advent of next generation sequencing represents a promising avenue for high-throughput genotyping of STRs. Indeed, recently several novel algorithms have been developed that allow STR genotypes to be called directly from whole genome sequencing data [Fondon et al. 2012; Gymrek et al. 2012; Highnam et al. 2013]. Due to the high rates of insertion/deletion polymorphism at many STR loci, these specialized bioinformatic tools utilize read alignments that are highly tolerant of gaps, and then select informative reads that are anchored in unique flanking sequence and span the entire repeat array to allow accurate genotype calls to be made. However, although the cost of whole genome sequencing is rapidly falling, it still remains costly for large population-scale studies.
Therefore, we have developed a method that allows targeted and multiplexed high-throughput sequencing of thousands of STRs in a single sequencing run. By performing minor modifications to standard sequence capture methods we are able to specifically enrich STR loci that are then sequenced and analyzed using a microsatellite-aware variant caller [Highnam et al. 2013]. Here, we demonstrate the feasibility of this approach for calling genotypes for thousands of STR loci at low cost, opening the door to more wide-spread investigation of the role of STRs in human phenotypes.
Material and Methods
Samples
Samples studied comprised DNA from five HapMap cell lines (Coriell Institute for Medical Research, NJ, USA), including two unrelated YRI individuals NA18501 and NA18502, and the YRI parent-offspring trio NA19238, NA19239 and NA19240. Two complete hydatidiform moles (CHM1 and CHM2) were analyzed to estimate the genotyping errors. DNA was extracted from HapMap lymphoblastoid cell lines by Coriell Institute for Medical Research, and from CHM tissue samples using salt:ethanol precipitation.
Library construction
2μg or 5μg of high molecular weight genomic DNA for PCR and PCR-free library respectively were sheared using a Covaris Acoustic Disruptor E210 using the following program: 10 duty cycles, 200 cycles per burst, intensity 4, 120 sec. Fragmented DNA was then purified using Agencourt AMPure XP purification beads (Beckman Coulter) and DNA fragment size checked on an Agilent DNA 1000 chip using an Agilent 2100 Bioanalyzer.
Library preparation was performed using the NEBNext DNA Sample Prep Reagent Set 1. Briefly, fragmented DNA was first end-repaired using NEBNext End Repair Reaction Buffer and NEBNext End Repair Enzyme mix (30 min at 20°C), and purified using Agencourt AMPure XP beads (Beckman Coulter). dA-tailing was then performed using NEBNext dA-Tailing Reaction Buffer and Klenow Fragment (3′–5′ exonuclease), for 30 min at 37°C and subsequently purified using Agencourt AMPure XP beads.
Two alternate protocols, either with or without PCR prior to hybridization with capture probes were used for adapter ligation: (i) For libraries prepared with subsequent PCR amplification Illumina universal adapters (Suppl. Table S1) were ligated onto DNA fragments using NEBNext Quick Ligation Reaction Buffer, 2μM adapters and Quick T4 DNA Ligase, for 15 min at 20°C. This reaction was then purified using the Agencourt AMPure XP system. (ii) For preparation of a PCR-free library, indexed adapters were annealed to the Illumina universal adapter (Suppl. Table S1) by denaturation at 97°C for 10 min followed by gradual cooling to room temperature to form a Y-structure. Ligations were performed with a 20:1 molar excess of adapters:dA-tailed DNA, using NEBNext Quick Ligation Reaction Buffer and Quick T4 DNA Ligase, at 20°C overnight. Ligation reactions were then purified using the Agencourt AMPure XP system. Ligated products were run on 2% agarose gel and a slice corresponding to fragments of size 300–450bp was cut using SafeXtractor (5 prime). Gel extraction was performed using the QIAQuick PCR purification kit (Qiagen).
For samples that were subject to PCR-amplification, PCR and indexing was performed simultaneously using KAPA HiFi HotStart ReadyMix (KAPA Biosystems), 0.5μM universal forward primer, 0.5μM indexed reverse primer and adapter-ligated DNA, with the following program: denaturation at 94°C for 10min, 13 cycles of 94°C for 1min, 62°C for 30sec and 72°C for 30sec, followed by elongation at 72°C for 10min. PCR products were then purified using the Agencourt AMPure XP system. Library quality and yield were assessed with an Agilent DNA 1000 chip using an Agilent 2100 Bioanalyzer. Libraries were pooled in equimolar amounts to ensure even sequence depth, totaling 1μg as recommended by the manufacturer.
Quantification of the ligated products in the PCR-free library
Given that the efficiency of ligation is potentially variable, we quantified the proportion of products that were ligated and therefore suitable for sequencing. We used the KAPA library quantification kit based on qPCR with primers targeting the adapters and a standard curve, following the manufacturer’s instructions. 2–3 PCR cycles were necessary for the samples CHM8986, NA19238, NA18501 and NA18502, using universal primers (Suppl. Table S1) and the same conditions as in the library with PCR. These libraries were pooled in equimolar amounts with the other libraries to ensure even sequence depth, totaling 1μg as recommended by the manufacturer.
Sequence capture of STR regions
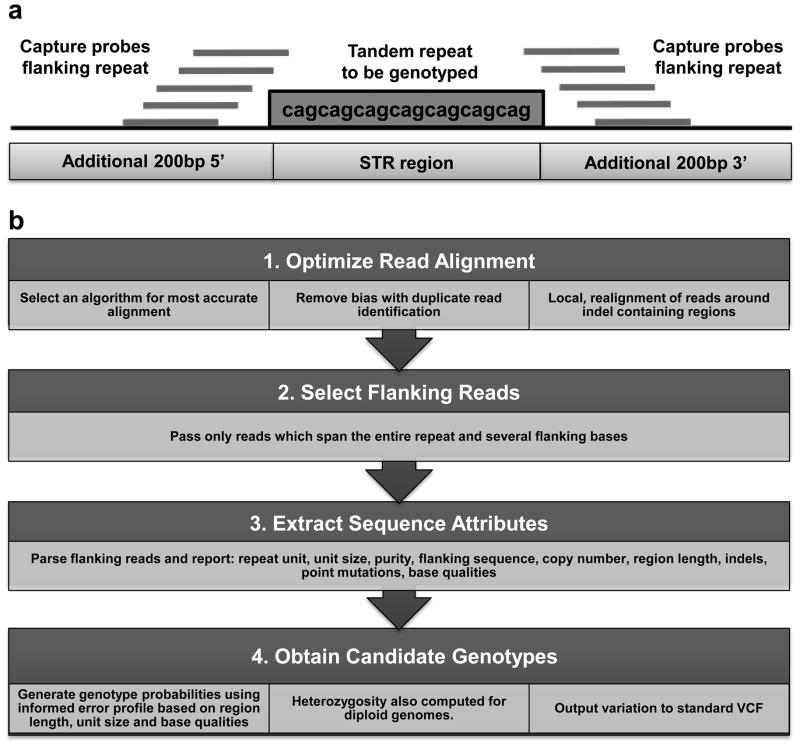
We designed a custom probe set to perform sequence capture of all STR loci that lie within 1kb of the transcriptional start site (TSS) of RefSeq genes. STRs were identified using Tandem Repeats Finder (downloaded from the Simple Repeats track in hg18 from the UCSC Genome Browser, http://genome.ucsc.edu/), without regard to motif size or imperfections (repeat span ranges from 25bp to 10,490bp in the hg18 assembly). Thus the capture design included not only microsatellites, but all types of tandemly repeated sequence. These were then merged down to a non-redundant set and intersected with 2kb windows centered on the TSSs of all Refseq genes using Galaxy (https://main.g2.bx.psu.edu/), identifying a total of 7,851 STR loci. In total 6,346 independent Refseq genes (27.3% of the total) contain one or more STRs within ±1kb of their transcription start site, with a mean of 1.2 STRs per gene. A Nimblegen SeqCap EZ Library was then designed to perform targeted enrichment of these regions. Given that due to their non-unique nature and propensity to form secondary structures many STRs are not suitable for direct placement of specific probes within them, we extended the target regions to include an additional 200bp both proximal and distal flanking each STR available for placement of capture probes (Figure 1a). Probes were designed based on uniqueness, assessed by SSAHA, with up to 5 insertions/deletions/mismatches allowed. Only probes with 1 such match in the genome were kept. Due to this design constraint, 7,353 of the 7,851 initial target loci (93.7%) were represented on the final capture design, which comprised 207,222 independent capture probes. The target STR regions are represented by a mean of 31.7 capture probes per locus (median 34, minimum 1, maximum 118), with a mean probe length of 73.3 bases, and a mean density of 62 probes per kb. In the design, 79.6% of capture probes are located in the 200bp regions flanking each STR, with only 20.4% of capture probes directly overlapping an STR (Suppl. Table S2).
Figure 1.
(a) Design of capture probes for STR loci. (b) Summary of the RepeatSeq analysis pipeline for producing genotype calls from Illumina sequencing reads.
Sequence capture was performed following the manufacturer’s instructions. Briefly, 5μg of yeast tRNA and 1μg of pooled input libraries, 1μl of each 1mM of forward and reverse primers used at the PCR step for blocking are mixed and dried using a DNA vacuum concentrator on high heat. Desicated DNA was resuspended in 7.5μl of 2x Hybridization Buffer and 3μl of Hybridization Component A. The whole mix was denatured for 10 min at 95°C and transferred to a 4.5ul aliquot of the capture probes and incubated for 64–72 hours at 47°C. The hybridized samples were then transferred to prepared Streptavidin Dynabeads, mixed thoroughly and incubated for 45 min at 47°C with vortexing every 15 min. Streptavidin Dynabeads plus bound DNA were then washed using the following buffers: once with Wash Buffer I heated to 47°C, twice with Stringent Wash Buffer heated to 47°C, once with Wash Buffer I at room temperature, once with Wash Buffer II at room temperature and once with Wash Buffer III at room temperature. Finally, 50μl PCR grade water were added to the bead-bound sequence capture samples.
Captured samples were amplified using Phusion High-Fidelity PCR Master Mix and 0.25μM IS5 and IS6 primers (Suppl. Table S1), using the following program: denaturation at 94°C for 10min, 12 cycles of 94°C for 1min, 62°C for 30sec and 72°C for 30sec, followed by elongation at 72°C for 10min. PCR products were then purified using the Agencourt AMPure XP system. Amplified sequence capture libraries were assessed using an Agilent high sensitivity DNA kit with the Agilent 2100 bioanalyzer.
Measurement of enrichment
Six pairs of primers specific to STRs targeted in our sequence capture design and four primer pairs targeting internal Nimblegen quality control loci were used in a qPCR experiment to assess the level of target enrichment within each sequence capture library (Suppl. Table S1). qPCR was performed using 0.85μM of primers and SYBR green PCR Master Mix (Applied Biosystems) on both the captured samples and the pre-capture samples (5.5ng), with the following program: denaturation at 95°C for 10 min, 40 cycles of 15 sec at 95°C and 1 min at 60°C.
High-throughput sequencing and genotype calling
After capture, samples were then sequenced with 100 base single-end reads using a HiSeq 2000 instrument (Illumina), or with 150 base paired-end reads using a MiSeq instrument (Illumina), following the manufacturer recommendations. In order to call STR genotypes we used the RepeatSeq genotyping software package using default parameters. In brief, reads were mapped to the hg18 human reference assembly using Novoalign, a local realignment performed and then reads that do not fully span the repeat were removed. A Bayesian model selection approach was then used to assign likelihoods to all possible genotypes supported by the spanning reads, with likelihoods influenced by a prior probability that incorporates an error model for different repeat classes. The algorithm and error model are described in detail by Highnam et al. [2013], and a summary of the analysis pipeline is shown in Figure 1. The minimum number of spanning reads required to call an STR genotype was two.
Sanger sequencing-based validation
Primers were designed to amplify STRs of interest and the amplicons were sequenced using an ABI3730 DNA Analyzer. For regions in which sequencing indicated potential heterozygosity, the amplicons were cloned into plasmids using the TA cloning kit (Invitrogen) and then sequenced.
Results
We achieved a median enrichment of 285-fold for the targeted regions (each STR±200bp) across all samples. On average, 38.7% of mappable reads were located within the capture regions (the 7,353 targeted STR loci ±200bp). However, an average of only 6.5% of these ‘on target’ reads (corresponding to 2.2% of all mapped reads) completely spanned an STR and thus yielded information that could be used to genotype STR length (Table 1). The remaining ‘on target’ reads either mapped to the flanking sequences of the target STRs, or only partially overlapped an STR tract and thus were not informative for STR length. Suppl. Figure S1 shows the distribution of on target and spanning reads obtained for each targeted STR locus. Suppl. Figure S2 shows the number of samples for which successful genotype calls were obtained in the 8 samples studied.
Table 1.
Summary of eight samples sequenced with 100 base reads after targeted enrichment of STR loci
| Sample Name | NA19240 | NA19240 | NA19238 | NA19239 | NA18501 | NA18502 | CHM1 | CHM2 |
|---|---|---|---|---|---|---|---|---|
| Number of PCR cycles used during library preparation | 0 | 13 | 3 | 0 | 2 | 3 | 3 | 13 |
| Total number of reads Produced, millions | 16.1 | 81.4 | 18.6 | 33.8 | 15.2 | 13.2 | 11.6 | 11.9 |
| Number of STR loci genotyped (% of target STRs with length ≤94bp) | 3,178 (50.9%) | 3,648 (58.4%) | 3,295 (52.8%) | 3,403 (54.5%) | 3,227 (51.7%) | 3,197 (51.2%) | 3,121 (50.0%) | 3,121 (50.0%) |
| Total number of on target reads, millions (STR±200bp) (% of total reads) | 6.52 (40.5%) | 30.95 (38.0%) | 7.31 (39.3%) | 12.59 (37.2%) | 5.65 (37.2%) | 5.00 (37.9%) | 4.37 (37.7% | 4.95 (41.6%) |
| Total number of informative reads that span STRs (% of total reads) | 363,534 (2.3%) | 1,766,728 (2.2%) | 425,012 (2.3%) | 691,640 (2.0%) | 325,541 (2.1%) | 294,007 (2.2%) | 249,844 (2.2%) | 271,351 (2.3%) |
| Percentage of STRs with ≥1 informative read | 54% | 67% | 56% | 58% | 55% | 55% | 53% | 54% |
| Percentage of STRs with ≥5 informative reads | 50% | 64% | 53% | 54% | 51% | 51% | 48% | 49% |
| Percentage of STRs with ≥10 informative reads | 47% | 61% | 50% | 51% | 47% | 47% | 45% | 45% |
| Percentage of STRs with ≥100 informative reads | 20% | 46% | 23% | 31% | 18% | 17% | 14% | 16% |
Consistent with the use of 100 base sequencing reads, the largest STR allele we were able to successfully genotype spanned 94bp. Thus, presuming no contractions of repeat length from that represented in the reference genome, STRs included in our capture design that have a span >94bp (n=1,218, or 15.5% of the total), could not be genotyped due to the inherent limitation imposed by read length of our experiment. In total we obtained at least one genotype for 54.5% of the STRs included in our capture design (Suppl. Figure S2). However, when considering only those STR loci of length ≤94bp, 4,212 of the 6,243 (67.5%) that were represented on our capture design were genotyped in one or more of the eight individuals studied (Suppl. Table S2). Using this same criterion, genotypes were obtained for 55% of sites in ≥4 individuals, 47.5% of sites in ≥6 individuals and 32.9% of sites in all 8 individuals.
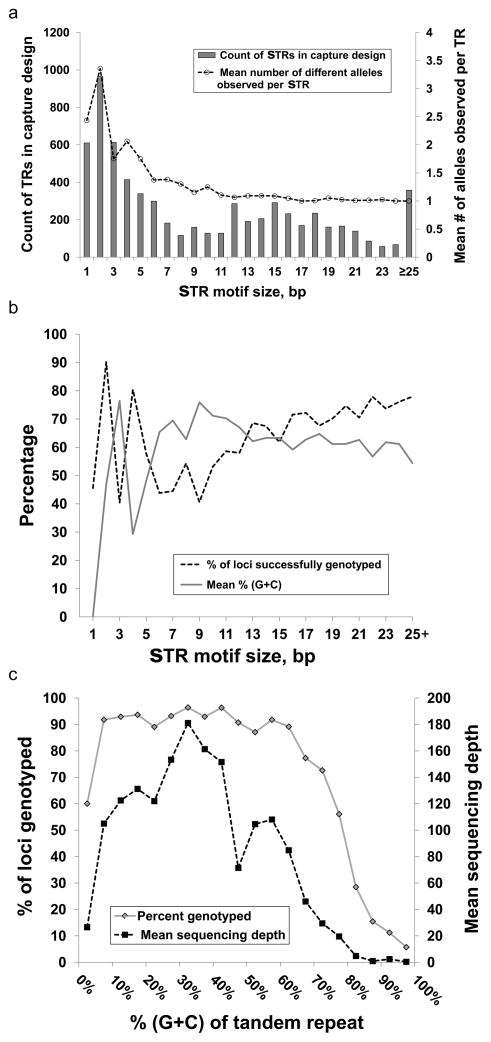
Although our sample size (10 independent alleles) is too limited to accurately determine levels of allelic diversity at any specific STR locus, we were still able to draw general inferences of levels of allelic variation by repeat class. Consistent with prior observations [Fondon et al. 2012], we observed an inverse relationship between STR motif size and allelic diversity. Dinucleotide and mononucleotide repeats showed by far the highest levels of variability, STRs with motif size 3–10bp showed moderate levels of variation, while most STRs with motif size >10bp tended to be invariant, at least in our small sample (Figure 2, Suppl. Table S2).
Figure 2.
(a) Distribution of motif size and levels of allelic diversity observed for 7,353 STRs targeted by our capture design. (b) Percentage of STRs successfully genotyped and their (G+C) content as a function of motif size. (c) Percentage of STRs successfully genotyped and mean depth of informative reads as a function of (G+C) content after grouping data into 5% bins. It should be noted that in addition to (G+C) content, repeat size also influences these data. For example, STRs with 45–50% (G+C) content typically have longer motif sizes (median 58bp) than other STRs targeted in our capture design (median 42bp). This increased size inevitably results in fewer informative reads that completely span the repeat tract, explaining the apparent dip in coverage here.
We observed a very strong dependence between capture/sequencing success of STRs and their (G+C) content. We obtained consistently high read depth and successfully genotyped >90% of STRs with moderate (10–60%) (G+C) content. In contrast average read depth was markedly reduced at extremes of (G+C) content, and we were able to genotype only ~60% of STRs comprised purely of (A+T), and only 12% of STR loci composed of ≥80% (G+C) (Figure 3, Suppl. Table S2).
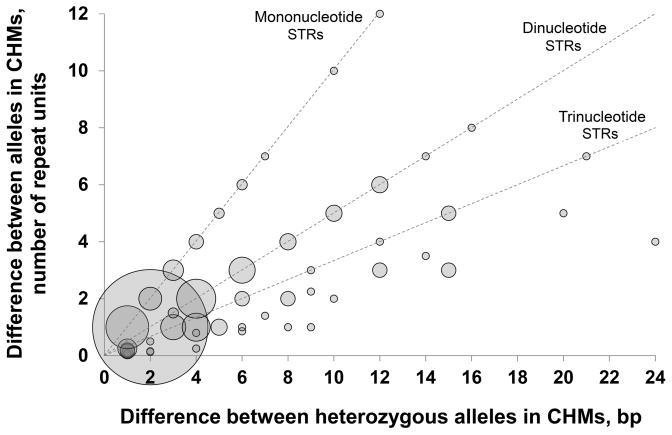
Figure 3. Assessment of genotyping errors using sites with heterozygous calls in two complete hydatidiform moles.
Repeat capture and sequencing was performed on two samples of Complete Hydatidiform Mole that show homozygosity across their entire genome. In this system, loci called as heterozygous must represent genotyping errors. Based on the presumption that one of two alleles called at “heterozygous” sites is correct, the difference in size between these two alleles corresponds to the magnitude of the error in the estimate of true TR length, thus enabling accurate measurement of the type of genotyping errors to be performed. The bubble plot shows the distribution of errors at 469 STR loci as a function of size in base pairs and number of TR units. 54% of all genotyping errors are differences of 2bp that occur at dinucleotide TRs, and thus represent a single repeat unit. Overall, 69% of all genotyping errors are ≤2bp, while 75% are ≤1 repeat unit. The area of each bubble is proportional to the number of events per category.
We investigated the degree of allelic bias for alleles of different lengths by measuring the ratio of spanning reads for the two alleles observed in heterozygous individuals. The use of PCR prior to sequence capture did not appear to adversely influence levels of allelic bias, with median allelic ratios of 1.4 with PCR and 1.5 without PCR in the two assays performed using NA19240. The median allelic bias observed across all heterozygous genotypes was 1.46 (range 1–4) (Suppl. Figure S3).
Assessment of genotyping accuracy
To assess the accuracy of STR genotypes produced using our targeted enrichment protocol, we utilized three different metrics to determine the genotyping error rate, as follows:
Measurement of Mendelian inconsistencies in a mother/father/offspring trio: We included a trio from family #117 of the YRI HapMap population, comprising samples NA19238 (mother), NA19239 (father), and NA19240 (daughter). In the PCR-free and with-PCR assays of the daughter that we performed, we observed 319 of 3,178 (10.0%) and 276 of 3,648 (7.6%) STRs that showed Mendelian inconsistencies respectively, indicating one or more individuals with genotyping errors within the trio (Suppl. Table S2). While not all genotyping errors will result in Mendelian inconsistencies, as each trio actually comprises three independent individuals that have been genotyped at that locus, it is also possible that the individual genotyping error rate may be much lower than this number.
Measurement of sites scored as heterozygous in two samples of complete hydatidiform mole (CHM): As sporadic CHMs arise by duplication of the paternal pronucleus in an anucleate oocyte, they show complete homozygosity genome-wide. We observed 210 of 3,121 (6.7%) STRs that were scored as heterozygous in CHM1, and 310 of 3,121 (10.0%) heterozygous STRs in CHM2, indicative of a genotyping error (Suppl. Table S2).
Concordance of genotype calls between two assays of the same individual: We performed capture and genotyping in two sequencing of the same HapMap individual NA19240 using two different protocols, either with or without PCR prior to sequence capture. 364 of the 2,942 (12.4%) STRs that were successfully genotyped in both assays showed discordant genotypes, indicating genotyping errors in one or both (Suppl. Table S2).
Based on the presumption that one of the two genotypes at all loci that were scored as heterozygous in the two CHM samples tested was erroneous, we performed an analysis of the magnitude and characteristics of STR loci associated with these genotyping errors (Figure 4). In total, 469 of 6,242 (7.5%) loci with genotypes in the two CHMs were scored as heterozygous. Figure 4 shows the distribution of error at these 469 loci as a function of size in base pairs and number of TR units. 54% of all genotyping errors are differences of 2bp that occur at dinucleotide TRs, and thus represent a single repeat unit. Of these, 88% occurred at CA/GT repeats. Overall, 69% of all genotyping errors are ≤2bp, while 75% are ≤1 repeat unit. As expected, we observed a strong inverse relationship between read depth and genotyping error rate (Suppl. Figure S4).
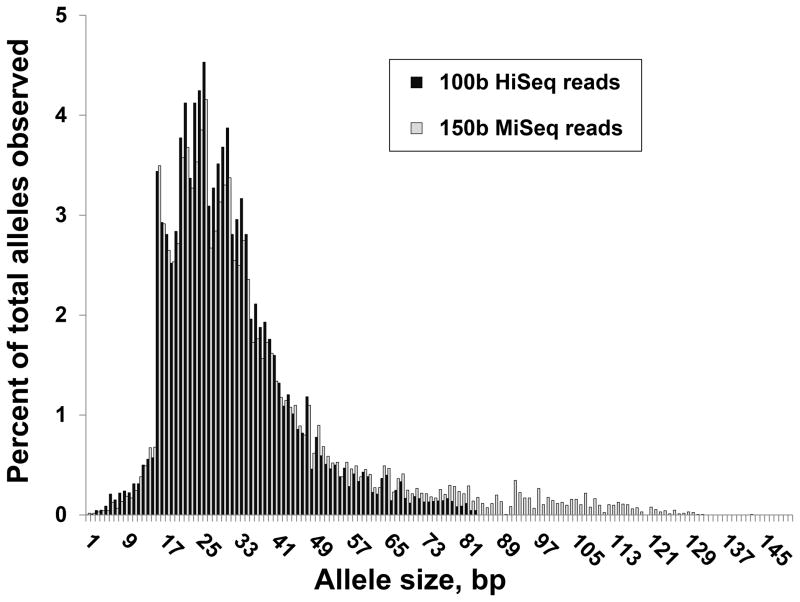
Figure 4. Distribution of observed STR allele sizes using 100 base HiSeq and 150 base MiSeq reads.
Using 100 base Illumina HiSeq reads, the longest alleles we were able to successfully genotype were 94bp, representing reads that span the entire STR locus with only an additional 3bp of anchoring sequence at each flank. Similarly using longer 150 base MiSeq reads, the longest STR allele we detected was 142bp in length.
Investigation of the use of longer read lengths
A fundamental limitation in genotyping STRs is the read length used at the sequencing step. We therefore also performed capture and sequencing of 4 HapMap samples multiplexed using a 150 base paired-end sequencing run generated using an Illumina MiSeq instrument. Although a typical run of a MiSeq produces about 1/10th of the total number of reads as a HiSeq, resulting in a reduced read depth per locus, the increased read length and the use of paired end sequences yielded in an improvement in the proportion of reads that could be successfully mapped, allowing an average of 3,628 STR loci to be genotyped per individual. However, despite the use of 50% longer reads compared to our original experiment using 100 base HiSeq reads, only a small fraction of loci for which we obtained genotypes were previously missed. In the four samples sequenced using 150 base MiSeq reads, only 4.5% of STRs genotyped had allele sizes >94bp (the largest alleles observed using 100 base reads), and the largest STR allele we observed spanned 142bp (Figure 4).
Genotype validation by Sanger sequencing
To confirm the genotype calls made by RepeatSeq using 100 base HiSeq reads of captured DNA, we performed validation experiments by amplifying and Sanger sequencing 26 STRs, including 11 loci that were called as heterozygous in one or both CHMs. These experiments confirmed 26 of 29 (90%) genotypes were correctly assigned by RepeatSeq in the HapMap trio (Suppl. Table S3). For one additional locus two replicates of Sanger sequencing were performed. One replicate agreed with the genotype called by RepeatSeq, while the other replicate was discordant.
In the two CHM samples, considering all genotypes tested, 26 of 39 (67%) were concordant between STR capture/RepeatSeq and Sanger sequencing (Suppl. Table S4). However, as we had specifically selected loci that were incorrectly genotyped based on their heterozygosity in CHMs, this figure represents an inflation of the true genotyping error rate. Considering instead only those loci that were called as homozygous in the CHMs, 24 of 25 (96%) were concordant between STR capture/RepeatSeq and Sanger sequencing.
For most of the loci that were called as heterozygous by RepeatSeq, the true allele length observed by Sanger sequencing corresponded to the larger of the two alleles defined by RepeatSeq, suggesting that the production of ‘stutter bands’ produced by polymerase slippage during STR amplification underlies these genotyping errors. Of a total of 12 errors made by RepeatSeq, 16 (75%) represented contractions compared to that reported by Sanger sequencing, with the majority being a single repeat unit.
Discussion
Although sequence capture, particularly exome sequencing, has become a widely used tool in modern genomics, high-copy repeats such as STRs are generally excluded from capture assays due to the constraints of probe design in non-unique regions. Here we show that by using a custom probe set comprising capture probes that, rather than directly overlapping, predominantly lie adjacent to STR loci, and hybridizing this to relatively large fragments of input DNA (250–400bp), we can take advantage of the fact that the DNA fragments recovered during sequence capture typically extend 100bp or more into the regions flanking each capture probe. Our results show that using this indirect method we are able to capture and perform multiplexed targeted sequencing of thousands of STRs across the genome. While a similar concept has been proposed before [Molla et al. 2009], this study represents the first to demonstrate that STRs are amenable to targeted sequencing. Although in theory rather than targeting the flanking sequence, an alternative approach would be to attempt to capture STRs using probes comprised solely of repetitive sequence, such approaches would be indiscriminant in that they would target all loci containing that sequence in the genome. Instead, using the approach we have taken we show that is it possible to capture a specific subset of STRs of interest from the genome – in this case STRs close to transcription start sites. However, it should also be noted that there are major technical problems associated with utilizing probes composed of highly repetitive sequences, such as their propensity to adopt secondary structures, the reaction kinetics of which would most likely strongly favor either intra-molecular or inter-molecular probe:probe pairing over inter-molecular probe:target hybridization. Thus, probes containing highly repetitive sequence would likely perform poorly in a capture experiment.
In contrast to more conventional target enrichment methods such as exome capture where current protocols typically yield high coverage for 80–90% of targeted loci [Ng et al. 2009], in our capture of STRs we observed a lower percentage of ‘on-target’ reads, a relatively low fraction of loci with informative spanning reads and a correspondingly lower rate of genotyping success (50–60% of targets). We propose several factors that likely contribute to this reduced performance. First, our use of ‘indirect capture’, where due to the constraints of probe design the majority of capture probes we used lie adjacent to rather than directly overlapping the STRs we wished to sequence. As a result, only a minority of mapped reads span the STRs of interest. Second, the unique challenge of genotyping STRs compared to SNPs requires reads that completely span the entire length of an STR in order to be informative. As a result, many ‘on-target’ reads that only partially overlap an STR do not provide useful information on STR length. Third, the highly variable and frequent extremes of (G+C) content found at many STRs. For example, 35% of all STRs we targeted comprised ≤10% or ≥80% (G+C) and these regions had consistently low coverage in our capture experiments. In contrast, for STRs with intermediate (G+C) content (10–60% G+C) we achieved >90% genotyping success. Fourth, reads that span an STR will by nature comprise a significant fraction of non-unique DNA, and thus the ability to uniquely map these within the genome is reduced, which adversely affects overall genotyping efficiency. Finally, the repetitive nature of many STRs means that they are capable of forming strong secondary structures that can inhibit capture and subsequent PCR amplification. Most likely this results from the propensity of such regions to preferentially form intra-strand base-pairs rather than capture probe:target hybrids, and/or the difficulty of melting these structures during PCR amplification that can inhibit polymerases. These secondary structures are often strongest in STRs that have motifs that are composed predominantly of combinations of G and C nucleotides, as when these become single stranded they allow frequent intra-strand complementary base-pairing to occur that can result in highly stable secondary structures such as guanine quadruplexes that can significantly impact molecular assays [Shanahan et al. 2012]. However, despite these problems with the use of barcoding this method allows multiple individuals to be combined in a single sequencing lane, potentially allowing dozens of genomes to be genotyped for thousands of STRs at low cost. Indeed, in recent experiments we have increased our multiplexing to 24 individuals per Illumina HiSeq2000 lane with only a modest drop in the number of loci genotyped per sample, enabling rapid genotyping of thousands of STR loci in large populations at reasonable cost (<$100/sample).
There might be several ways that our method could be optimized further to improve performance, such as modifications to the amplification to improve yields at (G+C) rich targets, the fragment size of input DNA, and probe density, placement and design parameters. However, even with further optimization, tandemly repeated sequences present a number of unique challenges for genotyping with short (50–250 bases) reads that are currently generated by most high-throughput sequencing platforms. First, their tandemly repeated nature means that only reads that completely span the entire STR locus and have unique anchoring sequence at both the 3′ and 5′ ends will be both mappable within the genome and informative for STR length. Thus, a fundamental limit of such experiments is dictated by the read length utilized, as this defines the upper bound of STRs that can be genotyped. In our analysis that primarily utilized 100 base Illumina HiSeq reads, the longest alleles we were able to successfully genotype were 94bp, representing reads that span the entire STR locus with only an additional 3bp of anchoring sequence at each flank. We also performed similar studies using longer read lengths of 150 bases, and here the largest STR allele we detected was 142bp in length. Thus, increased read lengths >100 bases do result in improved yields of STR genotypes, although such improvements are relatively modest. For example, even using read lengths of 150 bases, our studies showed that 92% of alleles observed at all STRs targeted in our design were ≤80bp in length, indicating that the use of 100 base reads is able to capture the diversity of alleles at the majority of STR loci in the human genome. The use of longer reads, and in particular paired-end sequencing, does however improve mapping efficiency in STR loci, and this fact alone facilitates read mapping and genotyping at some loci that are otherwise missed with the use of shorter single-end reads. However, it should be noted that due to the fundamental limitations of genotyping STRs imposed by read length, our method is not suitable for studying large repeat expansions typically seen in triplet repeat disorders such as Fragile X.
As capture protocols typically include two rounds of PCR amplification that might introduce bias into the DNA fragments available for sequencing, we performed three different analyses in an attempt to determine genotyping error rates at STR loci after capture and sequencing. Analysis of Mendelian inconsistencies in a trio, rates of heterozygous calls in genome-wide homozygous CHM samples, and concordance rates in repeat analyses of the same individual, all indicated genotyping error rates between 7.6–12.4%. Although this error rate is relatively high, we observed that three quarters of genotyping errors represent a single repeat unit, most of which occur at dinucleotide repeats. It is also worth noting that many of the loci at which these errors occurred were seen to be sites of recurrent error in multiple different analyses. For example, of the 297 STR loci that showed heterozygosity in CHMs and had genotypes in all three members of the HapMap trio, 151 of these (50.8%) also showed Mendelian errors in the HapMap trio we sequenced, representing a 3.4-fold increase above that expected by chance. This suggests that certain STRs are “error prone” in our system, and may represent loci that are particularly susceptible to polymerase slippage in vitro during PCR amplification. Most of these loci (79.5%) are dinucleotide repeats that are known to be one of the most polymorphic/unstable classes of STRs in the human genome. We suggest that these genotyping errors likely result from the typical polymerase slippage pattern that frequently generates so-called ‘stutter’ or ‘shadow’ bands after PCR amplification of STRs and microsatellites. It should also be noted that within RepeatSeq we applied a diploid STR genotyping model to make calls in the essentially haploid CHMs, and this occasionally results in the inappropriate consideration of heterozygosity. Finally, it is also possible that somatic mutation of STRs could occur, such that some loci are genuinely heterozygous in the CHM samples we utilized. The effect of this would be an over-estimation of the error rates in this system.
Although we included samples that were processed with varying amounts of PCR amplification prior to hybridization with the capture probes, we did not observe a clear relationship between the number of PCR cycles performed and either the genotyping error rate or the degree of allelic bias in heterozygotes. In fact, for the two assays performed for NA19240 we actually observed a stronger allelic bias and a higher rate of Mendelian inconsistencies in the library produced without PCR prior to the capture, although it should be noted that there was also a large difference in the number of mapped reads between these two assays that likely confounds this interpretation. However all samples did undergo PCR amplification post-capture, and thus with current technology we were unable to completely avoid PCR amplification prior to sequencing. We also note that even for sequencing libraries prepared without any PCR, an inherent part of current Illumina sequencing protocols is cluster generation that involves amplification of the template library molecules. Thus, without the use of alternative sequencing technologies that completely avoid amplification we predict that the problem of elevated genotyping error rates at certain highly mutable STRs will persist.
In summary, here we demonstrate for the first time the targeted capture, sequencing and genotyping of STR loci in the human genome. Although there are clear limitations in the capture of STRs showing extremes of (A+T) or (G+C) content, our methodology provides a highly multiplexable solution by which thousands of STR loci can be genotyped at low cost, opening the door for large-scale population studies of STR variation.
Supplementary Material
Suppl. Figure S1. Median number of on-target and STR-spanning reads obtained for 7,851 STRs included in the custom capture design. STR loci are sorted by median number of on-target reads (reads mapping within ±200bp of the target STR, red bars, right axis). Blue bars show the number of spanning reads per STR (left axis). The black line shows a 20-point moving average of the number of spanning reads.
Suppl. Figure S2. The number of successful genotype calls obtained in the 8 samples studied. Black bars show data based on all 7,851 STRs included in the custom capture design, while grey bars show results based on those STRs with span ≤94bp, corresponding to the maximum allele size observed using 100 base reads. We obtained genotypes for 55% of sites in ≥4 individuals, 47.5% of sites in ≥6 individuals and 32.9% of sites in all 8 individuals.
Suppl. Figure S3. Distribution of allelic bias observed in heterozygous samples. Using all heterozygous genotypes obtained across STR capture assays we calculated the relative ratio of reads spanning the two observed alleles (Suppl. Table S2). Allele ratios in the range 1–3 are shown in bins of width 0.1, with the two final bins showing STRs with allelic ratios of 3–3.5 and >3.5–4.
Suppl. Figure S4. An inverse relationship between read depth and STR genotyping error rate. We performed three different analyses to investigate the prevalence of genotyping errors of STR length after sequence capture. (a) Measurement of concordance of genotype calls between two assays of the same individual, (b) measurement of sites scored as heterozygous in two CHM samples that are homozygous for every chromosome, and (c) measurement of Mendelian inconsistencies in a HapMap trio. In all three analyses, there is a strong inverse relationship between genotyping error rates and the number of spanning informative reads, indicating that as expected, increased read depth yields more accurate genotyping. To ensure robustness, for sites with 20–50x coverage error rates shown are the average after binning into groups of two adjacent data points, and for 50–200x coverage represent the average after binning into groups of five adjacent data points.
Suppl. Table S1. List of PCR primers and sequencing adapters used in the study.
Suppl. Table S2. Summary of 7,851 STRs included in the custom capture design and results of sequencing using 100 base Illumina reads.
Sanger-sequencing was used to genotype a subset of repeat regions from NA19238, NA19239, and NA19240, a parent/offspring trio. Alleles are reported as the length of the repeat tract for the hg18 reference genome, RepeatSeq calls, and Sanger-based sequencing. Discordance between RepeatSeq and Sanger is indicated in bold/outline. For the asterisked sample (NA19240, chr5:140,541,360–140,541,397) two replicates of Sanger sequencing were performed. One replicate agreed with the genotype called by RepeatSeq, while the other replicate was discordant.
Sanger-validated genotypes for a subset of repeat regions from the CHM1 and CHM2 hydatidiform moles. Alleles are reported as the length of the repeat tract for the hg18 reference genome, RepeatSeq calls, and Sanger-based sequencing of PCR products. Discordance between RepeatSeq and Sanger is indicated in bold/outline.
Acknowledgments
We would like to thank Dr. Omar Jabado, Zuly Peralta and Stephen Xanthoudakis of the Genomics core at Mount Sinai School of Medicine for advice and technical assistance. This work was supported by the National Institute of Health [grants 1R01DA033660, 1R01HG006696 and 1R03HD073731 to A.J.S.; NS079926 to D.M.] and the Alzheimer’s Association [grant 2012ALZNIRG69983 to A.J.S.].
Footnotes
Disclosure Statement: The authors declare no conflict of interest.
References
- Borel C, Migliavacca E, Letourneau A, Gagnebin M, Béna F, Sailani MR, Dermitzakis ET, Sharp AJ, Antonarakis SE. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum Mutat. 2012;33:1302–9. doi: 10.1002/humu.22115. [DOI] [PubMed] [Google Scholar]
- Burgner D, Rockett K, Ackerman H, Hull J, Usen S, Pinder M, Kwiatkowski DP. Haplotypic relationship between SNP and microsatellite markers at the NOS2A locus in two populations. Genes and Immunity. 2003;4:506–514. doi: 10.1038/sj.gene.6364022. [DOI] [PubMed] [Google Scholar]
- Campbell CD, Chong JX, Malig M, Ko A, Dumont BL, Han L, Vives L, O’Roak BJ, Sudmant PH, Shendure J, Abney M, Ober C, Eichler EE. Estimating the human mutation rate using autozygosity in a founder population. Nat Genet. 2012;44:1277–81. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Microsatellites: simple sequences with complex evolution. Nature Reviews Genetics. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- Fondon JW, 3rd, Martin A, Richards S, Gibbs RA, Mittelman D. Analysis of microsatellite variation in Drosophila melanogaster with population-scale genome sequencing. PloS One. 2012;7:e33036. doi: 10.1371/journal.pone.0033036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonville NC, Ward RM, Mittelman D. Stress-induced modulators of repeat instability and genome evolution. J Mol Microbiol Biotechnol. 2011;21:36–44. doi: 10.1159/000332748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annual Review of Genetics. 2010;44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- Gymrek M, Golan D, Rosset S, Erlich Y. lobSTR: A short tandem repeat profiler for personal genomes. Genome Research. 2012;22:1154–1162. doi: 10.1101/gr.135780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highnam G, Franck C, Martin A, Stephens C, Puthige A, Mittelman D. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Research. 2013;41:e32. doi: 10.1093/nar/gks981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Human Mutation. 2003;21:12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- Molla M, Delcher A, Sunyaev S, Cantor C, Kasif S. Triplet repeat length bias and variation in the human transcriptome. Proc Natl Acad Sci USA. 2009;106:17095–100. doi: 10.1073/pnas.0907112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang AW, Migita O, Macdonald JR, Feuk L, Scherer SW. Mechanisms of formation of structural variation in a fully sequenced human genome. Human Mutation. 2013;34:345–354. doi: 10.1002/humu.22240. [DOI] [PubMed] [Google Scholar]
- Shanahan HP, Memon FN, Upton GJ, Harrison AP. Normalized Affymetrix expression data are biased by G-quadruplex formation. Nucleic Acids Res. 2012;40:3307–15. doi: 10.1093/nar/gkr1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwach P, Ganesh S. Tandem repeats in human disorders: mechanisms and evolution. Frontiers in Bioscience. 2008;13:4467–4484. doi: 10.2741/3017. [DOI] [PubMed] [Google Scholar]
- Sun JX, Helgason A, Masson G, Ebenesersdottir SS, Li H, Mallick S, Gnerre S, Patterson N, Kong A, Reich D, et al. A direct characterization of human mutation based on microsatellites. Nature Genetics. 2012;44:1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. Unstable tandem repeats in promoters confer transcriptional evolvability. Science. 2009;324:1213–1216. doi: 10.1126/science.1170097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorbij AM, van Steenbeek FG, Vos-Loohuis M, Martens EE, Hanson-Nilsson JM, van Oost BA, Kooistra HS, Leegwater PA. A contracted DNA repeat in LHX3 intron 5 is associated with aberrant splicing and pituitary dwarfism in German shepherd dogs. 2011 doi: 10.1371/journal.pone.0027940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Wong C. Mutation of human short tandem repeats. Human Molecular Genetics. 1993;2:1123–1128. doi: 10.1093/hmg/2.8.1123. [DOI] [PubMed] [Google Scholar]
- Xu X, Peng M, Fang Z. The direction of microsatellite mutations is dependent upon allele length. Nature Genetics. 2000;24:396–399. doi: 10.1038/74238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Figure S1. Median number of on-target and STR-spanning reads obtained for 7,851 STRs included in the custom capture design. STR loci are sorted by median number of on-target reads (reads mapping within ±200bp of the target STR, red bars, right axis). Blue bars show the number of spanning reads per STR (left axis). The black line shows a 20-point moving average of the number of spanning reads.
Suppl. Figure S2. The number of successful genotype calls obtained in the 8 samples studied. Black bars show data based on all 7,851 STRs included in the custom capture design, while grey bars show results based on those STRs with span ≤94bp, corresponding to the maximum allele size observed using 100 base reads. We obtained genotypes for 55% of sites in ≥4 individuals, 47.5% of sites in ≥6 individuals and 32.9% of sites in all 8 individuals.
Suppl. Figure S3. Distribution of allelic bias observed in heterozygous samples. Using all heterozygous genotypes obtained across STR capture assays we calculated the relative ratio of reads spanning the two observed alleles (Suppl. Table S2). Allele ratios in the range 1–3 are shown in bins of width 0.1, with the two final bins showing STRs with allelic ratios of 3–3.5 and >3.5–4.
Suppl. Figure S4. An inverse relationship between read depth and STR genotyping error rate. We performed three different analyses to investigate the prevalence of genotyping errors of STR length after sequence capture. (a) Measurement of concordance of genotype calls between two assays of the same individual, (b) measurement of sites scored as heterozygous in two CHM samples that are homozygous for every chromosome, and (c) measurement of Mendelian inconsistencies in a HapMap trio. In all three analyses, there is a strong inverse relationship between genotyping error rates and the number of spanning informative reads, indicating that as expected, increased read depth yields more accurate genotyping. To ensure robustness, for sites with 20–50x coverage error rates shown are the average after binning into groups of two adjacent data points, and for 50–200x coverage represent the average after binning into groups of five adjacent data points.
Suppl. Table S1. List of PCR primers and sequencing adapters used in the study.
Suppl. Table S2. Summary of 7,851 STRs included in the custom capture design and results of sequencing using 100 base Illumina reads.
Sanger-sequencing was used to genotype a subset of repeat regions from NA19238, NA19239, and NA19240, a parent/offspring trio. Alleles are reported as the length of the repeat tract for the hg18 reference genome, RepeatSeq calls, and Sanger-based sequencing. Discordance between RepeatSeq and Sanger is indicated in bold/outline. For the asterisked sample (NA19240, chr5:140,541,360–140,541,397) two replicates of Sanger sequencing were performed. One replicate agreed with the genotype called by RepeatSeq, while the other replicate was discordant.
Sanger-validated genotypes for a subset of repeat regions from the CHM1 and CHM2 hydatidiform moles. Alleles are reported as the length of the repeat tract for the hg18 reference genome, RepeatSeq calls, and Sanger-based sequencing of PCR products. Discordance between RepeatSeq and Sanger is indicated in bold/outline.